Abstract
Provided that generalist viruses will have access to potentially unlimited hosts, the question is why most viruses specialize in few hosts. It has been suggested that selection should favor specialists because there are tradeoffs limiting the fitness of generalists in any of the alternative hosts or because evolution proceeds faster with narrower niches. Here we review experiments showing that virus adaptation to a specific host is often coupled with fitness losses in alternative ones. In most instances, mutations beneficial in one host are detrimental in another. This antagonistic pleiotropy should limit the range of adaptation and promote the evolution of specialization. However, when hosts fluctuate in time or space, selective pressures are different and generalist viruses may evolve as well.
INTRODUCTION
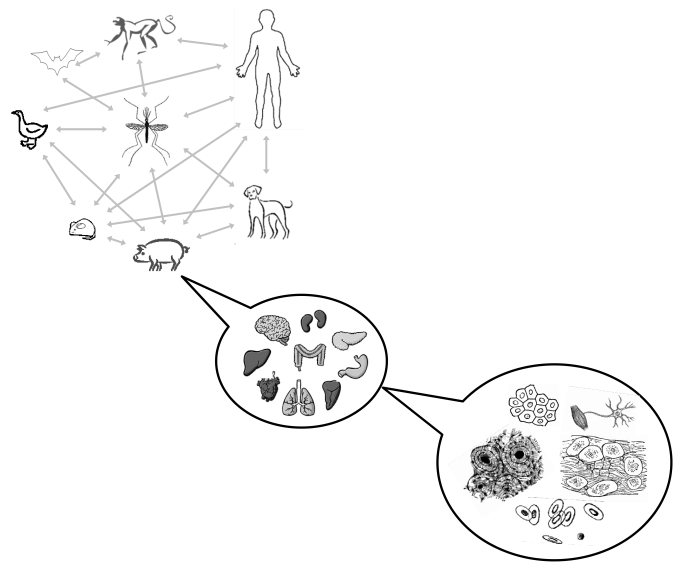
Viruses live in an always fluctuating world (Fig. 1). They move from host to host, sometimes being air- or water-borne but sometimes using vectors (e.g. insects) in which they may also replicate. Within an individual host, viruses face multiple tissues and cell types that differ in physiological and biochemical properties and they are constantly challenged by a diversity of antiviral immune responses and perhaps even by drugs. Some viruses have evolved to become specialized in infecting one or very few host species whereas others, especially plant viruses, are generalists and successfully infect hosts from different species and even from higher taxonomical units. Examples of specialists are dengue and mumps viruses, whose only known mammalian host are humans. Examples of generalist viruses are Cucumber mosaic virus, that infects more than 1000 species including monocots and dicots, herbaceous plants, shrubs and trees, and Influenza A virus, which infects birds and several different species of mammals. Also, some viruses show strong cell and tissue tropisms and are only able of replicating in very limited cell types whereas others can infect and replicate in several different cell types. The intrinsic evolvability of viruses, owed to their large population sizes, short generation times, and high mutation rates, can facilitate host range changes that may eventually lead to epidemics of emergent new viruses [1].
Fig. (1).
Viruses face an always changing world. The figure illustrates the changing environments in which many viruses live. Viruses may move between different hosts, in some instances using insect vectors into which they may even replicate (as it is the case for arboviruses). Within an infected host, in some instances viruses may show strong tropisms and only replicate in a limited number of tissues and cell types, whereas in other cases, viruses can colonize and successfully replicate into several different tissues and cell types. Although the image only shows animals, a similar scheme can be drawn for the case of plant viruses.
Hereafter, the term “host” will be used in a loosely way and it will means different host species, different host genotypes or different cell types within an individual host. By specializing in a single host, viruses may reduce interespecific competition at the cost of accessing a more limited set of available resources [2]. In stark contrast, the advantages of generalism are more obvious: a generalist virus would be able of exploiting multiple hosts thus enhancing its fitness. Since generalist viruses are not the norm, it is generally assumed that generalism comes with a cost, in keeping with the adage that a “jack-of-all-trades” is a master of none [3]. It has been suggested that evolution should favor specialists because there are tradeoffs that limit the fitness of generalists in any of the alternative hosts or because evolution proceeds faster with narrower niches [3, 4] (Fig. 2a). Tradeoffs can be generated by different mechanisms, being antagonistic pleiotropy the simplest and most intuitive one. Antagonistic pleiotropy means that mutations that are beneficial in one host may be deleterious in an alternative one [5]. A second mechanism that promotes tradeoffs is mutation accumulation, in which neutral mutations accumulate by drift in genes that are useless in the actual host but may be essential in a future new one [6]. Although both mechanisms involve differences in mutational fitness effects across hosts, it is necessary to stress out that by no means they are equivalent phenomena: while natural selection is the only reason for the tradeoff in the former mechanism, genetic drift is so in the latter. The examples provided in the next section will drive us to the conclusion that when a single host is available, viruses become specialist.
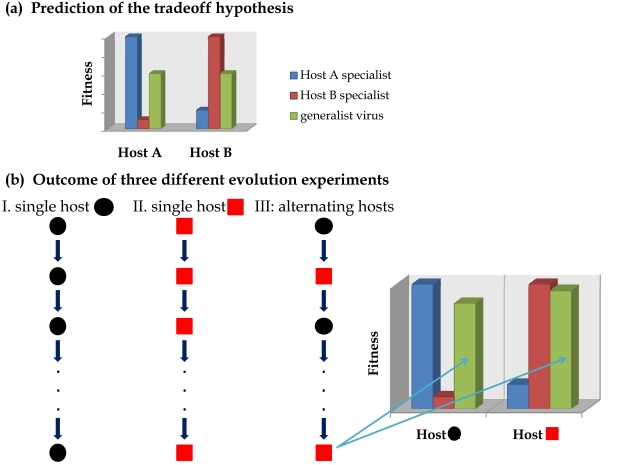
Fig. (2).
Fitness tradeoffs across hosts. (a) Expected fitness for specialist and generalist viruses if a tradeoff exists. Although both specialist genotypes perform well in their respective hosts, each one is poorly adapted in the other host. The green bars illustrate the behavior of a generalist virus that performs fairly well in both hosts but has lower fitness than either specialist in its preferred host. According with this picture, a specialist virus will always outcompete a generalist one on its host but if hosts vary in time or space, the generalist may have an overall advantage. (b) Outcome of three evolution experiments. Virus evolved in single host become specialists on their respective hosts; by contrast, a virus evolved in a fluctuating host landscape becomes generalist and improves fitness in both hosts at the same time (green bars).
What are the evolutionary mechanisms that determine host-range for viruses? How common are tradeoffs and when do they arise? Why some viruses may opt for specialization whereas others opt for a generalist strategy? What generates these tradeoffs? These are the questions we are going to address here by reviewing empirical evidences collated from several viral systems during the last few years.
VIRUSES BECOME SPECIALISTS WHEN FACING A SINGLE HOST
Bacteriophage ØX174 natural host is Escherichia coli. Crill et al. [7] undertook the task of expanding this phage’s host range by evolving it throughout serial transfers in Salmonella enterica. After 11 days of selection, the replicative fitness of the Salmonella-evolved phages was evaluated in the new bacterial host and it was almost 700-fold higher than the value estimated for the ancestral virus, proving that adaptation took place during the experiment. More interestingly, this tremendous fitness improvement was not costless: the replicative fitness of the evolved lineages back in the original E. coli host was almost zero. This work provides first evidence that adaptation to a new host comes with a decrease in fitness within the ancestral host.
However, most of the relevant information concerning the problem of virus specialization and host-range expansion has been obtained by in vitro evolution experiments in which RNA arboviruses such as Vesicular stomatitis virus (VSV) [8-11], Eastern equine encephalitis virus (EEEV) [12, 13], Sindbis virus (SINV) [14], and Venezuelan equine encephalitis virus (VEEV) [15] or even a retrovirus such as the Human immunodeficiency virus type 1 [16] were evolved in and adapted to different lineages of animal cells in in vitro cultures. A common result of all these studies, in agreement with the above ØX174 results, is that viral populations evolved on a single cell host type became specialists: they increase replicative fitness in the new host and pay a costs in any alternative host cell type, including the original one (Fig. 2b).
There are many studies in the plant virology literature in which the qualitative costs associated with viruses expanding their host range from sensitive to resistant plant genotypes had been explored. For example, Jenner et al. [17] quantified the replicative fitness penalty on wildtype plants paid by Turnip mosaic virus (TuMV) after expanding its host range from wildtype turnips to plants bearing the TuRB01 resistance gene. The replicative fitness of three resistance-breaking TuMV genotypes was evaluated on wildtype turnips by multi-day competition assays against the wildtype-specialist isolate. The replicative fitness costs associated with host-range were widely variable and ranged from ~32% to 100%. In a second recent study, Wallis et al. [18] have shown that following serial passages in an herbaceous host (Pisum sativum), Plum pox virus (PPV) increased its infectivity, viral load and virulence in the new host with a concomitant reduction in transmission efficiency in peach trees, the original host, suggesting that host-range expansion was a costly trait for PPV. Similar results have been reported recently by Agudelo-Romero et al. [19] using Tobacco etch virus (TEV). Independent evolutionary lineages of TEV were maintained by serial passages in two different hosts. While TEV lineages maintained in the original tobacco host showed no increase neither in viral load nor virulence, lineages evolved in the new host pepper showed increases in both traits. However, these increases were specific of the pepper host, and the pepper-adapted lineages did not show any replicative fitness increment in the ancestral tobacco host.
All these examples have delineated an image suggesting that in a homogenous host landscape, selection promotes the evolution of specialist viruses and the existence of a fitness burden associated with host-range expansion. Nonetheless, it is fair mentioning that exceptions to this tradeoff rule exist. For example, it has been described that Foot-and-mouth disease virus adapted to fibroblasts from hamster kidney (BHK) resulted in expanded host range. BHK-adapted FMDV acquired the ability to successfully infect cells from monkeys and humans [20]. Also, Novella et al. [21] reported for VSV that adaptation to acute infections of either BHK or sandfly cells resulted in generalist populations with improved fitness in both hosts.
ALTERNATION AMONG HOSTS RESULTS IN GENERALIST VIRUSES THAT MAY OR MAY NOT EXPERIENCE A FITNESS TRADEOFF
Several in vitro studies using VSV have also explored the effect of temporal host heterogeneity. Despite methodological differences and the use of different cell types for the experiments, most of these studies came up to a common observation: when the viral population alternated in time between two cell types, natural selection improved fitness in every type to a similar extent as when adaptation happened to each one individually, that is, VSV populations became generalists without paying fitness costs in any of the alternative hosts [9, 11, 21]. However, a significant cost was paid by these generalist viruses in the ancestral host cell type not included in the fluctuation treatment [9]. The same observation was made for EEEV populations evolved in two alternating cell types (hamster and mosquito): EEEV reached replicative fitness values on each cell type similar to those reached by viral lineages evolved only on single cell types [12]. Therefore, all these results suggest that no fitness tradeoff exists when the host landscape fluctuates fast, since the replicative fitness in both environmental extremes is maximized (Fig. 2b).
However, the observation of a lack of fitness tradeoff seems not to be ubiquitous. For example, for some but not all SINV lineages alternatively passaged in mosquito and hamster cells [14], the replicative fitnesses on each alternative host were lower than those reached by SINV lineages evolved on each host hold constant. This result is still compatible with the existence of a fitness tradeoff across hosts. The fact that not all SINV generalist lineages showed the tradeoff might be explained by some lineages overcoming the tradeoff by finding the right combination of mutations whereas the lineages still showing the tradeoff did not found such combinations. The question that remains is whether they will eventually find the optimal solution if enough time is allowed.
In the case of complex host organisms, a virus faces different compartments within individual hosts: different tissues and various barriers among them, as well as being sampled and mixed by the circulatory system. The question is then how this spatial heterogeneity affects viral adaptive dynamics? This question was experimentally addressed by Cuevas et al. [22], who found that the extent in which VSV adapts to diverse host cell types strongly depends on the migration rate among cell types. Increasing migration rate among heterogeneous cell types selects for generalist viruses with increased replicative fitness in all the alternative hosts (Fig. 3). By contrast, in the absence of migration, viral populations become specialized for their host cell type (Fig. 3). This result supports the general view that migration among hosts must be sufficiently low relative to the strength of selection to generate local adaptation to each host [23-25]. Indeed, the conditions for the coexistence of specialist viruses in a heterogeneous host environment are very restrictive. If the selective differences among hosts are not so large the balance of production from each host must be roughly equal in order to maintain diversity [26, 27]. This implies that there must be lots of opportunities for generalists to evolve in heterogeneous environments, even if selection favors in the short term specialization to the host wherein virus productivity is maximized.
Fig. (3).
Effect of migration rate among different hosts in the fraction of generalist viruses that can be maintained in the population. Migration rate increases from left to right. Increasing migration rate among different cell types favors generalist genotypes, whereas a reduced migration rate would favor viral genotypes specialized on each host. In the absence of migration, specialist viruses (black bars) dominate the population. Frequency of generalist viruses (red bars) increases with migration rate.
One caveat of all the above studies is that they have been performed in a very particular set of in vitro conditions, hence, one may argue against their generality in the real world. In a recent study, Coffey et al. [15] tried to overcome this problem by evolving independent lineages of VEEV either in Aedes aegypti mosquitoes, rodents or alternating between both animals. As expected by the tradeoff hypothesis, serial in vivo mosquito passages resulted in enhancement of mosquito infectivity but at the cost of reduced replication ability in rodents. Consistently, VEEV populations serially passaged in rodents showed increased replication rate in the vertebrate host but reduced infectivity in mosquitoes. Interestingly, alternating in vivo passages between mosquito and rodents did not significantly increased in VEEV replicative fitness in either host. This result illustrates the evolutionary constraint imposed to arboviruses by their obligatory transmission among radically different animal hosts.
Therefore, the fitness tradeoff described in the previous section does not necessarily hold when viruses face rapid fluctuations in host landscape.
ANTAGONISTIC PLEIOTROPY VS MUTATION ACCUMULATION
At the beginning of this review, it was stated that two mutually non-exclusive explanations may hold to justify the existence of across host fitness tradeoffs: mutation accumulation and antagonistic pleiotropy. Given the compactness of virus genomes, with many cases of overlapping reading frames and multifunctional proteins, the latter is expected to be a more plausible explanation. Indeed, the experimental results reviewed in the following paragraphs overwhelmingly support this as the reason for fitness tradeoffs across hosts. Here we are not exhaustively reviewing the endless list of relevant references but have simply chosen a few representative cases.
A first example of antagonistic pleiotropy was reported in the above mentioned work by Crill et al. [7]. The genome of the Salmonella-evolved ØX174 phages was fully sequenced and the same two or three substitutions in the major capsid gene were recurrently identified in the different lineages. The fact that independent lineages fixed the same mutation provides strong support for the selective advantage conferred by these mutations in the new host. Indeed, when the Salmonella-adapted virus was evolved back on E. coli, these mutations quickly reverted to the ancestral stage. Reversion was the outcome rather than second-site compensatory mutations, thus confirming that these mutations had an antagonistic pleiotropic effect in the E. coli host. In a second example, Duffy et al. [28] isolated nine different phage ø6 genotypes each carrying a single nonsynomymous substitution in the P3 attachment protein that allowed them to infect at least one (one case) but mostly two (eight cases) new Pseudomonas spp that were not susceptible to the ancestral unmutated virus. In other words, these mutations conferred Ø6 with a wider host range. Then, the authors measured the replicative fitness of all these nine genotypes into the ancestral P. syringae pv. phaseolicola host and found that in seven instances the replicative fitness was significantly reduced, thus confirming that antagonistic pleiotropy, although certainly not unique, was a major issue associated with host range expansion. These results were further confirmed by Ferris et al. [29] with a larger sample size. Fifteen out of sixteen mutations identified in the P3 attachment protein that allowed successful infection of a new host P. syringae pv. glycinea suffered from significant replicative fitness loss in the ancestral host phaseolicola.
Some of the most remarkable recent examples of antagonistic pleiotropy driving host specialization come from plant viruses. For example, five independent lineages of Hibiscus chlorotic ringspot virus (HCRSV) were evolved by serial transfers into the local lesion host Chenopodium quinoa [30]. After evolution in the novel host, HCRSV virulence on its natural host was dramatically reduced, in agreement with the tradeoff hypothesis. Interestingly, all five lineages fixed exactly the same eight amino acid changes in non-contiguous sites of the coat protein (CP). A similar example was reported for Pelargonium flower break virus (PFBV) populations adapted to C. quinoa [31]. PFBV isolates maintained for long time on C. quinoa leaves had five specific non-contiguous amino acid substitutions in the CP, which were not present in other natural isolates. The C. quinoa-specific pattern of amino acids at the relevant sites was VFYII. When a wildtype isolate from geranium, containing the amino acid pattern ASHMV, was mechanically inoculated onto C. quinoa leaves, the viral population generated right after the first passage had already fixed two of the C. quinoa-specific changes (ASYMI), and only four serial passages were necessary to restore the entire VFYII C. quinoa-specific pattern [31]. The fact that this pattern has never been found in the natural host, not even incomplete, suggests that it may impose a strong burden for viral replicative fitness on the natural host geranium.
So far, the most exhaustive study of the relative contribution of antagonistic pleiotropy and mutation accumulation to the evolution of host specialization has been done by Remold et al. [11]. In a previous work, independent lineages of VSV were evolved on human cancer cells (HeLa), dog kidney fibroblast (MDCK) or alternating among both cell types [9]. Replicative fitness was quantified on each new host as well as in the ancestral host type BHK. Viruses evolved in HeLa cells increased their replicative fitness in this cell type but showed significantly lower replicative fitness in MDCK than the ancestral virus. Viruses evolved in MDCK significantly improved replicative fitness on this cell type but experience no replicative fitness effect on the HeLa host when compared with the ancestral virus. The viruses evolved by the fluctuating treatment always improved replicative fitness in HeLa whereas only half of them had increased replicative fitness in MDCK [9, 11]. To explore the molecular basis of this host-range expansion, full genomic sequences were obtained for each evolved lineage after ~95 generations of viral replication. In short, alleles shared among all the lineages adapted to human cells were not present in lineages evolved on the alternative host. This observation is consistent with antagonistic pleiotropy as the mechanism responsible for the low replicative fitness of the HeLa-adapted virus on MDCK cells. By contrast, MDCK-evolved and alternating host-evolved populations shared many more mutations. Authors’ interpretation of this observation was that perhaps in this case mutation-accumulation, rather than antagonistic pleiotropy, was a better explanation [11]. In a very recent similar study, it has been confirmed that whenever a tradeoff exists in the replicative fitness of VSV on BHK and sandfly cells, antagonistic pleiotropy is the cause [32]. Interestingly, this study suggests that among the mutations putatively responsible for the tradeoff, some are affecting the regulatory sequences at the 3’ end of the viral RNA genome, which may result in changes in the regulation of RNA synthesis [32].
In conclusion, as a consequence of the compact genomes of RNA viruses, antagonistic pleiotropy seems to be the most general, although certainly not universal, explanation for the widely observed fitness tradeoffs across hosts.
CONCLUSIONS AND FUTURE PERSPECTIVES
The experiments here reviewed, the tip of a much larger set, suggest that whenever a virus switches from hosts, acquiring the ability of replicating in a new host imposes a fitness burden in the original host. This may be a consequence of the different selective requirements characteristic of different hosts. However, some evidences also suggest that the fitness of a virus simultaneously facing multiple hosts is either constrained by the most restrictive one or there is no tradeoff at all. In this respect, the extent to which generalism evolves depends on the frequency at which viruses transmit among heterologous hosts [33]. When transmission among heterologous hosts represents an infrequent event, the viral population essentially adapts to the current host. However, if heterologous transmissions are frequent, the viral population behaves as if the fitness landscape did not change at all but was the average of the changing landscapes [33]. The behavior at intermediate oscillation frequencies rests between these two extremes.
What are the causes for fitness tradeoff across hosts? Most of the accumulated evidences suggest that antagonistic pleiotropy is the principal, although certainly not the only reason. Antagonistic pleiotropy may be an unavoidable consequence of the small size of viral genomes, which in many instances contain overlapping genes and encode for multifunctional proteins, making extremely difficult to optimize one function without jeopardizing another.
Studies seeking to characterize the molecular basis of host switch or trying to disentangle the importance of antagonistic pleiotropy vs mutation accumulation as the cause of fitness tradeoffs across hosts have been suffering from the problem that, in many instances, the number of mutations fixed was large, thus making hard to decide which mutation was really responsible for the host range expansion. However, the property of having compact genomes also imposes restrictions to the number of possible evolutionary solutions reachable, therefore evolutionary convergences are common. When the same mutation pervasively appears in independent lineages, it is an excellent candidate to be responsible for adaptation to the new host. Nowadays, it is relatively simple to construct infectious cDNA clones for many viruses. The use of reverse genetic analysis allows testing directly the adaptive value of observed mutations one by one or in specific combinations. This type of studies will provide the raw material necessary to really weight the contribution of different antagonistic pleiotropy or mutation-accumulation in the evolution of host specialization.
Finally, the results here reviewed are of relevance for our understanding of emerging (and re-emerging) viral infections, since it is expected that generalist viruses would more likely be able of jumping over the species boundary and infect new potential hosts [1].
ACKNOWLEDGEMENT
This work was supported by a Spanish Ministerio de Ciencia e Innovación grant (BFU2006-14819-C02-01/BMC).
REFERENCES
- 1.Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg Infect Dis. 2005;11:1842–7. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Futuyma DJ, Moreno G. The evolution of ecological specialization. Annu Rev Ecol Syst. 1988;19:207–33. [Google Scholar]
- 3.Whitlock MC. The Red Queen beats the jack-of-all-trades: the limitations on the evolution of phenotypic plasticity and niche breadth. Am Nat. 1996;148:S65–S77. [Google Scholar]
- 4.Woolhouse MEJ, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–12. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 5.Fry JD. The evolution of host specialization: are tradeoffs overrated? Am Nat. 1996;148:S84–S107. [Google Scholar]
- 6.Kawecki TJ. Accumulation of deleterious mutations and the evolutionary cost of being a generalist. Am Nat. 1994;144:833–8. [Google Scholar]
- 7.Crill WD, Wichman HA, Bull JJ. Evolutionary reversals during viral adaptation to alternating hosts. Genetics. 2000;154:27–37. doi: 10.1093/genetics/154.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland JJ, de la Torre JC, Clarke DK, Duarte EA. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J Virol. 1991;65:2960–7. doi: 10.1128/jvi.65.6.2960-2967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner PE, Elena SF. Cost of host radiation in an RNA virus. Genetics. 2000;156:1465–70. doi: 10.1093/genetics/156.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zárate S, Novella IS. Vesicular stomatitis virus evolution during alternation between persistent infection in insect cells and acute infection in mammalian cells is dominated by the persistence phase. J Virol. 2004;78:12236–42. doi: 10.1128/JVI.78.22.12236-12242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remold SK, Rambaut A, Turner PE. Evolutionary genomics of host adaptation in vesicular stomatitis virus. Mol Biol Evol. 2008;25:1138–47. doi: 10.1093/molbev/msn059. [DOI] [PubMed] [Google Scholar]
- 12.Weaver SC, Brault AC, Kang W, Holland JJ. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J Virol. 1999;73:4316–26. doi: 10.1128/jvi.73.5.4316-4326.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper LA, Scott TW. Differential evolution of eastern equine encephalitis virus populations in response to host cell type. Genetics. 2001;157:1403–12. doi: 10.1093/genetics/157.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greene IP, Wang E, Deardorff ER, Milleron R, Domingo E, Weaver SC. Effect of alternating passage on adaptation of Sindbis virus to vertebrate and invertebrate cells. J Virol. 2005;79:14253–60. doi: 10.1128/JVI.79.22.14253-14260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coffey LL, Vasilakis N, Brault AC, Powers AM, Tripet F, Weaver SC. Arbovirus evolution in vivo is constrained by host alternation. Proc Natl Acad Sci USA. 2008;105:6970–5. doi: 10.1073/pnas.0712130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Opijnen T, de Ronde A, Boerlijts MC, Berkhout B. Adaptation of HIV-1 depends on the host-cell environment. PLoS ONE. 2007;3:e271. doi: 10.1371/journal.pone.0000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenner CE, Wang X, Ponz F, Walsh JA. A fitness cost for Turnip mosaic virus to overcome host resistance. Virus Res. 2002;86:1–6. doi: 10.1016/s0168-1702(02)00031-x. [DOI] [PubMed] [Google Scholar]
- 18.Wallis CM, Stone AL, Sherman DJ, Damsteegt VD, Gildow FE, Schneider WL. Adaptation of Plum pox virus to a herbaceous host (Pisum sativum) following serial passages. J Gen Virol. 2007;88:2839–45. doi: 10.1099/vir.0.82814-0. [DOI] [PubMed] [Google Scholar]
- 19.Agudelo-Romero P, de la Iglesia F, Elena SF. The pleiotropic cost of host-specialization in Tobacco etch potyvirus. Infect Genet Evol. 2008;8:806–14. doi: 10.1016/j.meegid.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz-Jarabo CM, Pariente N, Baranowski E, Dávila M, Gómez-Mariano G, Domingo E. Expansion of host-cell tropism of foot-and-mouth disease virus despite replication in a constant environment. J Gen Virol. 2004;85:2289–97. doi: 10.1099/vir.0.80126-0. [DOI] [PubMed] [Google Scholar]
- 21.Novella IS, Hershey CL, Escarmís C, Domingo E, Holland JJ. Lack of evolutionary stasis during alternating replication of an arbovirus in insect and mammalian cells. J Mol Biol. 1999;287:459–65. doi: 10.1006/jmbi.1999.2635. [DOI] [PubMed] [Google Scholar]
- 22.Cuevas JM, Moya A, Elena SF. Evolution of RNA virus in spatially structured heterogeneous environments. J Evol Biol. 2003;16:456–66. doi: 10.1046/j.1420-9101.2003.00547.x. [DOI] [PubMed] [Google Scholar]
- 23.Brown JS, Pavlovic NB. Evolution in heterogeneous environments: effects of migration on habitat specialization. Evol Ecol. 1992;6:360–82. [Google Scholar]
- 24.Holt RD. Adaptive evolution in source-sink environments: direct and indirect fitness effects of density-dependence on niche evolution. Oikos. 1996;75:182–92. [Google Scholar]
- 25.Kawecki TJ. Adaptation to marginal habitats: contrasting influence of the dispersal rate on the fate of alleles with small and large effects. Proc R Soc Lond B. 2000;267:1315–20. doi: 10.1098/rspb.2000.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maynard Smith J, Hoekstra R. Polymorphism in a varied environment: how robust are the models? Genet Res. 1980;35:45–57. doi: 10.1017/s0016672300013926. [DOI] [PubMed] [Google Scholar]
- 27.van Tienderen PH. The evolution of generalists and specialists in spatially structured heterogeneous environments. Evolution. 1991;45:1317–31. doi: 10.1111/j.1558-5646.1991.tb02638.x. [DOI] [PubMed] [Google Scholar]
- 28.Duffy S, Turner PE, Burch CL. Pleiotropic costs of niche expansion in the RNA bacteriophage ø6. Genetics. 2006;172:751–7. doi: 10.1534/genetics.105.051136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feris MT, Joyce P, Burch CL. High frequency of mutations that expand the host range of an RNA virus. Genetics. 2007;176:1013–22. doi: 10.1534/genetics.106.064634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang XZ, Lee BTK, Wong SM. Covariation in the capsid protein of Hibiscus chlorotic ringspot virus induced by serial passaging in a host that restricts movement leads to avirulence in its systemic host. J Virol. 2002;76:12320–4. doi: 10.1128/JVI.76.23.12320-12324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rico P, Ivars P, Elena SF, Hernández C. Insights into the selective pressures restricting Pelargonium flower break virus genome variability: evidence for host adaptation. J Virol. 2006;80:8124–32. doi: 10.1128/JVI.00603-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Presloid JB, Ebendick-Corpus BE, Zárate S, Novella IS. Antagonistic pleiotropy involving promoter sequences in a virus. J Mol Biol. 2008;382:342–52. doi: 10.1016/j.jmb.2008.06.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilke CO, Forster R, Novella IS. Quasispecies in time-dependent environments. Curr Top Microbiol Immunol. 2006;299:33–50. doi: 10.1007/3-540-26397-7_2. [DOI] [PubMed] [Google Scholar]