Abstract
Objective
To develop a genotype exercise to improve pharmacy students’ comprehension of pharmacogenetic principles that apply to patient care.
Design
Deoxyribonucleic acid (DNA) was collected during class from 10 student volunteers and subjected to genotype analysis. The results were presented to the class and discussed in the context of a patient genetic counseling session. Students completed a survey instrument regarding their attitudes toward this learning experience.
Assessment
Students indicated that the exercise engaged them with the course content and would positively influence their ability to apply pharmacogenetic principles to patient care.
Conclusion
An applied genotype exercise enhanced learning of pharmacogenetic principles. Based on these findings, conducting a genotype exercise in a large classroom setting is feasible in terms of time and expense, and meaningful in terms of student satisfaction.
Keywords: genotype, polymerase chain reaction, cardiovascular disease, angiotensin converting enzyme, pharmacogenetics
INTRODUCTION
The ability to precisely predict a patient's pharmacologic response to a medication using genetic information, called pharmacogenetics and pharmacogenomics, is changing pharmacy practice.1,2 In 2002, the Academic Affairs Committee of the American Association of Colleges of Pharmacy (AACP) recommended that pharmacy curricula incorporate a comprehensive instructional approach to prepare future practitioners in this evolving field.3
A new 3-credit hour course in clinical pharmacogenetics and pharmacogenomics was designed and administered to third-year doctor of pharmacy (PharmD) students during winter quarter in 2007 at The Ohio State University College of Pharmacy. Development of this new course was a direct result of recommendations in our last self-study for accreditation conducted in 2004. The primary objective of this course is to instruct students on the principles needed to understand the role of human genetic variation and using patient-specific information to predict the pharmacokinetic and pharmacodynamic response to medications.
A survey was conducted at the conclusion of the first course offering (winter 2007). In general, students perceived pharmacogenetics as a potentially important aspect of pharmacy practice; however, many had a difficult time envisioning how they would utilize this information within their future practice. In addition, many students found the technology associated with identifying patient genotypes in pharmacogenetic-based clinical trials and clinical practice difficult to grasp. Finally, a majority of students suggested that the addition of workshops or exercises to the course work would enhance their overall learning experience. Following careful consideration of this valuable feedback, a decision was made to integrate an exercise into the course that would comprehensively address these major concerns. As a result, a longitudinal genotyping exercise that connected across multiple lectures from the first week of the course to the last week was designed. The genotype exercise was administered at the time of the second course offering. The primary objective of the exercise was to provide students the opportunity to participate and engage in an applied activity that covered major aspects of pharmacogenetics and included use of terminology and technology, interpretation of genetic information, and application to patient care. The details and results of this novel, large classroom instructional activity are reported here.
DESIGN
The Clinical Pharmacogenetics course originally consisted of two 1½-hour lectures per week taught by multiple instructors from the Division of Pharmaceutics and Division of Pharmacy Practice and Administration in a large classroom setting with 115 students. The course was divided into 3 major didactic components that began with background lectures on genetics, molecular biology, terminology, and technology. The second section involved lectures on the influence of polymorphic genetic variation as they pertain to drug metabolism, drug transport, and drug target receptors. The remaining portion of the course was devoted to major disease and organ systems in which pharmacogenetics has, or is beginning to have, a clinical impact on patient care.
The expected outcomes of the course were that, upon completion, students would be able to analyze and apply patient-related pharmacogenetic data in order to synthesize an action plan for personalized patient care. To better achieve this outcome, we developed and added a genotype exercise that was incorporated throughout the course (Table 1). A significant challenge during the planning stage was to identify an “ideal” gene that would yield clinically meaningful information and be relevant to aspects of the course content, as well as be deliverable in a large classroom setting. In addition, the gene had to be chosen with careful consideration to the safety and well-being of the student volunteers.
Table 1.
Lectures Presented in an Elective Clinical Pharmacogenetics Course Relevant to Conducting Genotyping
PCR = polymerase-chain-reaction
ACE = angiotensin converting enzyme
Knowing that identification of polymorphic variation among patients (eg, the patient genotype) serves as a foundation for the majority of all patient-related interventions in the context of applied pharmacogenetics, we chose to engage students in a genotype-related exercise. The technology, level of sophistication, and expense associated with genotyping varies significantly. Therefore, we had to evaluate different options as we developed the exercise so that it would have intellectual value, yet be feasible, economical, and reproducible in a large classroom setting. Our original intention was to identify a clinically relevant gene that was discussed in the course so that the exercise would integrate well with the didactic components involving technology and applied pharmacogenetics. In addition, we wanted to identify a gene that had a high degree of heterogeneity so that we would observe differences between the genotypes of student volunteers within a small group (sample size, n = 10). Following consultation with our institutional review board (IRB), we were informed that the gene preferably should not be associated with substantial adverse consequences so as not to jeopardize or traumatize students upon revealing their specific genotype in a large classroom setting.
After these considerations, we chose to use the angiotensin converting enzyme (ACE) gene for the genotype exercise. The product of the ACE gene converts the inactive decapeptide, angiotensin I, into the active octapeptide and potent vasoconstrictor antiotensin II, which is the main active product of the renin-angiotensin system (RAS).4 In turn, the RAS controls long-term regulation of blood pressure and blood volume. Therefore, this gene has important physiologic, pathophysiologic, and therapeutic implications. Furthermore, the discovery that circulating ACE levels are under genetic control is well established.5 The ACE gene, and particularly the insertion/deletion polymorphism, has been one of the most extensively studied since the advent of genomics and pharmacogenetics. Given the clinical relevance and polymorphic nature of the ACE gene, its discussion was incorporated into the pharmacogenetics of cardiovascular disease section of the course, thereby satisfying one of our requirements.
One of the first polymorphisms identified in the ACE gene was found to code for the presence (insertion, I) or absence (deletion, D) of a 287 base pair sequence of DNA in intron 16.5 Taking advantage of the significant difference in size between the insertion and deletion alleles, Lindpaintner and colleagues first reported the development of a convenient polymerase chain reaction (PCR)-based assay that allows rapid delineation of the ACE genotype at this location. Furthermore, the 3 possible genotypes (II and DD homozygotes, and ID heterozygotes) all commonly occur within the general population. The protocol is standardized, readily available, and highly reproducible, and incorporates the use of PCR, a technology covered in the course material. Furthermore, the high degree of variability commonly expressed at this location within the ACE gene predicted a high likelihood that we would identify DD, II, and ID carriers in a small sample size. The specific details of the adapted genotype procedure are provided below.
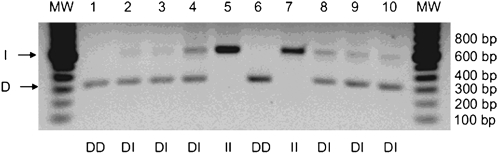
The D and I alleles were identified on the basis of polymerase-chain-reaction (PCR) amplification of the respective fragments from intron 16 of the ACE gene and size fractionation and visualization by electrophoresis. All laboratory work was conducted in the laboratory of the corresponding author by an experienced research technician. One microliter of whole blood was added to 5 μl of GeneReleaser (Bioventures, Murfreesboro, TN) and taken through 2 cycles of heating to 97ºC and cooling to 8ºC, according to the manufacturer's recommendations. After incubation at 80ºC for 30 minutes, 20 μl of a PCR master mix containing 1 μM primers, 200 μM deoxynucleotide triphosphates, 1.3 mM magnesium chloride, 50 mM potassium chloride, 10 mM TRIS–hydrochloric acid (pH 8.4 at 25ºC), 0.1 % Triton X-100, and 0.35 unit of Thermus aquaticus DNA polymerase was added. An optimized primer pair was used to amplify the D and I alleles, resulting in 319-bp and 597-bp amplicons, respectively (ACE(56)-F: 5-GCCCTGCAGGTGTCTGCAGCATGT-3 and ACE(56)-R: 5-GGATGGCTCTCCCCGCCTTGTCTC-3). The thermocycling procedure (PTC 100 apparatus, MJ Research, Watertown, MA) consisted of denaturation at 94ºC for 30 seconds, annealing at 56ºC for 45 seconds, and extension at 72ºC for 2 minutes, repeated for 35 cycles, followed by a final extension at 72ºC for 7 minutes. After the addition of 5 μl of a glycerol-based loading buffer, 7 μl of the mixture was loaded onto a 1.5 % submarine agarose slab (FMC, Rockland, ME) containing 40 mM TRIS acetate, 2 mM EDTA, and 1 μg of ethidium bromide per milliliter of solution and fractionated according to size at 5 V per centimeter. The amplification products of the D and I alleles were identified by 300-nm ultraviolet transillumination as distinct bands; in heterozygous samples a third band, assumed to represent a heteroduplex DNA product, was commonly seen (Figure 1).
Figure 1.
Genotype analysis of the angiontensin converting enzyme (ACE) gene in ten student volunteers. Genomic DNA was isolated from 10 de-identified buccal swab specimens obtained from student volunteers. Purified DNA was then subject to standard PCR to identify the presence (insertion, I) or absence (deletion, D) of a 287 base pair sequence of the ACE gene within intron 16. As shown, both the insertion (I) and deletion (D) alleles were observed across the ten samples. Further, the frequency of II, ID, and DD carriers was exactly within the expected pattern of distribution. The results, as shown, were presented during a large class workshop in the context of a patient-counseling workshop.
Our original intention was to have a large workshop discussion that emphasized the clinical implications of genotype results. Based on this, an assumption was made that demonstration of genetic variability in student volunteers would have higher impact and provide a basis for a more thorough and engaging discussion. Our plan was to incorporate these components into a simulated patient counseling experience during the final workshop session. Finally, following careful consideration and in consultation with the IRB, we concluded that the I/D genotypes were not associated with serious consequences sufficient to prevent their use as a suitable gene for the exercise. As a result, IRB exempt status was granted. Based on all of these considerations and the fact that PCR-based reactions for genotype analyses were covered in detail during the technology section of the course, it was determined that the ACE gene was well suited for the classroom exercise.
The educational environment consisted of a large classroom setting. On the first day of class, we introduced the organizational outline of the genotype exercise and then requested 10 student volunteers. In addition, students were informed that a buccal swab would be administered to the volunteers at the beginning of the second lecture in order to initiate the process of DNA isolation. All 10 volunteers were present and participated in the buccal swab procedure, which required approximately 10 minutes of class time. Samples were immediately de-identified to protect the privacy of volunteers. However, students were provided the opportunity to record their sample identities in order to determine their ACE genotype upon completion of the exercise. Even though this was not an authentic clinical diagnostic test, the IRB ruled that personal information could not be withheld from volunteers. Importantly, provisions were put in place to offer genetic counseling to student volunteers. A 20-minute lecture on the technical aspects of the procedure for DNA isolation and PCR-based genotype analysis then followed, and students were informed that test results would be introduced and discussed in a workshop format during the final week of class. The content of the workshop was developed in collaboration with a clinical genetic counselor from our own institution. The exercise was designed as a simulated counseling encounter that included obtaining patient consent, acquiring a DNA sample, interpreting findings, and developing an action plan for the patient.
On the day of the buccal swab procedure, volunteer samples were transported immediately after lecture to the laboratory of a course instructor. The genomic DNA from each sample was extracted on the same day, using a commercial kit (BuccalAmp DNA Extraction Kit; Epicentre Biotechnologies, Madison, WI). The samples were then stored in a −80°C freezer until further analysis. A PCR-based procedure was then used to amplify a specific region of the ACE gene, thereby revealing the genotype of each volunteer sample. The reaction products were resolved using an agarose gel and then photographed for presentation in the workshop (Figure 1). The sample isolation and PCR analysis procedures for all samples took approximately 5 hours to complete. The cost for all reagents, excluding personnel time, was approximately $400. Major equipment required for the exercise included a thermal-cycler for PCR, a spectrophotometer for DNA quantitation, and small equipment that would be commonly found in a standard molecular biology laboratory, including a power supply, a submarine electrophoresis chamber, and an ultraviolet visualization chamber.
In week 3, 2 lectures on technology were given in which the instructor introduced PCR technology and how the technique is used to conduct genotype analysis. The procedure for taking the volunteer samples was then highlighted in class. In addition, the original scientific citation that identified the PCR-genotype procedure was posted online as supplemental reading material.5 Cardiovascular pharmacogenetics was covered during the last week of class over a span of 1.5 lectures. During this time, the ACE gene and related clinical studies were discussed. On the final half-day of class, the workshop revealing the results of the analysis was presented. A brief slide presentation detailing the structure of the exercise was revisited before revealing the results of the genotype analysis. The results of genotype analysis were then presented to the entire class (Figure 1) and discussed in the context of a patient-oriented genetic counseling session. Instructor preparation for this component of the workshop involved a detailed discussion with a certified genetic counselor at our institution. This counselor, who routinely engaged in all aspects of individual genetic counseling, helped structure our exercise based on the context of an actual patient encounter. The classroom experience incorporated aspects of the entire counseling process, including pretest and posttest decision-making.
EVALUATION AND ASSESSMENT
At the conclusion of the course, student opinions were gathered via a survey instrument from our online course management system, which is built on the Desire2Learn platform. The survey consisted of 4 Likert-scale questions and 5 open-ended questions. Students were offered 2 additional points that would be factored into their overall course grade as incentive for completing the survey instrument. The survey function in Desire2Learn allowed respondents to anonymously provide feedback, which the course instructors then collated.
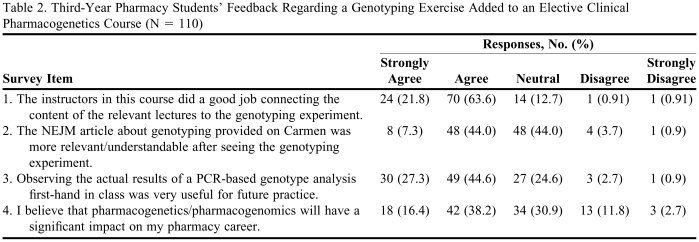
Of the 115 students enrolled in the Clinical Pharmacogenetics course offered in winter 2008, 110 (96%) completed the genotyping exercise survey. The survey consisted of four 5-point (strongly agree to strongly disagree) Likert-scale type questions and an additional 5 open-ended questions relating to both the genotyping exercise and the class in general. Survey questions and responses based on the Likert scale are shown in Table 2. Eighty-five percent of the students agreed or strongly agreed that the genotyping exercise was beneficial in terms of helping them better connect to course content, while only 2% found the exercise not useful. More than half of the respondents felt that the exercise helped them understand a scientific article that focused on genotyping, while 4 % did not. Although more than 70% of the respondents felt that the exercise would benefit them in future practice, only 44% felt that pharmacogenomics would impact their pharmacy careers. Furthermore, when asked “On a scale of 1-10, where 1 = not at all useful and 10 = very useful, how would you rate the genotyping experiment?” the average response was a 7, and only 9 students answered with a value below 5.
Table 2.
Third-Year Pharmacy Students’ Feedback Regarding a Genotyping Exercise Added to an Elective Clinical Pharmacogenetics Course (N = 110)
Abbreviations: NEJM = New England Journal of Medicine; PCR = polymerase-chain-reaction
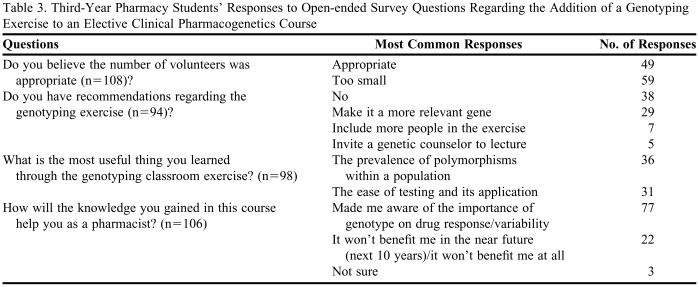
Open-ended questions related to the genotyping exercise along with a representative sample of the major themes from the most prevalent responses are presented in Table 3. The majority of respondents understood that the number of students used in the exercise was sufficient to retrieve a sample size capable of distinguishing among the 3 major ACE polymorphisms. However, most students wanted to increase the sample size. The following is a student quote that reflects one of the consensus opinions that emerged: “The number was large enough to mimic the normal population distribution of that polymorphism, but I think it should have included more (students) because it is fun to think of this technology and how we can get involved—literally.”Although most students did not have any recommendations to improve the personal relevance of the exercise, over 30% of respondents wanted to genotype a clinically relevant gene that directly effects medication decision making, as reflected in these student comments: “I think it should be an influential gene,”and “Pick a SNP that has clinical impact.”
Table 3.
Third-Year Pharmacy Students’ Responses to Open-ended Survey Questions Regarding the Addition of a Genotyping Exercise to an Elective Clinical Pharmacogenetics Course
The prevalence of a given polymorphism in the population and the ease of detecting candidate polymorphisms were 2 major themes that appeared when students were asked: “What is the most useful thing you learned through the genotyping classroom exercise?” One response stated: “It helped me realize that genetic differences really do exist and that you don't have to look very far to recognize them;” “Now I know how quickly genetic testing can be performed and its implications for drug therapy.” Finally, when asked “How will the knowledge you gained in this course help you as a pharmacist?” 73% of the students responding indicated that pharmacogenetic knowledge would make them more aware of drug response variability due to different genetic profiles, which in turn could lead to a change in dosing paradigms, as typified in this response: “It will make me more aware of genotype and drug response in patients.” Nonetheless, more than 20% of respondents indicated that pharmacogenomics will not play a role early in their pharmacy careers, as typified in these responses: “I don't see it being relevant in the next 10-20 years at least,”and “I feel that this is something that won't apply to pharmacy for at least 10 more years.” In sharp contrast, a number of students described that the exercise helped to clarify how pharmacogenetics will be used in their future practice, as expressed in this response: “It was nice to see visually the differences within our classroom. By actually participating and seeing the results, it seems more real than if we had not done the genotyping exercise. I definitely think you need to keep this exercise in the curriculum.”
The time required to perform the analysis outside of class time for DNA extraction and PCR analysis was approximately 5 hours to process 10 samples. Genotype analysis was conducted by an instructor with the assistance of 1 laboratory technician. Both individuals had prior experience working with DNA and conducting PCR-related analysis. The supplies included one BuccalAmp DNA Extraction sufficient for 15 extractions kit (BQ0901S; Epicentre Technology, Madision, WI, at $61 per kit), DNA primers for PCR analysis at an approximate cost of $20 per primer, and the reagents necessary for PCR amplification (Invitrogen Supermix II, $279), for a total cost of approximately $360. The Invitrogen supermix II kit is a ready-for-use reagent. Although use of this reagent decreased laboratory preparation time, it was more expensive to purchase compared to the summative cost of purchasing individual reagents. The equipment necessary to conduct the PCR reaction and genotype analysis included a standard PCR thermocycler for PCR, a horizontal DNA submersion gel chamber, a bench-top power supply to run the DNA agarose gel, and a fluorescence imaging processor to visualize and photograph the agarose gel. All of the equipment is standard in most cellular and molecular biology laboratories, and it is reasonable to speculate that these items would be available at many institutions. If this were not the case, cost would likely be prohibitive, especially if the equipment were acquired only for the genotype exercise.
DISCUSSION
Upon completion of the second offering of this course, an online survey that focused on the genotyping exercise was administered to students. Overall, student feedback was positive, indicating that the exercise was useful and would help in preparation for the use of pharmacogenetic-related information in practice. Based on feedback from students and faculty members, the genotype exercise will be incorporated into future course offerings.
A major challenge of developing a large class genotype exercise was to carefully identify a suitable gene and then plan the logistics of the exercise. As described above, the genotype exercise required IRB review before the exercise was administered to students. In addition to the approval process, the dialogue with IRB was valuable during the planning stage and helped to identify suitable candidate genes. At the onset, it was not obvious that the ACE gene would be ideal, but this gene met the requirements of the IRB because it is not associated with serious health consequences. Although other genes met this criteria, the ACE gene was more favorable since the high frequency of allelic variation would appear in a small sample size of volunteers (n = 10). Upon review of our protocol, the IRB considered the exercise to be exempt and no further approval was required. Their conclusion was based of the fact that the gene was not associated with a detrimental phenotype that would have potential adverse consequences to the student volunteers. Also, as a result, students were able to determine their individual genotype at the conclusion of the exercise, although confidentiality was maintained in the classroom setting.
A major factor in the overall assessment of the curricular design was the ability of the teaching team to execute the genotyping exercise. Although the original exercise was restricted to 10 student volunteers, it is reasonable to consider expanding the exercise to involve more students, a consideration consistently recommended by students. Further, the number of volunteers substantially exceeded the original request of 10 students; thus, a larger sample size would have been readily obtainable.
In the end, student feedback indicated that identification of heterogeneity in classmates was a powerful finding that had a positive impact on their view of pharmacogenetics. In fact, a significant number of students recommended that the analysis be conducted on all students. When considering the time and cost associated with processing 115 samples, it is currently prohibitive at our institution to accommodate this request. Expanding the number of volunteers would require adding a laboratory component to the course and additional personnel, or require that PCR-based analysis be conducted with more commonly used automated technologies that facilitate high through-put analysis. As described, there are considerable costs associated with the equipment required for PCR-based genotype analysis. We anticipate that these items would be available at most pharmacy programs associated with research intensive institutions, however, if this were not the case, the cost associated with equipment purchase could be prohibitive. Our approach was to use a straightforward approach involving gel electorphoresis-based analysis. A benefit of this technique is that students are able to directly visualize the amplified DNA product using an economically feasible platform. On the other hand, this technique is outdated and not commonly employed in the clinical laboratory setting. It is also not amenable to high through-put processing. Given these limitations, others have developed and utilized virtual Web-based activities to facilitate higher learning in a pharmacogenetics/pharmacogenomics course.6,7
Although the majority of US pharmacy schools provide some instruction in pharmacogenetics/pharmacogenomics and plan to increase it in the coming years,8 many do not provide the depth of instruction recommended by an AACP academic affairs committee.3 In our experience, a clinically applied pharmacogenetic course was designed, guided by AACP recommendations, and delivered to third-year PharmD students. Student feedback after the first offering indicated a disconnect in student perception of the practical use and application of the course, as well as difficulty grasping the technology used to conduct laboratory testing. In response to these deficits, we designed a longitudinal genotype exercise. Our experience with this exercise is that it served the intended purposes of helping students apply genetic information in the context of medication management and become more comfortable with application of information and technology in patient care. Key to success with this exercise was developing an integrated, as opposed to isolated, exercise that involved course content over the span of the entire course. The capstone workshop that brought together fundamental elements across the entire pharmacogenetics spectrum (terminology, sample acquisition, analysis, interpretation, and counseling) enhanced the learning experience in a manner that we contend provided the students with more confidence to utilize pharmacogenetic skills with patients in their future practices. Utilization of a genotype-based exercise in the classroom is not entirely novel,9 but we believe ours is the first report of such an exercise conducted in a pharmacy classroom setting. Most important, as more programs develop pharmacogenetic/pharmacogenomic-based courses, we hope that this experience will serve as a useful guide when considering the use of a hands-on genotyping or related exercise to enhance student learning.
SUMMARY
A longitudinal genotyping exercise was developed and administered during the second course offering of a new clinical pharmacogenetic and pharmacogenomic course. The primary objective of the exercise was to provide students with the opportunity to participate and engage in an applied activity that involved fundamental aspects of pharmacogenetics, including DNA sample acquisition, analysis, and interpretation. A secondary motive was to develop a longitudinal experience that was dispersed across the course so that fundamental components of the exercise could be strategically incorporated into the course content. This applied genotype exercise enhanced learning of pharmacogenetic principles, and this paper shows that conducting a genotype exercise in a large classroom setting is feasible, in terms of time and expense, and meaningful, in terms of student satisfaction.
ACKNOWLEDGEMENTS
Special thanks to Julie A. Johnson and Taimour Langaee for consultation during the early planning stages of this educational endeavor.
REFERENCES
- 1.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–91. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 2.Evans WE, McLeod HL. Pharmacogenomics—drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–49. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JA, Bootman LJ, Evans WE, Hudson RA, Knoell D, Simmons L, Straubinger RM, Meyer SM. Pharmacogenomics: A scientific revolution in pharmaceutical sciences and pharmacy practice. Report of the 2001-2002 academic affairs committee. Am J Pharm Sci. 2002;66:12S–15S. [Google Scholar]
- 4.Sayed-Tabatabaei FA, Oostra BA, Isaacs A, van Duijn CM, Witteman JC. ACE polymorphisms. Circ Res. 2006;98:1123–33. doi: 10.1161/01.RES.0000223145.74217.e7. [DOI] [PubMed] [Google Scholar]
- 5.Lindpaintner K, Pfeffer MA, Kreutz R, et al. A prospective evaluation of an angiotensin-converting-enzyme gene polymorphism and the risk of ischemic heart disease. N Engl J Med. 1995;332:706–11. doi: 10.1056/NEJM199503163321103. [DOI] [PubMed] [Google Scholar]
- 6.Kirkpatrick G, Orvis K, Pittendrigh B. A teaching model for biotechnology and genomics education. J Biol Educ. 2002;37:31–5. [Google Scholar]
- 7.Busstra MC, Hartog R, Kersten S, Muller M. Design guidelines for the development of digital nutrigenomics learning material for heterogeneous target groups. Adv Physiol Educ. 2007;31:67–75. doi: 10.1152/advan.00090.2006. [DOI] [PubMed] [Google Scholar]
- 8.Latif DA. Pharmacogenetics and pharmacogenomics instruction in schools of pharmacy in the USA: is it adequate? Pharmacogenomics. 2005;6:317–9. doi: 10.1517/14622416.6.4.317. [DOI] [PubMed] [Google Scholar]
- 9.Levis-Fitzgerald M, Denson N, Kerfeld CA. Undergraduate students conducting research in the life sciences: opportunities for connected learning. Online Submission. Paper presented at the Annual Meeting of the Association for the Study of Higher Education; November 17 – 19, 2005; Philadelphia, PA. [Google Scholar]