SUMMARY
Diversity-generating retroelements (DGRs) introduce vast amounts of sequence diversity into target genes. During mutagenic homing, adenine residues are converted to random nucleotides in a unidirectional, reverse transcriptase-dependent transposition process from a donor template repeat (TR) to a recipient variable repeat (VR). Using a Bordetella bacteriophage DGR as a model, we demonstrate that homing occurs through a TR-containing RNA intermediate and is RecA-independent. Marker transfer studies show that cDNA integration at the 3′ end of VR occurs within a (G/C)14 element, and deletion analysis demonstrates that the reaction is independent of 5′-end cDNA integration. cDNA integration at the 5′ end of VR requires only short stretches of sequence homology. We propose that homing occurs through a unique target DNA-primed reverse transcription (TPRT) mechanism that precisely regenerates target sequences. This non-proliferative, “copy and replace” mechanism enables repeated rounds of protein diversification and optimization of ligand-receptor interactions.
INTRODUCTION
Retroelements are genetic elements that encode reverse transcriptases (RTs) and function through RNA intermediates (Eickbush, 1994; Temin, 1989). They include not only retroviruses, but also diverse retrotransposons, retrons and retroplasmids (Galligan and Kennell, 2007). Retroelements are widely distributed in eukaryotic and prokaryotic organisms and the viruses that infect them. A unique class of retroelements, diversity-generating retroelements (DGRs), have recently been found in bacteria and phages (Doulatov et al., 2004). DGRs are capable of conferring overt selective advantages to their host genomes via their ability to diversify protein-encoding DNA sequences (Medhekar and Miller, 2007).
The prototypic DGR was discovered in a bacteriophage, BPP-1, which infects the mammalian respiratory pathogen Bordetella bronchiseptica (Liu et al., 2002). The infectious cycle of this organism is controlled by the BvgAS signal transduction system which alternates between active (Bvg+) and inactive (Bvg−) states to regulate expression of cell surface and secreted factors that mediate respiratory infection or ex vivo survival, respectively (Cotter and Jones, 2003). The BPP-1 phage receptor is pertactin, a Bvg+ phase-specific outer membrane protein which serves as a virulence factor and a protective immunogen (Liu et al., 2002). The observation that BPP-1 is capable of producing variants that preferentially infect Bvg− phase Bordetella led to the discovery that receptor tropism can be altered by the activity of a phage-encoded element, the DGR. The range of receptors that can be recognized by DGR-diversified BPP-1 variants appears to be vast, allowing phage adaptation to the highly dynamic surface of their bacterial hosts. The potential utility of DGRs is illustrated by the identification of over 40 related elements in bacterial, phage, and plasmid genomes. DGRs are present in human pathogens (Treponema, Legionella spp.), human commensals (Bacteroides, Bifidobacterium spp.), green sulfur bacteria (Chlorobium, Prosthecochloris spp.), cyanobacteria (Trichodesmium, Nostoc spp.), magnetotactic bacteria (Magnetospirillum spp.), and many other diverse species. (Medhekar and Miller, 2007; Gingery et al., unpublished data).
BPP-1 tropism switching is mediated by a phage-encoded DGR which introduces nucleotide substitutions in a 134 base pair (bp) variable repeat (VR) located at the 3′ end of mtd, which encodes the phage receptor binding protein (Figure 1A), (Liu et al., 2002; McMahon et al., 2005). Variable sites in VR correspond to adenine residues in the homologous template repeat (TR), which is invariant and essential for DGR function. As silent substitutions in TR are transmitted to VR during tropism switching, TR supplies the raw sequence information for variability. Information transfer occurs through a unidirectional mechanism in which TR adenine residues are converted to random deoxyribonucleotides which appear at the corresponding positions in VR, a process called mutagenic homing (Doulatov et al., 2004; Liu et al., 2002). In addition to TR and VR, homing requires a unique RT encoded by the brt locus, and atd, a small open reading frame (ORF) of unknown function (Medhekar and Miller, 2007). The BPP-1 DGR TR has 23 adenine residues subject to mutagenesis, yielding a library of 423, or about 1014 different VR DNA sequences. This corresponds to a theoretical diversity of ~1013 polypeptides, rivaling the repertoires of mammalian T cell receptors (Liu et al., 2002; Medhekar and Miller, 2007). The structures of five, distinct Mtd variants have recently been determined (McMahon et al., 2005). VR-encoded variable amino acids are presented by a C-type lectin fold at the C-terminus of the Mtd protein. Variable residues are all solvent exposed, forming three discrete binding surfaces on the bottom face of tetrahedron-shaped Mtd homotrimers, two of which are positioned at the distal tips of each of the six phage tail fibers.
Figure 1. DGR Homing Occurs through an RNA Intermediate.
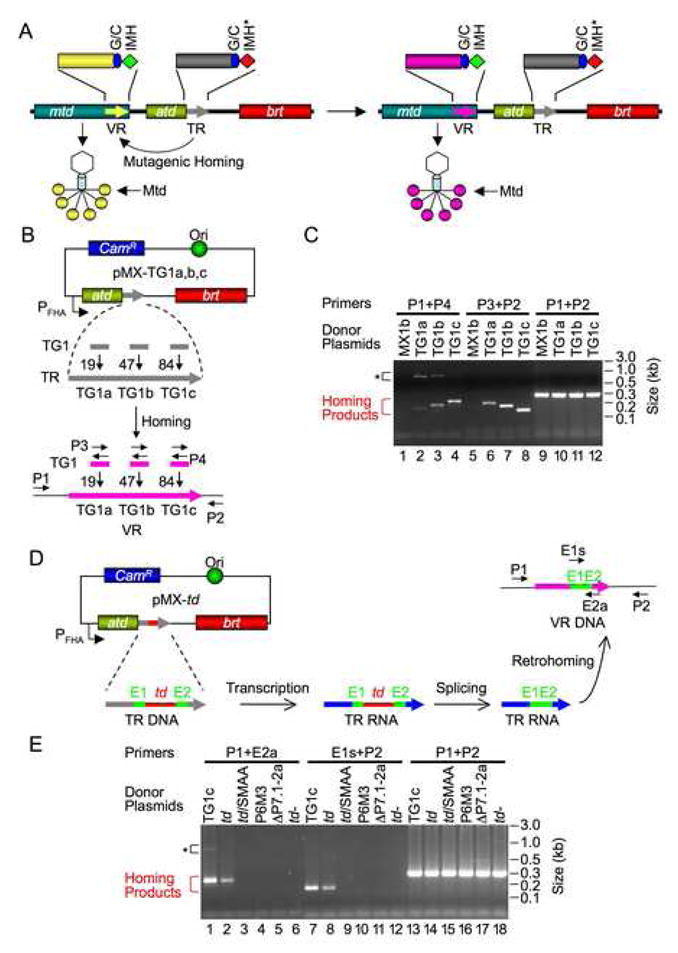
(A) Bordetella phage DGR cassette undergoes mutagenic homing and facilitates tropism switching. mtd, atd, brt and the two repeats (VR and TR) are indicated. VR and TR are expanded to show the (G/C)14, IMH and IMH* elements. DGR homing leads to VR diversification, resulting in progeny phages with altered Mtd trimers at the distal ends of tail fibers.
(B) PCR-based DGR homing assays. Plasmids pMX-TG1a, b, c are derived from pMX1b, which carries the BPP-1 atd-TR-brt region placed downstream of the BvgAS-regulated fhaB promoter. pMX-TG1a, b, c contain 36 bp inserts (TG1), corresponding to ligated exons of the T4 td group I intron flanked by SalI sites, at TR positions 19 (TG1a), 47 (TG1b) or 84 (TG1c), respectively. Grey and pink arrows represent TR and VR, respectively. Small horizontal arrows indicate primers used for homing assays: P1, sense-strand primer located in mtd; P2, antisense-strand primer downstream of VR; P3 and P4 are sense- and antisense-strand primers, respectively, that anneal to ligated td exons. CamR, chloramphenicol resistance gene.
(C) pMX-TG1a, b, c are functional donors for DGR homing. Assays were performed following BPP-1d single-cycle lytic infection of B. bronchiseptica RB50 cells transformed with the indicated donor plasmids. pMX1b (negative control) lacks the TG1 insert. Larger PCR products (*) in lanes 2–4 are Brt-independent PCR artifacts (Figure 3, lanes 1&2 and data not shown). They contain sequences from mtd to TR, have not undergone adenine mutagenesis, and were likely generated by PCR template switching between BPP-1d DNA and contaminating donor plasmids in phage DNA preparations. P1+P2 product levels demonstrate that approximately equal amounts of input DNA were used for PCR homing assays.
(D) Testing the DGR retrohoming hypothesis. The T4 td group I intron (td) and flanking exons (E1, E2) were inserted at position 84 in TR. Following transcription and intron splicing, ligated exons will be retained in a subset of TR-containing transcripts. If homing occurs via an RNA intermediate (retrohoming), some VRs will acquire ligated exons. Primers used for PCR homing assays are indicated by small horizontal arrows: P1 and P2 are described in (B); E1s, sense-strand primer annealing to E1; E2a, antisense-strand primer annealing to E2.
(E) Precisely ligated td exons are transferred to VR during homing. Products from PCR homing assays with indicated primer pairs and donor plasmids are shown. TG1c, pMX-TG1c positive control; td, pMX-td donor plasmid; td/SMAA, RT-deficient pMX-td derivative; P6M3 and ΔP7.1-2a, splicing-defective pMX-td derivatives; td-, pMX1b with the td intron and flanking exons inserted in inverted orientation at TR position 84. *, Brt-independent PCR artifacts.
Mutagenic homing by the BPP-1 DGR requires a 21 bp sequence located at the 3′ end of VR, designated the Initiation of Mutagenic Homing (IMH) site (Doulatov et al., 2004). It differs from its counterpart at the 3′ end of TR, IMH*, by only 5 bp. This subtle difference prevents TR from becoming a homing recipient, and a simple substitution of IMH* with IMH converts TR into a recipient of diversified sequence information. Thus, IMH* plays a critical role in preserving the mutagenesis capability of the DGR by preventing deadeninization of TR. Immediately upstream of IMH and IMH* are identical 14 bp G/C-only sequences [(G/C)14]. Since IMH is never observed to vary in the wild-type DGR, VR mutagenesis is confined to sequences upstream of the (G/C)14/IHM elements (Doulatov et al., 2004).
At present, mechanisms of DGR homing and adenine-specific mutagenesis are not well characterized. Based on the requirement for an RT activity, homing was hypothesized to occur through an RNA intermediate (Liu et al., 2002; Medhekar and Miller, 2007). Using a plasmid donor system expressing the BPP-1 atd, TR, and brt loci in trans (Xu et al., unpublished data), we show that TR can accommodate insertions of heterologous sequences and that adenine mutagenesis occurs efficiently during transfer of inserted sequences to VR. By engineering a self-splicing group I intron into TR, we provide conclusive evidence that DGR homing occurs via an RNA intermediate and is RT-dependent. We also identified regions of the TR-containing RNA transcript that are important for homing. Interestingly, although VR and TR share significant homology, homing was found to be RecA-independent. A marker coconversion assay showed that cDNA initiation likely occurs within the (G/C)14 element of VR, and further analysis demonstrated that cDNA initiation does not require VR sequences upstream of the (G/C)14 region, but does require IMH. cDNA integration upstream of the (G/C)14 element requires only short stretches of homology between VR and cDNA, and is otherwise sequence-independent. On the basis of these and other results, we propose that DGR homing initiates via a specialized target DNA-primed reverse transcription (TPRT) mechanism. Complete cDNA integration may occur through cDNA template switching or strand displacement. This model renders mechanistic insights into the unique, non-proliferative, “copy and replace” pathway of DGR homing, and accounts for the ability of the DGR to regenerate target sequences in a manner that enables repeated rounds of homing and VR diversification. In addition to shedding light on this recently discovered family of unusual retroelements, our results suggest new approaches for DGR-based genetic engineering.
RESULTS
Multiple Sites in TR Can Tolerate Heterologous Sequence Insertions
We initially set out to tag the BPP-1 TR with a self-splicing group I intron, which could then be used to determine whether DGR homing occurs through an RNA intermediate. Intron tagging is a classic method for verifying retrotransposition of mobile genetic elements (Boeke et al., 1985; Cousineau et al., 1998; Guo et al., 2000; Moran et al., 1996). As a prerequisite, the ability of TR to tolerate DNA insertions was first assessed. We inserted a 36 bp fragment at three different positions in TR on plasmid pMX1b, which expresses atd, TR, and brt from the BvgAS activated fhaB promoter (Figure 1B) (Jacob-Dubuisson et al., 2000). The 36 bp fragment contains the ligated exons of the phage T4 td group I intron, flanked by SalI sites (Cousineau et al., 1998; Guo et al., 2000). The resulting plasmids pMX-TG1a, pMX-TG1b, and pMX-TG1c, and the parental plasmid pMX1b, were transformed into B. bronchiseptica strain RB50. Transformed cells were induced to express pertactin, the BPP-1 phage receptor, and to activate the fhaB promoter. Following single-cycle lytic infection with a derivative of BPP-1 containing null mutations in TR and brt (BPP-1d), a homing assay was performed on DNA isolated from progeny phages. Although BPP-1d is defective for tropism switching and DGR activity, it can be efficiently complemented by pMX1b in trans. Using the 36 bp insert as a tag (TG1), we devised a PCR-based assay to detect TR-derived TG1’s transferred to VR as a result of homing. As shown in Figure 1B, three sets of primers were used: primers P1/P4 amplify TG1’s transferred to VR along with upstream sequences; primers P2/P3 amplify TG1’s transferred to VR along with downstream sequences; and primers P1/P2 amplify VR and flanking sequences to confirm equal input of phage DNA in PCR reactions.
Using primer pairs P1/P4, and P2/P3, we detected PCR products of predicted sizes resulting from complementation with all of the constructs containing TG1 (Figure 1C). No products were detected with the same primers following complementation with pMX1b, which lacks TG1, or with the brt-deficient plasmid pMX-TG1c/SMAA (data not shown; Figure 3). Sequence analysis of PCR products demonstrated adenine mutagenesis of TG1 inserts as well as flanking VR sequences (Figures S1–3 and data not shown), confirming that they were derived from homing events. Of the three TR insertion sites, position 84 appeared to retain the highest homing activity. These results demonstrate that TR can tolerate heterologous sequence insertions and that inserted sequences are transferred to VR and are subject to adenine mutagenesis. The PCR assay shown in Figure 1B provides a sensitive and specific means to detect DGR homing products in a manner that is independent of tropism switching or phage infectivity.
Figure 3. Homing Does Not Require RecA.
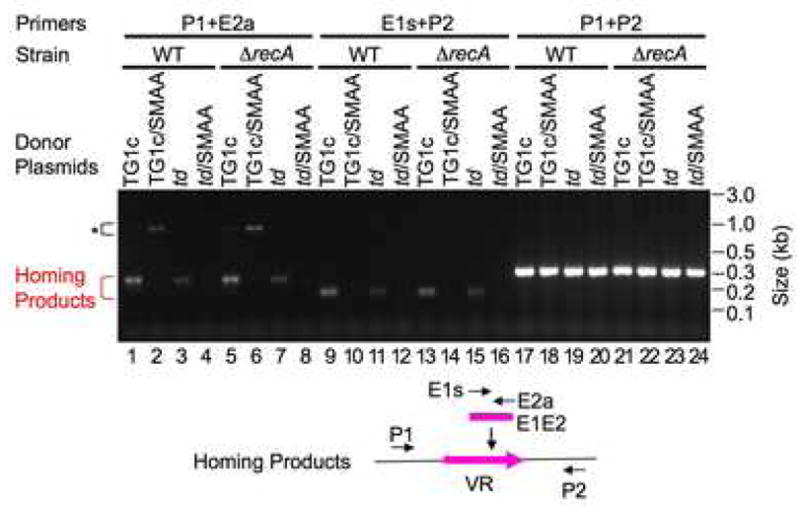
DGR homing assays were performed following BPP-1d single-cycle lytic infection of RB50 or RB50ΔrecA cells transformed with the indicated plasmids. TG1c/SMAA, RT-deficient mutant of pMX-TG1c (Figure 1B); other donor plasmids have been described (Figure 1E). Primers used in homing assays are shown below the gel. *, Brt-independent PCR artifacts.
DGR Homing Occurs through an RNA Intermediate
To test the hypothesis that DGR homing occurs through a TR-containing RNA intermediate, we used a modified group I intron, tdΔ1–3, which lacks most of the intron ORF but retains self-splicing activity (Cousineau et al., 1998; Guo et al., 2000). Plasmid pMX-td contains the modified td intron inserted into TR at position 84 (Figure 1D). Following intron splicing, pMX-td transcripts will only retain ligated exons. If our hypothesis is correct, some VRs that have undergone mutagenic homing should acquire precisely ligated exons from the intron-bearing TR.
The td intron was verified to be capable of RNA splicing in Bordetella through both reverse transcription-polymerase chain reaction (RT-PCR) and primer extension-termination assays (Figure S4) (Zhang et al., 1995). No adenine substitutions were detected in RT-PCR products generated from spliced transcripts, suggesting that nucleotide alterations do not occur at the RNA level. We next set out to determine whether the intron-tagged TR is capable of homing and to characterize the homing products. RB50 cells transformed with pMX-td or control plasmids were infected with BPP-1d, and DNA isolated from progeny phages was subjected to PCR analysis. Two td exon primers were used to detect homing products: E1s is a sense-strand primer annealing to exon 1 (E1), and E2a is an antisense-strand primer annealing to exon 2 (E2) (Figure 1D). With primers P1/E2a, and primers E1s/P2, we observed PCR products generated with pMX-td that were identical in size to those from the positive control, pMX-TG1c, which contains ligated exons inserted at TR position 84 (Figure 1E). This suggested that spliced transcripts had been used for homing. The lower amount of homing products observed with pMX-td compared to pMX-TG1c correlates with the observation that the spliced RNA product produced by pMX-td corresponds to ~18% of the pMX-TG1c transcripts containing ligated exons (Figure S4B). Characterization of homing products confirmed that the td intron was precisely excised, and that adenine mutagenesis occurred in the ligated exons and flanking sequences (Figure S5).
Consistent with a retrohoming process, detection of ligated-exon products required a functional td intron, as splicing-defective mutants (P6M3 and ΔP7.1-2a; Figure S4) (Mohr et al., 1992), and a construct with an inverted intron (td−), failed to generate homing products (Figure 1E). Interestingly, we did not detect transfer of intron sequences from TR to VR, and this was true for functional as well as nonfunctional derivatives of the td intron. The failure to detect products containing unspliced or unsplicible introns appears to be due to a size limitation for heterologous sequences inserted into TR at position 84. TR can only tolerate insertions of up to ~200 bp at this site (Tse et al., unpublished), and this limit is exceeded by inserts containing splicing-competent (429 bp) or defective (397–429 bp) td introns. As expected, transmission of ligated exons to VR is RT-dependent, as an RT-deficient mutant (td/SMAA) failed to support detectable homing. Taken together, our results demonstrate that DGR homing is a retrotransposition process which occurs via a TR-containing RNA intermediate.
Regions of the RNA Transcript Important for Homing
To identify sequence requirements for the RNA intermediate involved in homing, we constructed a series of donor constructs containing deletions within TR and a unique 32 bp tag (TG2) at the deletion junctions (Figure 2A). Using PCR-based homing assays with primers P1/P6 and P5/P2, we discovered that most of TR is dispensable (Figure 2). Only ~10 bp at the 5′ end, and ~38 bp at the 3′ end are required for homing. The 3′ sequence requirements correspond almost entirely to the (G/C)14 and IMH* elements. Interestingly, deletion constructs ΔTR23-84, ΔTR33-84, and ΔTR33-96 consistently generated a higher abundance of homing products than the donor with a full-length TR (FL in Figure 2C), suggesting that shorter TRs may support more efficient homing. Consistent with these results, TR sequences between the TG1a and TG1c insertions (positions 20 to 84) were found to be dispensable, and their deletion increased DGR homing activity (Figure S7). Although 10 bp were sufficient for homing, optimal activity requires 10–19 bp at the 5′ end of TR. Sequence analysis showed that PCR products from functional TR deletion derivatives underwent adenine mutagenesis, confirming that they were generated by mutagenic homing (data not shown).
Figure 2. Internal TR Sequences Are Dispensable for Homing.
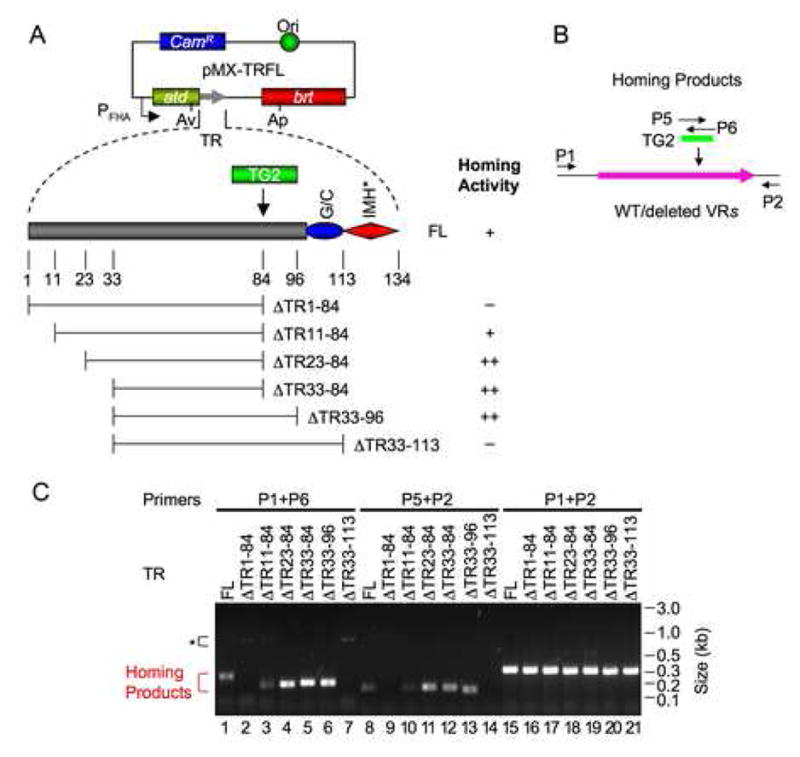
(A) TR deletion constructs and homing activities. All donor plasmids are derivatives of pMX1b, containing AvrII (Av) and ApaI (Ap) sites in atd and brt, respectively, as well as a sequence tag (TG2). AvrII and ApaI sites were introduced via silent mutations and do not affect DGR homing (Figure S6). FL (full-length) is a positive control with TG2 inserted at position 84. ΔTR1-84, ΔTR11-84, ΔTR23-84, ΔTR33-84, ΔTR33-96 and ΔTR33-113 contain TR deletions with TG2 at deletion junctions. Results from PCR homing assays are summarized: +, homing efficiency similar to the FL positive control; ++, homing activity greater than FL; −, no homing activity detected.
(B) Primers used for homing assays. P5 and P6 are sense- and antisense-strand primers annealing to TG2, respectively; primers P1 and P2 are described in Figure 1.
(C) PCR homing assays with TR deletion constructs. Homing assays were performed following BPP-1d single-cycle lytic infection of RB50 cells transformed with the indicated plasmids. *, Brt-independent PCR artifacts.
Consistent with the above data, mutations at positions 8–11 of TR, and in the (G/C)14 and IMH* elements, were deleterious (Figure S8). Furthermore, silent mutations at the 3′ end of atd and a substitution in the intergenic region between TR and brt (mutation T5 in Figure S8) also significantly decreased homing efficiency. These results demonstrate that internal sequences are largely dispensable, and TR function requires the ends of the repeat and flanking upstream and downstream sequences. These may be required for the integrity of an RNA structure important for homing.
Retrohoming is RecA Independent
RecA is required for repairing UV-induced DNA damage and homologous recombination (Courcelle and Hanawalt, 2003). Considering the extent of TR/VR homology, it is important to evaluate the role of RecA in DGR homing. We generated a B. bronchiseptica RB50ΔrecA strain by allelic exchange. The recA knockout was verified by PCR, sequencing, and loss of RecA-dependent DNA repair in a UV-sensitivity assay (data not shown). We analyzed the ability of donor plasmids pMX-TG1c, pMX-td, and their RT-deficient mutants to complement BPP-1d in wild-type RB50 and its isogenic recA-deficient derivative (Figure 3). The relative yields of homing products generated from DGR-competent plasmids in wild-type and recA mutant strains were indistinguishable, indicating that DGR retrotransposition is RecA-independent. Consistent with these results, tropism switching by BPP-1d was also found to be RecA-independent (Figure S9).
Marker Coconversion Boundaries
As DGR homing occurs through an RNA intermediate and is RecA-independent, we suspected that cDNA integration at the 3′ end of VR might occur through a TPRT reaction as opposed to homologous recombination following cDNA synthesis. To identify potential priming site(s) for TPRT, we determined if there is a specific boundary at the 3′ end of TR for sequence transfer to VR (Figure 4A). Such a boundary could indicate a site at which cDNA synthesis initiates. We also wished to determine if there is a boundary at the 5′ end of TR for sequence transfer, which could define a potential cDNA integration site at the 5′ end of VR (Figure 4A). We generated a series of donor constructs with single C to T substitutions at multiple positions in TR, and inserted TG1 at position 84 to facilitate PCR homing assays (Figure 4B). C to T substitutions were chosen to minimize disruption of potential RNA structures since G residues can form both G-C base pairs and G-U wobble pairs, and single markers were introduced into individual constructs to minimize disruption of homology. Since thymidine residues are not altered during DGR homing, marker coconversion assays should allow a precise mapping of VR sequences that have been acquired from TR. From the boundaries of marker coconversion, sites of cDNA initiation at the 3′ end and integration at the 5′ end of VR can be inferred.
Figure 4. Marker Conconversion Analysis of DGR Retrohoming.
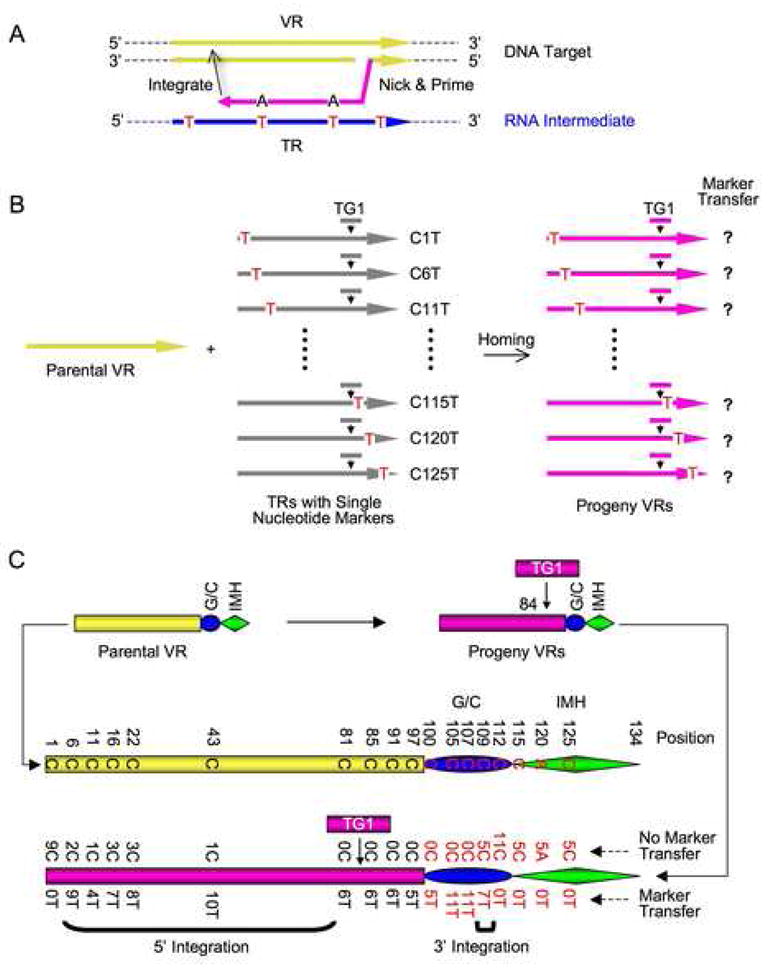
(A) Strategy to identify cDNA integration sites at the 3′ and 5′ ends of VR via marker coconversion. Markers introduced into TR (red Ts) are transferred to VR only if they are located between 3′ cDNA priming and 5′ cDNA integration sites.
(B) Schematic of coconversion experiments. Single C to T markers in donor TRs are indicated. All TRs are tagged with TG1 at position 84.
(C) Homing assays were performed following BPP-1d single-cycle lytic infection of RB50 cells transformed with marked donor plasmids. Results from PCR homing assays are shown in Figure S10 and marker coconversion data are summarized here. Nucleotides at relevant positions in the wild type parental VR are shown in the center. At bottom, the number of progeny VRs with a transferred marker, or no transferred marker at specific sites are shown. Deduced cDNA integration regions at the 3′ and 5′ ends of VR are indicated by brackets.
19 donor constructs, with C to T substitutions at TR positions upstream or downstream of the TG1 insertion, were generated and tested for homing into the VR of BPP-1d (Figure 4B; Figure S10). Although some substitutions in the (G/C)14 and IMH* elements slightly decreased homing activity, all constructs supported sufficient homing to allow marker coconversion analysis. DGR homing products were cloned and sequences were determined for multiple independent clones derived from each donor construct. As summarized in Figure 4C, marker coconversion downstream of TG1 occurred with 100% efficiency at positions 85 to 107. At position 109, 5 clones showed marker transfer while 7 did not. No marker transfer occurred at position 112 or at positions further downstream. These data identify a boundary for marker transfer from TR to VR, located within the (G/C)14 element between positions 107 and 112. This coconversion boundary is likely to represent sites for cDNA initiation during homing. The heterogeneity observed at position 109 suggests that initiation can occur at multiple sites between positions 107 and 112.
Substitutions in the 5′ region of TR displayed a more diffuse pattern of marker transfer. The marker immediately upstream of TG1 (C81T) was transmitted with 100% efficiency. This was expected, since PCR homing assays select for TG1 transfer with nearby sequences subject to co-selection. At greater distances upstream of the tag, coconversion was observed for the majority of clones, with the exception of the marker at position 1. The observation that markers at positions 6, 11, 16, 22 and 43 were partially transferred suggests that cDNA integration can occur before extending to the 5′ end of TR. Template switching or strand displacement mediated by VR/TR homology could account for these results. Sequences from PCR assays displayed adenine mutagenesis, confirming their assignment as DGR homing products.
cDNA Integration at the 3′ End of VR Is Independent of 5′-End Integration
Results from marker coconversion assays suggest that homing could take place in a sequential manner, with cDNA integration at the 3′ end of VR occurring first through a TPRT reaction, followed by cDNA integration at the 5′ end mediated by VR/TR homology. We sought to determine whether cDNA integration at the 3′ end of VR could occur independently of 5′-end integration by deleting all VR sequences upstream of the (G/C)14 and IMH elements, creating prophage BPP-1d ΔVR1-99 which lacks the first 99 bp of VR (Figure 5A). If our hypothesis is correct, the VR1-99 deletion should eliminate cDNA integration at the 5′ end, preventing complete homing but allowing 3′-end cDNA integration. As a negative control, we replaced IMH with IMH* to generate prophage BPP-1dΔVR1-99IMH*.
Figure 5. cDNA Integration at the 3′ End of VR.
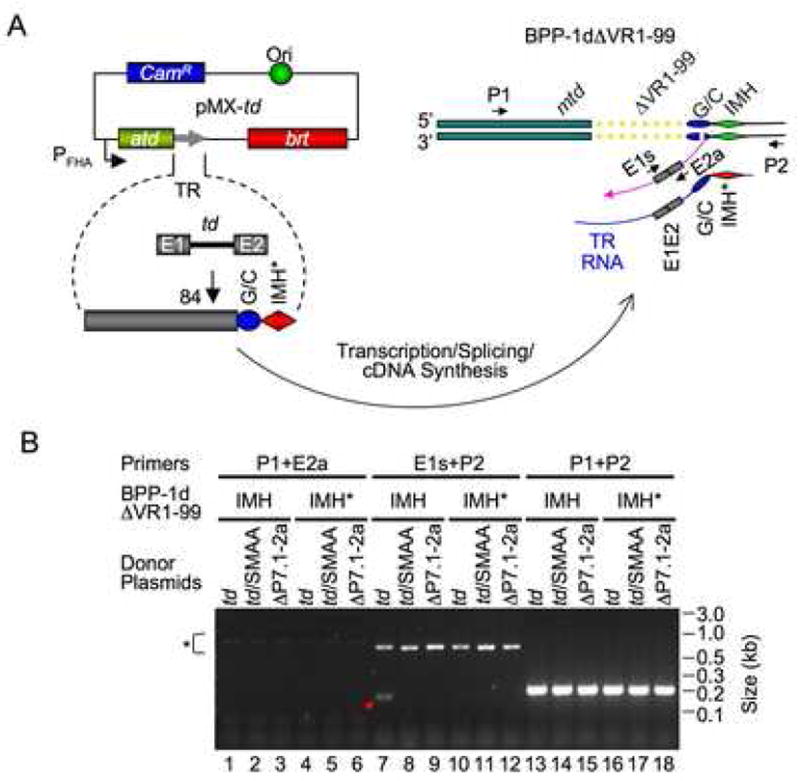
(A) Experimental design. Donor plasmid pMX-td contains the td group I intron at position 84 in TR, and prophage BPP-1dΔVR1-99 lacks the first 99 bp of VR. cDNA integration at the 3′ or 5′ end of VR was assessed in PCR assays using the same primer pairs described in Figure 1D. (B) VR lacking the first 99 bp supports cDNA integration at the 3′ end, but not the 5′ end. BPP-1dΔVR1-99 (IMH) and BPP-1dΔVR1-99IMH* (IMH*) lysogens transformed with the indicated donor plasmids (described in Figure 1E) were induced with mitomycin C for 2 hrs. Total nucleic acids isolated from induced cultures were assayed for cDNA integration by PCR using primers shown in (A). Pink arrow, cDNA intermediate. *, Brt-independent PCR artifacts.
The td intron-tagged pMX-td (Figure 1D), and negative control derivatives pMX-td/SMAA (RT-deficient) and pMX-td/ΔP7.1-2a (splicing-defective), were used to complement mutant prophage in homing assays. By using an intron-tagged TR, we can identify true cDNA products since they will acquire ligated exons. With primers E1s and P2, a specific product of the expected size (167 bp) was detected in DNA isolated from pMX-td transformed BPP-1dΔVR1-99 lysogens (Figure 5B, lane 7). This product was not detected in samples from the same lysogen complemented with Brt-deficient or splicing-defective donor plasmids, nor in samples from BPP-1dΔVR1-99IMH* lysogens transformed with pMX-td. These results suggested that the 167 bp product was the result of a Brt- and IMH-dependent retrohoming reaction. This was confirmed by sequence analysis, which demonstrated precise exon ligation and adenine mutagenesis (Figure S11). In contrast, PCR assays with primers P1 and E2a did not detect a specific product (Figure 5B, lanes 1–6). This was not due to primer failure, since this primer pair efficiently amplified a product of correct size (240 bp) in progeny phages when pMX-TG1c transformants were infected with BPP-1d (data not shown; Figure 1E and 3). The product detected in Figure 5B (lane 7, arrow) appears to represent a cDNA intermediate “trapped” in the homing reaction, as depicted in Figure 5A.
Taken together, these results demonstrate that cDNA integration at the 3′ end of VR can occur independently of 5′-end integration, consistent with the hypothesis that DGR homing initiates through a TPRT mechanism. cDNA initiation at the 3′ end of VR requires IMH, but is independent of VR sequences or VR/TR homology upstream of the (G/C)14 element.
cDNA Integration at the 5′ End of VR
We hypothesized that cDNA integration at the 5′ end of VR requires only TR/VR homology upstream of the (G/C)14 and IMH elements, as opposed to specific sequences. To test this, we determined whether the homing defect resulting from the VR1-99 deletion could be rescued by inserting a 50 bp segment of mtd (M50), derived from sequences upstream of VR, into the TR of plasmid pMX-ΔTR23-84 (Figure 2A). The resulting plasmid, pMX-M50 (Figure 6A), and control constructs pMX-M50/SMAA (Brt-deficient) and pMX-ΔTR23-84 (lacking the M50 insert) were evaluated for their ability to support homing in BPP-1d and BPP-1dΔVR1-99 lysogens.
Figure 6. cDNA Integration at the 5′ End of VR.
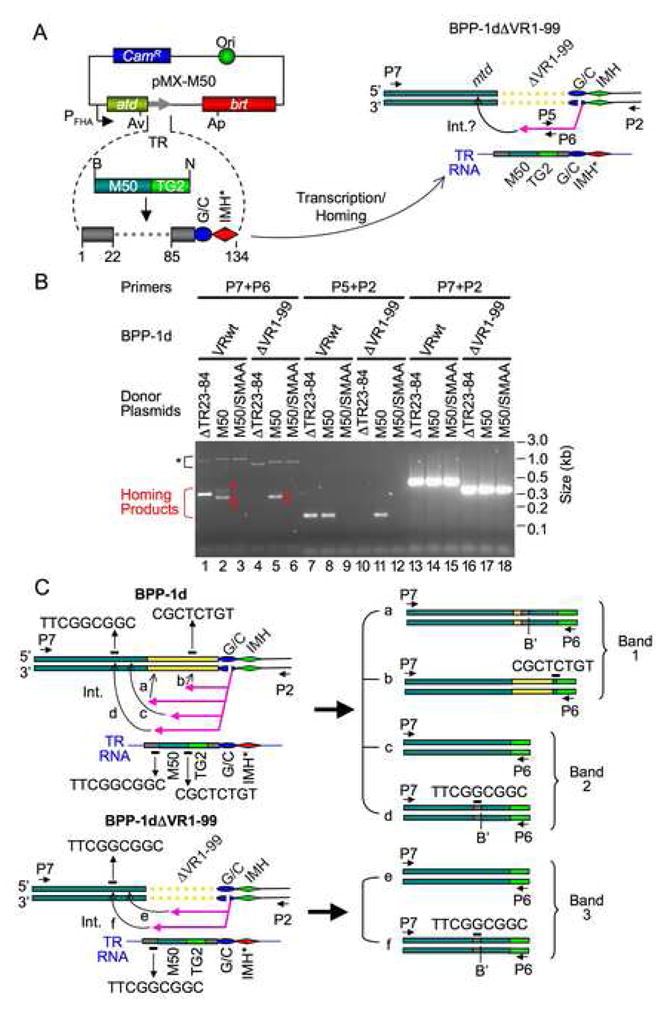
(A) Strategy to identify requirements for cDNA integration at the 5′ end of VR. Donor plasmid pMX-M50 is a derivative of pMX-ΔTR23-84 (Figure 2A) containing an insert in TR consisting of a 50 bp mtd segment (M50, derived from sequences located upstream of VR). The M50 insert in TR provides homology to the mtd locus in prophage BPP-1dΔVR1-99. Primers are indicated as small horizontal arrows: P7 is a sense-strand primer annealing to mtd; P2, P5 and P6 are described in Figure 1 and Figure 2B. Int., integration.
(B) The M50 insert in TR restores homing into sequences upstream of VR in BPP-1dΔVR1-99. PCR assays were performed on DNA isolated from intact phage particles produced following induction by mitomycin C. RB50 lysogens carried BPP-1d prophage genomes with wild type VR sequences (VRwt) or the VR1-99 deletion (ΔVR1-99). Donor plasmids were as follows: ΔTR23-84, pMX-ΔTR23-84; M50, pMX-M50; M50/SMAA, an RT-deficient mutant of pMX-M50. Product bands 1 and 2 in lane 2 and band 3 in lane 5 are labeled. *, Brt-independent PCR artifacts. (C) Major products derived from bands 1, 2 and 3 in (B) are shown and described in the text. Yellow bars, VR1-99 or portions thereof.
As shown in Figure 6B, pMX-ΔTR23-84 and pMX-M50 supported homing into the VR of BPP-1d with similar efficiencies, while the Brt-deficient donor did not. Interestingly, pMX-M50 yielded two PCR products in the expected size range with primers P7/P6 (Figure 6B, lane 2), both of which were cloned and characterized. We suspected that band 1 corresponded to cDNA integration within the first 22 bp region of VR mediated by TR/VR homology, generating a slightly larger product than observed with pMX-ΔTR23-84 due to the M50 insert (Figure 6C, a). Out of 15 clones analyzed, 3 had this structure, and adenine mutagenesis patterns showed they were the products of mutagenic homing events (Figure S12A). A majority (8/15) of the clones obtained from band 1, however, corresponded to cDNA integration at VR positions 60–67 via a homologous 8 nucleotide (nt) sequence located at the junction of the M50 insert and the TG2 tag (Figure 6C, b; Figure S12B). These also appeared to be derived from true homing products, as they were the major species from band 1, which was not detected with the Brt-deficient mutant pMX-M50/SMAA (Figure 6, lane 2 vs. 3). Integration most likely occurred through cDNA template switching from the engineered TR to VR within the 8 nt homologous sequence. In addition, 4 minor species of clones were detected, each of which can also be accounted for by cDNA integration through template switching between short stretches (4–13 bp) of sequence homology (data not shown).
Sequence analysis of clones from band 2 revealed two products of the same size. One (Figure 6C, c; Figure S13A) corresponds to 5′ cDNA integration through the M50 insert in TR. The other product (Figure 6C, d; Figure S13B) corresponds to cDNA integration via yet another fortuitously occurring short stretch of sequence homology (9 nt), located 6 bp upstream of the M50 insert on the donor plasmid and within mtd on the phage genome. All clones displayed adenine mutagenesis within the M50 sequence (Figure S13), confirming that they resulted from DGR homing. Adenine-mutagenized homing products of the expected size resulting from complementation with pMX-M50 were also detected using primers P5/P2 (Figure S14).
When tested in BPP-1dΔVR1-99 lysogens, pMX-ΔTR23-84 did not generate detectable homing products (Figure 6B, lanes 4&10). In contrast to the experiment shown in Figure 5, in which total DNA was used to identify homing intermediates, the assays in Figure 6 used DNA isolated from intact phage particles to eliminate abortive products. Although pMX-ΔTR23-84 is capable of cDNA integration at the 3′ end of VR, the lack of integration at the 5′ end appears to prevent packaging. In contrast, the presence of the 50 bp mtd segment in pMX-M50 efficiently restored Brt-dependent homing (Figure 6B, lanes 5&6, 11&12). Sequence analysis of pMX-M50 homing products amplified with primers P7/P6 (Figure 6B, lane 5, band 3) revealed two species of identical size. One corresponds to cDNA integration within the M50 region of mtd due to homology (Figure 6C, e; Figure S15A). The other species (Figure 6C, f; Figure S15B) resulted from cDNA integration at the same 9 nt sequence implicated in the generation of product d. Adenine mutagenesis was observed in the 50 bp mtd sequence in the majority of homing products derived from pMX-M50 (Figure S15). Taken together, these results demonstrate that cDNA integration at the 5′ end of VR, upstream of the (G/C)14 element, is homology-driven. Furthermore, only short stretches of nucleotide identity between the cDNA and target sequences are required to complete the homing reaction.
DISCUSSION
DGRs are a unique family of retroelements that use RT-mediated mobility to generate diversity in protein-encoding DNA sequences (Doulatov et al., 2004). Our results suggest that DGRs have evolved an adaptation of TPRT which is site-specific, and capable of precisely regenerating target sequences. This “copy and replace” pathway allows continuous rounds of protein diversification and the creation of new binding specificities for ligand-receptor interactions.
The RNA Intermediate
We demonstrated that the BPP-1 TR can accommodate sequence insertions at multiple sites. Inserted sequences were not only transferred to VR, but they also underwent adenine mutagenesis. The observation that heterologous sequences can be diversified by a DGR has practical applications as discussed below. In the course of these experiments, we developed a sensitive and selective PCR assay that allows the identification and characterization of VR sequences that have specifically undergone mutagenic homing.
By engineering a self-splicing group I intron into the BPP-1 DGR TR, we were able to show that homing occurs through a TR-containing RNA intermediate. This conclusion is based on the observation that precisely spliced, adenine-mutagenized exons were transferred to VR. Initially used in yeast to demonstrate retrotransposition of Ty1 (Boeke et al., 1985), intron tagging has become a “gold standard” for identifying retrotransposition (Cousineau et al., 1998; Guo et al., 2000; Moran et al., 1996). Further experiments revealed sequence requirements for the RNA intermediate. Surprisingly, deletion analysis showed that most internal sequences in TR are nonessential, with only ~10 bp at the 5′ end and ~38 bp at the 3′ end required for homing. The 10 bp at the 5′ end of TR could form part of an essential RNA structure and/or provide homology for cDNA integration into VR. The 3′-end requirements are almost entirely composed of the (G/C)14 and IMH* elements. Although sequences internal to TR are largely dispensable, regions of the RNA transcript important for homing extend upstream and downstream of TR. Synonymous mutations used here (Figure S8), and an accompanying analysis by Xu et al. (unpublished data), show that sequences extending into the 3′ end of atd (~42 bp upstream of TR), and the 5′ end of brt (~194 bp downstream of TR) form the boundaries of the RNA transcript required for maximum activity. Work is currently in progress to characterize RNA sequences and structures that are important for homing.
Mechanisms of cDNA Integration
We developed a marker coconversion assay, based on the introduction of single-nucleotide markers in TR, to genetically map cDNA integration sites at the 3′ and 5′ ends of VR. Sequence analysis of homing products revealed a narrow boundary for marker coconversion at the 3′ end, occurring within the (G/C)14 element (Figure 4C). We interpret this to represent the sites at which cDNA synthesis initiates during homing. Priming is likely to occur following DNA cleavage, generating a 3′-OH for TPRT. A TPRT mechanism for cDNA integration within the (G/C)14 element explains our previous observation that mutagenesis only occurs upstream of this element; adenines in IMH* never cause mutations in IMH, and IMH coconversion to IMH* is never observed (Doulatov et al., 2004; Liu et al., 2002). In contrast to the 3′ end, the boundary of marker coconversion at the 5′ end of VR was diffuse, suggesting that cDNA integration can occur at virtually any position upstream of the point of initiation.
Our results predict a sequential process in which cDNA integration initially occurs within the (G/C)14 element, followed by 5′-end integration driven by TR/VR homology. To test this we generated a recipient phage lacking the first 99 bp of VR. This deletion includes all VR sequences located upstream of the (G/C)14 and IMH elements. PCR homing assays detected 3′-, but not 5′-end integration products, indicating that cDNA integration at the 3′ end of VR is independent of integration at the 5′ end. The ability to detect 3′-end integration was IMH-dependent, indicating that IMH dictates the unidirectional nature of sequence transfer by mediating 3′-end cDNA initiation. As no 5′-end integration products were identified, the 3′ integration products most likely resulted from amplification of cDNA intermediates “trapped” in the homing reaction.
Complete homing into a BPP-1 derivative missing the first 99 bp of VR could be restored by inserting a 50 bp homologous segment of mtd into TR (Figure 6B). These data show that cDNA integration at the 5′ end of VR is homology-driven and does not depend on specific VR sequences. Interestingly, by analyzing numerous products from this and other homing events, we discovered that short stretches of sequence homology as small as 4–12 bp are sufficient to mediate cDNA integration at sites upstream of the (G/C)14 element. As expected, transposition of the mtd segment from TR to VR was accompanied by efficient adenine mutagenesis of the heterologous sequences. RecA-mediated homologous recombination machinery is not required for DGR function, as a recA deletion had no effect on mutagenic homing or phage tropism switching. In this respect, DGR homing resembles group II intron homing in Escherichia coli and cDNA-mediated gene conversion by the Ty1 retroelement in Saccharomyces cerevisiae, which occur in the absence of RecA or its yeast homologs Rad51, Rad55 and Rad57, respectively (Cousineau et al., 1998; Derr, 1998).
A Model for DGR Function
Our results support the model for DGR-mediated diversity generation outlined in Figure 7. DGR homing is a unique, site-specific retrotransposition process that does not lead to a copy number increase in either TR or VR. It most likely initiates through a TPRT mechanism primed with either a single-stranded nick in the antisense strand, or a double-stranded break within the (G/C)14 element. This leads to a narrow boundary of marker coconversion at the 3′ end of VR. Our model predicts that cDNA initiation involves sequence-specific recognition of the (G/C)14 and IMH elements and, potentially, adjacent downstream sequences. It also predicts the existence of an endonuclease activity which could be provided by a protein and/or a catalytic RNA. Although cDNA initiation occurs within a narrow boundary, the heterogeneity observed at position 109 (Figure 4C) suggests a slight relaxation in the specificity of the proposed endonuclease. The model proposed in Figure 7 is consistent with past and present data, and with the close evolutionary relationships between DGRs and group II introns which are known to use TPRT for retrohoming (Doulatov et al., 2004; Lambowitz and Zimmerly, 2004).
Figure 7. A Model for DGR Mutagenic Homing.
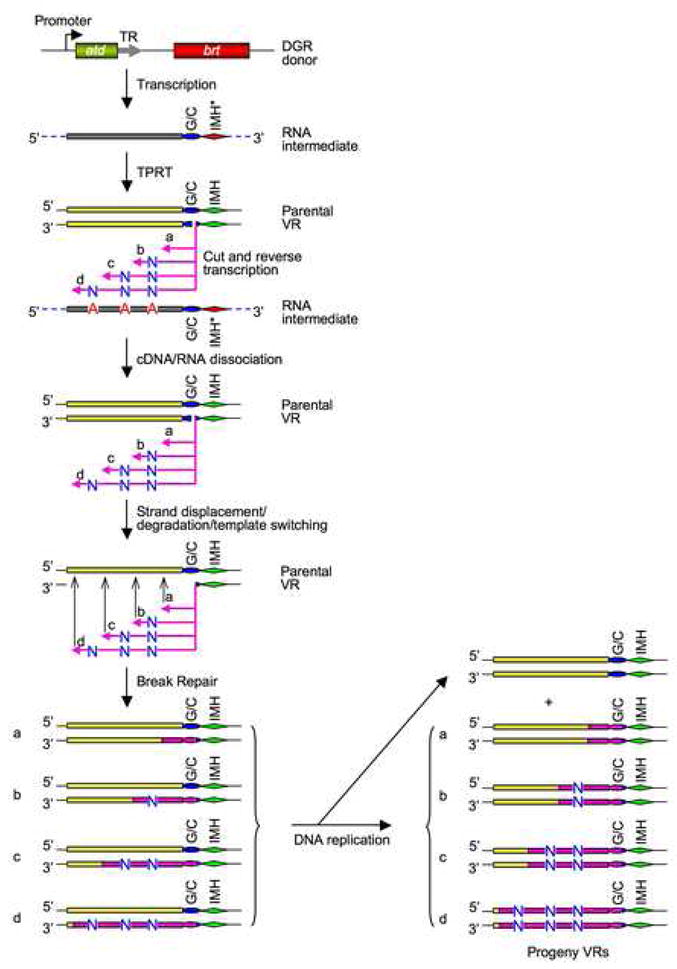
DGR homing occurs via a unique, “copy and replace” pathway that substitutes parental VR sequences with diversified cDNA copies of TR transcripts. cDNA integration at the 3′ end of VR is proposed to occur via a TPRT mechanism, while integration at the 5′ end of VR requires short stretches of TR/VR sequence homology and occurs through template-switching and/or strand displacement. Adenine mutagenesis most likely occurs during minus-strand cDNA synthesis by the DGR-encoded RT. The resulting mismatches in VR DNA strands could be resolved via DNA replication. Pink arrows and bars, cDNAs and cDNA-derived sequences. “N”, any of the four deoxyribonucleotides. Dashed lines, TR flanking sequences. See the text for details.
cDNA integration into VR sequences upstream of the (G/C)14 element is homology-dependent and could occur through template switching or strand displacement (Figure 7). The observation that 5′-end integration can be mediated by very short stretches of sequence identity suggests that template switching could be the primary pathway for cDNA integration into the 5′ end of VR. Such a process would not require RecA, and cDNA integration into VR before extending to the end of the TR RNA would account for the diffuse boundary of marker coconversion observed at the 5′ end of VR (Figure 4C). Once integration has occurred, the nascent minus-strand cDNA is predicted to contain mismatches at specific positions due to adenine mutagenesis. These could be resolved via DNA replication to separate the two strands. Although its precise function is unknown, atd appears to encode an RNA-binding protein with an essential role in the homing reaction (Guo et al., unpublished data).
Although the precise mechanism of adenine mutagenesis remains to be determined, our results reveal several key features of the process. Sequence analysis of RT-PCR products derived from precisely spliced RNA transcripts of pMX-td, a functional donor plasmid containing a group I intron inserted in TR, demonstrated the presence of intact adenines in the RNA intermediate required for mutagenic homing. This, and the lack of identifiable sequences in DGRs that could potentially encode RNA modifying enzymes, argue against RNA editing as the basis for adenine mutagenesis. In contrast, analysis of cDNA intermediates shown in Figure 5A provided clear evidence of nucleotide substitutions at positions corresponding to adenines in TR (Figure S11). This indicates that adenine mutagenesis occurs early in homing, most likely during minus-strand cDNA synthesis initiated at the 3′ end of VR, before integration at the 5′ end. Our data also allow us to estimate the efficiency of adenine mutagenesis relative to homing. When sequence tags in TR are used to amplify VR sequences that have undergone homing, no selection is imposed for adenine mutagenesis. Nonetheless, over 90% of the products collectively analyzed contained substitutions at positions corresponding to adenines in TR. This indicates that adenine mutagenesis accompanies nearly every homing event, and that the limiting step in generating diversity is the initiation or completion of the homing reaction. Taken together, our results are consistent with the hypothesis that adenine mutagenesis is an inherent property of the BPP-1 encoded Brt protein, and we predict the same is true for RTs encoded by other DGRs.
Our models for TPRT at the 3′ end of VR, and short homology-mediated cDNA integration at the 5′ end, bear similarities to the site-specific retrotransposition mechanism of the R2 element of Bombyx mori (R2Bm) (Eickbush et al., 2000; Eickbush, 1994). A seminal difference, however, is that retrotransposition of R2Bm and similar elements leads to destruction of their target sites. In contrast, DGR activity precisely regenerates sequences essential for both 3′- and 5′-end cDNA integration, thus preserving the ability to undergo repeated cycles of mutagenic homing and protein diversification. This is crucial to the beneficial nature of DGRs.
To date, over 40 DGRs have been identified in bacterial, phage and plasmid genomes, and it appears that nature has adapted these elements to perform a diverse array of functions (Medhekar and Miller, 2007). The potential also exists for engineering DGRs to evolve proteins with new and desired properties. We have demonstrated that the Bordetella phage DGR is flexible and can tolerate sequence insertions in TR. Most importantly, transposition of heterologous sequences is accompanied by efficient adenine mutagenesis. Furthermore, cDNA initiation at the 3′ end of VR is sequence-specific and requires the (G/C)14 and IMH elements, while cDNA integration upstream of these elements is homology-driven, but sequence-independent. These properties suggest that DGR-mediated targeted evolution could be directed to desirable heterologous sequences by placing them upstream of the (G/C)14 and IMH elements and by appropriately constructing hybrid TRs. As DGRs are continually capable of generating vast amounts of diversity, they may pose significant advantages over synthetic library-based approaches for generating diverse protein repertoires and new protein functions.
EXPERIMENTAL PROCEDURES
Phage Production for DGR Homing Assays
For single-cycle lytic infection, B. bronchiseptica RB50 cells (Liu et al., 2004) transformed with appropriate donor plasmids were grown overnight at 37°C in Luria-Bertani (LB) media containing 25 μg/ml of chloramphenicol (Cam), 20 μg/ml streptomycin (Str) and 10 mM nicotinic acid (NA). A 200 μl aliquot of cells was pelleted, rinsed, and resuspended in 1.2 ml Stainer Scholte (SS) medium (Stainer and Scholte, 1970) containing 25 μg/ml Cam and 20 μg/ml Str (SS+Cam+Str). Cultures were grown for 3 hours at 37°C to modulate bacteria to the Bvg+ phase, phages were added at a multiplicity of infection (MOI) of ~2.0 and incubated at 37°C for 1 hour to allow phage absorption. Infected cells were pelleted and resuspended in 1 ml fresh, pre-warmed SS+Cam+Str media and incubated at 37°C for 3 hours total post phage addition to allow completion of a single cycle of phage development. Progeny phages were harvested through chloroform extraction.
For phage production from lysogens, RB50 derivatives carrying prophage and plasmids of interest were grown and modulated to the Bvg+ phase as in single-cycle lytic infections. Phage production was induced with 2 μg/ml mitomycin for 3 hours at 37°C. Progeny phages were harvested through chloroform extraction.
PCR-Based DGR Homing Assay
Sequence insertions in TR that are transferred to VR during DGR homing are used as tags for PCR-based detection of homing products. Standard assays were carried out in a volume of 50 μl containing 60 mM Tris-SO4 (pH 9.1), 18 mM (NH4)2SO4, 2 mM MgSO4, 200 μM dNTPs, 5% DMSO, 6 ng/μl each of appropriate primers, 0.5 μl Elongase Enzyme Mix (Invitrogen) and ~2–50 × 105 copies of phage DNA. PCR reactions were performed as follows: 1× (94°C, 2 minutes); 30–35× (94°C, 30 seconds; 50–55°C, 30 seconds; 72°C, 1 minute); 1× (72°C, 10 minutes); 1× (4°C, hold). 5 to 20 μl samples were analyzed on 2% agarose gels.
Supplementary Material
Acknowledgments
We thank members of our laboratory for constructive input. This work was supported by NIH grants AI071204 and AI061598 to J.F.M. J.F.M is a founder of AvidBiotics corporation and a member of its scientific advisory board.
Footnotes
Supplemental Data include 16 figures, Supplemental Experimental Procedures and References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Jones AM. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 2003;11:367–373. doi: 10.1016/s0966-842x(03)00156-2. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet. 2003;37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, Mills D, Manias D, Dunny G, Lambowitz AM, Belfort M. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- Derr LK. The involvement of cellular recombination and repair genes in RNA-mediated recombination in Saccharomyces cerevisiae. Genetics. 1998;148:937–945. doi: 10.1093/genetics/148.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Hodes A, Dai L, Mandhana N, Liu M, Deora R, Simons RW, Zimmerly S, Miller JF. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- Eickbush DG, Luan DD, Eickbush TH. Integration of Bombyx mori R2 sequences into the 28S ribosomal RNA genes of Drosophila melanogaster. Mol Cell Biol. 2000;20:213–223. doi: 10.1128/mcb.20.1.213-223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH. Origin and Evolutionary Relationships of Retroelements. In: Morse SS, editor. Evolutionary Biology of Viruses. New York: Raven Press, Ltd; 1994. pp. 121–157. [Google Scholar]
- Galligan JT, Kennell JC. Retroplasmids: Linear and circular plasmids that replicate via reverse transcription. In: Meinhardt F, Klassen R, editors. Microbial Linear Plasmids. Berlin/Heidelberg: Springer; 2007. pp. 163–185. [Google Scholar]
- Guo H, Karberg M, Long M, Jones JP, 3rd, Sullenger B, Lambowitz AM. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science. 2000;289:452–457. doi: 10.1126/science.289.5478.452. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Kehoe B, Willery E, Reveneau N, Locht C, Relman DA. Molecular characterization of Bordetella bronchiseptica filamentous haemagglutinin and its secretion machinery. Microbiology. 2000;146:1211–1221. doi: 10.1099/00221287-146-5-1211. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- Liu M, Deora R, Doulatov SR, Gingery M, Eiserling FA, Preston A, Maskell DJ, Simons RW, Cotter PA, Parkhill J, Miller JF. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002;295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- Liu M, Gingery M, Doulatov SR, Liu Y, Hodes A, Baker S, Davis P, Simmonds M, Churcher C, Mungall K, et al. Genomic and genetic analysis of Bordetella bacteriophages encoding reverse transcriptase-mediated tropism-switching cassettes. J Bacteriol. 2004;186:1503–1517. doi: 10.1128/JB.186.5.1503-1517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SA, Miller JL, Lawton JA, Kerkow DE, Hodes A, Marti-Renom MA, Doulatov S, Narayanan E, Sali A, Miller JF, Ghosh P. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat Struct Mol Biol. 2005;12:886–892. doi: 10.1038/nsmb992. [DOI] [PubMed] [Google Scholar]
- Medhekar B, Miller JF. Diversity-generating retroelements. Curr Opin Microbiol. 2007;10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr G, Zhang A, Gianelos JA, Belfort M, Lambowitz AM. The Neurospora CYT-18 protein suppresses defects in the phage T4 td intron by stabilizing the catalytically active structure of the intron core. Cell. 1992;69:483–494. doi: 10.1016/0092-8674(92)90449-m. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Stainer DW, Scholte MJ. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol. 1970;63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- Temin HM. Reverse transcriptases. Retrons in bacteria Nature. 1989;339:254–255. doi: 10.1038/339254a0. [DOI] [PubMed] [Google Scholar]
- Zhang A, Derbyshire V, Salvo JL, Belfort M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


