Abstract
Food intake of humans is governed by the food's nutritional value and pleasing taste, but also by other factors such as food cost and availability, cultural imperatives, and social status. The biological determinants of human food intake are not easily parsed from these other factors, making them hard to study against the whirligig aspects of human life in a modern age. The study of animals provides a useful alternative. Humans have a history of studying animal food intake, for agricultural reasons (e.g., pigs and cows), and for personal reasons (e.g., dogs and cats), and these practical concerns have been joined with the appreciation that other models can teach us the principles of behavior, genetics, and nutrition. Thus there is a steady use of the traditional animal models in this type of research, as well as growth in the use of other systems such as worms and flies. Rats and mice occupy a special niche as animal models for two reasons; first, they share with humans a love of the same types of food, and second, they are the target of a number of well-developed genetic tools. The available genetic tools that make mice a popular model include a well-annotated genome (Mouse Build 37), profiles of RNA expression from many tissues, a diverse panel of inbred strains, and the ability to manipulate genes in the whole animal, including removing a gene only in specific tissues (e.g., Cre-lox system). Mice have been harnessed to find genotypes that contribute to sweet-liking, and other studies are underway to understand how genetic variation might at least partially explain other puzzles of human appetites. Animal models provide a way to study the genetic determinants of food selection with experimental rigor and therefore complement human genetics studies.
The human food environment has changed dramatically in the last few hundred years, and some humans now have the broadest range of food choices ever in the history of the species. This increase in the number of available foods has led to a concomitant increase in the number of variables that can determine food selection. These influences include subject characteristics such as sex and age, as well as more subtle environmental and experiential variables, such as the easy availability of certain foods, and expectations about food created by advertising and branding (1,2). Another contributor to human food preferences is genetic makeup (3). Genetic variation among people can lead to differences in taste perception, digestion, and metabolism (4), but how these differences translate into food selection are not well understood. The study of human food intake has been handicapped by the unavoidable problems of accurate measurement of behavior among free-living people. It is possible to study human food intake in a controlled environment, but laboratory measures may not generalize to the world outside. There are further difficulties in the study of human behavior because each person has idiosyncratic experiences and beliefs about food. To circumvent these difficulties, animal models have been developed to try to understand how food preferences are formed. These models are desirable because the environment and experiences of an animal can be controlled and studied over the life span. The theme of this article is how genetic variation influences food selection, and how animal models can be used to understand the details of genotype–phenotype relationships.
Definition of Genotype
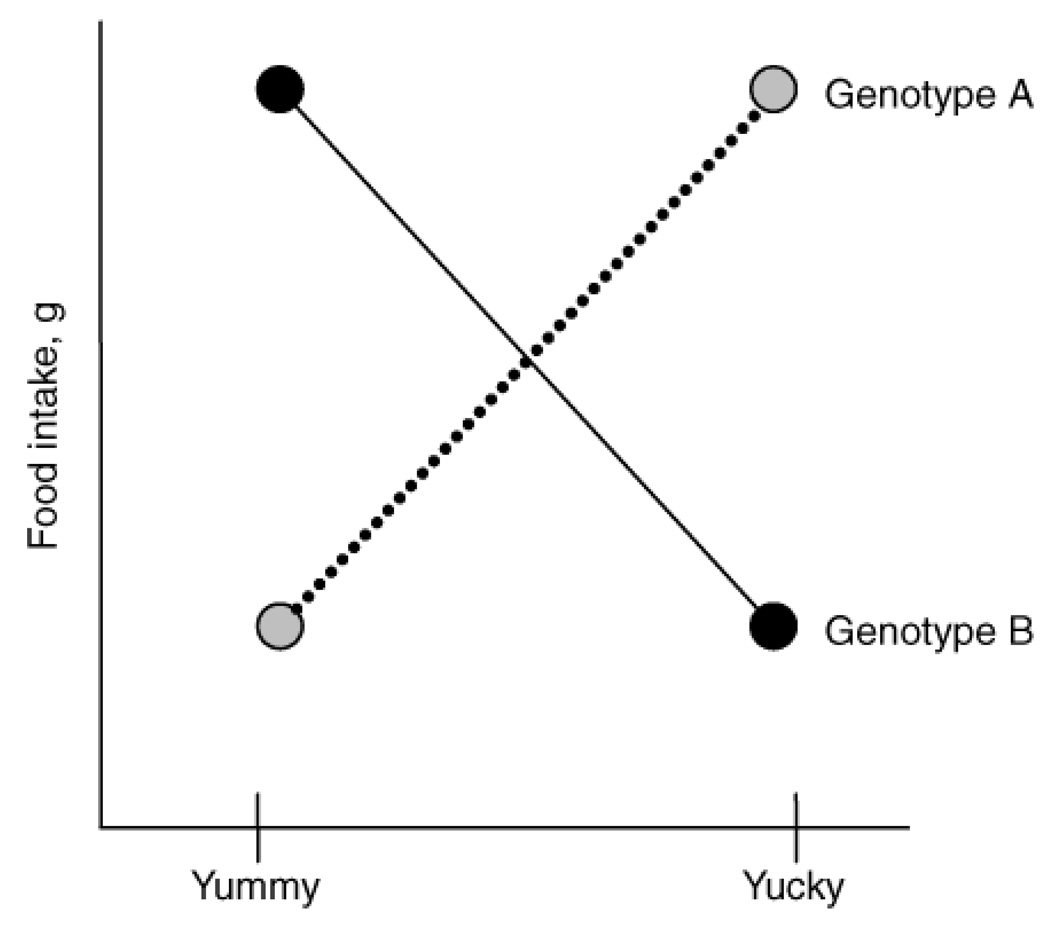
The use of vocabulary in genetics is changing, and so it is useful to define what is meant by the term "genotype." The DNA inside the nucleus of cells provides the template to be transcribed into a messenger molecule, which is translated into a particular protein, and this template is stored inside cells in two copies, the paired autosomal chromosomes. A region of transcribed genomic DNA that results in a protein or functional RNA product is called a gene. The DNA sequence of the same gene may differ between people, and these regions are called alleles or polymorphisms. Differences among individuals in DNA sequence may result in RNA or proteins that differ in function. The consequences of these changes in function differ depending on the specific gene and the specific allele; some changes are undetectable, whereas others cause a total loss of function of the protein. Still others create unusual products that interfere with pathways not normally associated with the gene. Besides protein coding and other known RNA genes, we have recently learned that some regions of genomic DNA are transcribed but not translated to protein, and are therefore known as noncoding RNA (5). Although the function of these noncoding RNAs is not well understood, they may regulate the expression of protein-coding genes (5). Genotype has two meanings. First, it is used as a general term that refers to the genetic makeup of an individual, and this concept is sometimes called "genetic background." Genotype also refers to the specific combination of alleles at a particular location. A common method of studying the role of a specific genotype on behavior is to group subjects according to their particular genotype, and compare the groups for a dependent variable such as food intake. In Figure 1, there is a schematic illustrating an interaction between genotype and the type of food preferred by an individual. A recent (November 2007) query of the largest repository of human genetic variation suggests that there are approximately 11,751,216 polymorphisms in the human genome (see Electronic Resource 1). Each of these polymorphisms is a potential genotypic site worth studying, but the effect of these millions of alleles and their influence on normal behavior largely unexplored.
Figure 1.
The point illustrated is the hypothetical effect of particular genotypes on the intake of certain types of food. "Yummy" refers to food that is regarded by most people as especially desirable to eat, like sweet foods or savory meats, whereas "Yucky" refers to food that is regarded by most people as less desirable, like bitter vegetables or unripe fruit. A definition of "genotype" is found in the text.
The point illustrated is the hypothetical effect of particular genotypes on the intake of certain types of food. "Yummy" refers to food that is regarded by most people as especially desirable to eat, like sweet foods or savory meats, whereas "Yucky" refers to food that is regarded by most people as less desirable, like bitter vegetables or unripe fruit. A definition of "genotype" is found in the text.
Popular Animal Models for Nutrition and Genetics Research
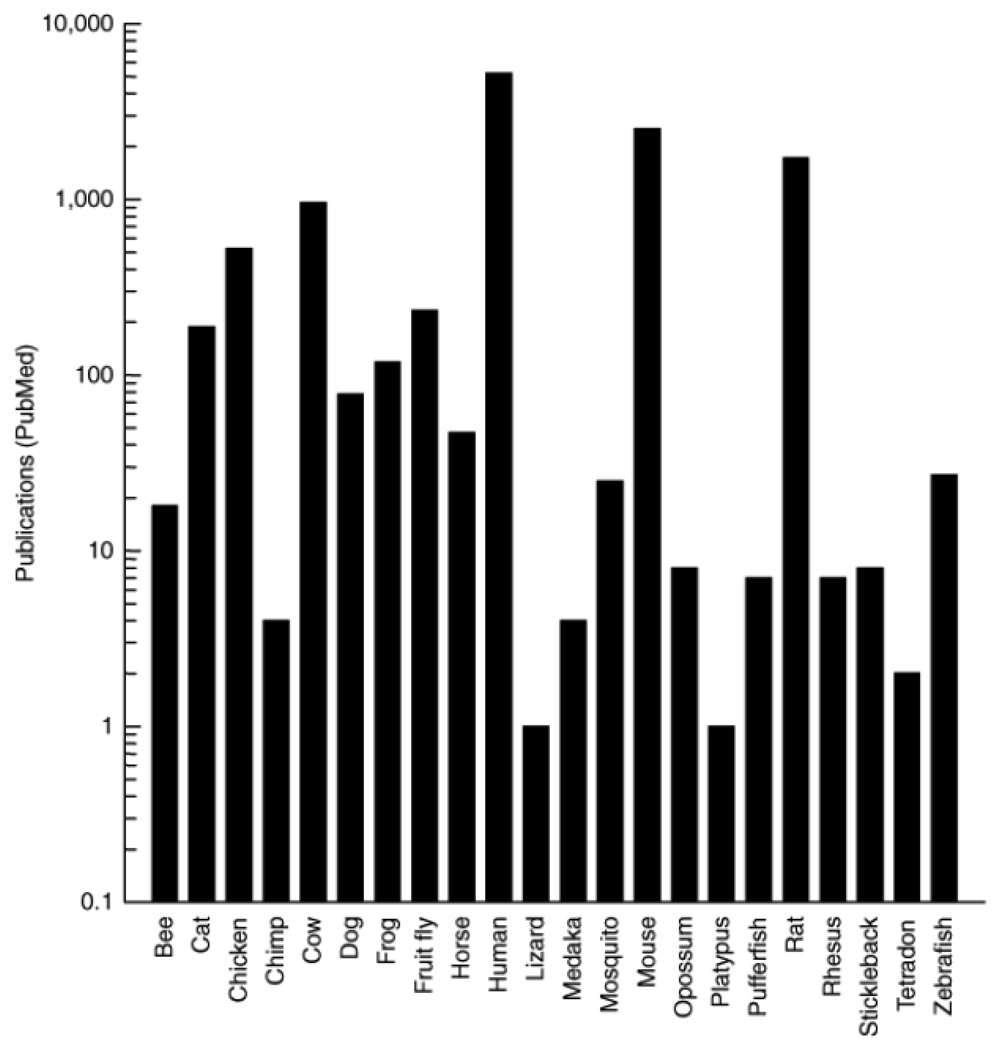
Given the complexity of human behavior and the complexity of the human genome, the study of animals provides a useful option to learn about genetic influences on food selection. The use of animals to study food intake is a natural extension of long-standing interest in animal nutrition for practical reasons, like raising animals for use as food (e.g., dairy cows), animals that do farm work (e.g., sheepdogs), or as companion animals (e.g., cats). Therefore the use of animals for food intake research was a natural outgrowth of the applied study of animal nutrition. One prerequisite for the genetic analysis of behavior is that the animal model should have a well-characterized genome; indeed, the number of model organisms with sequenced genomes is large and continues to grow. Some animals are frequently used in food intake research and others are used infrequently, although all models have some advantages. A summary of the number of studies of animals with sequenced genomes and their popularity for study in the genetics of nutrition is shown in Figure 2 (and Electronic Resource 2). Humans are the most studied species, followed by the workhorses of laboratory research (mice and rats), then animals used for human food (pigs and cows), and fruit flies. By comparing the genomes of species as disparate as flies and humans, we have learned that many genes are conserved in their DNA sequence and their function, and so flies and worms are becoming well-accepted model systems to conduct translational research (6,7,8).
Figure 2.
The results of a query of a database of biomedical research (PubMed, see Electronic Resources) using the common species name (e.g., human or cow), "nutrition" and "genetics" as keywords. The number of publications returned from the query is shown on the Y-axis. Species are displayed alphabetically on the X-axis. Note the log scale.
The results of a query of a database of biomedical research (PubMed, see Electronic Resources) using the common species name (e.g., human or cow), "nutrition" and "genetics" as keywords. The number of publications returned from the query is shown on the Y-axis. Species are displayed alphabetically on the X-axis. Note the log scale.
Mice and Rats are Well Suited as Models for Human Food Selection
Rats and mice live commensurably with humans and exploit the tendency of humans to store food. The colloquial name for the type of mouse used most often in laboratories is the "house mouse, " so it was a small step from living in our homes and eating our food to living in the laboratory and being fed by humans. House mice are common in almost all places that humans live, and according to one source, there is a specific word for "house mouse" in almost all human dialects (9). Mice and rats also get fat when offered human junk food (the term "junk food" is employed to describe processed foods high in fat and calories with added sugar and salt). Laboratory animals are fed junk food to make them fat, a paradigm known as the "cafeteria" or "supermarket" diet (10). This situation may seem unremarkable, but consider whether a diet of human junk food would make other animals fat, e.g., cats or snakes. Rats and mice are partially apt models for human food selection because they are close to us in food likes and dislikes, and this useful property of mice and rats is apt to be overlooked and unappreciated.
Studies in Mice Can Exploit Extensive Genetic Resources
The mouse is second only to the human in the largess of the genetic toolbox for study of variation and behavior. The mouse was the first animal genome to be described by comprehensive sequencing (11). Gene targeting methods now allow us to remove a particular gene or add a particular gene (12). For instance, human genes can be added into the mouse genome, resulting in the "humanization" of taste behavior so that these mice can taste a compound that humans can but normal mice cannot (13). New technologies also allow genes to be expressed at a time controlled by the experimenter (14) or using other methods, genes can be deleted only from certain tissues (15). Another key genetic tool is the creation of inbred mice strains, which are genetically identical at each locus. There is little or no heterozygosity within these inbred lines, so every mouse of a specific strain is genetically identical. If we compare two or more inbred strains for a trait, variation within a strain will be due to nongenetic factors whereas the difference between strains is ascribed to the genetic differences (16). Therefore the creation of more than 180 inbred mouse strains, each with a different behavioral repertoire, physiology, and set of traits is a valuable resource (see Electronic Resources 3). There is a program to comprehensively examine inbred mouse strains and to assess their genetic and behavioral differences, and these data are collected and organized in the Mouse Phenome Database (17). The availability of these strains and the organization of the data into a comprehensive database is a tool that can be used in the study of food selection in mice.
Experimental Approaches to Find Genes that Influence Behavior such as Food Selection
Inbred strains of mice can be used to study the genetics of food selection. As a first step in a genetic analysis, differences among inbred strains are identified. For instance, some inbred strains of mice like saccharin much more than other inbred strains (18). Once these strain differences were identified, the details about which genotypes account for the behavioral differences could be studied by interbreeding the parental strains of mice, and then testing the offspring. Some regions of DNA inherited from the parents will be shared among mice that drink more saccharin, and these regions will not be shared by mice that drink less saccharin. These genomic segments contain genes and the genetic variation that account for the strain differences. In the example of saccharin preference, we found that regions of chromosome 4 were shared by the sweet-preferring grandchildren (or the F2, a term used by mouse geneticists to describe the second filial generation) (19). Examining the DNA sequence on chromosome 4, a new gene was identified that later proved to be a component of the sweet receptor (20). This approach to finding genes that influence a particular trait is called quantitative trait loci (QTL) mapping.
Strain Surveys and QTL Mapping for Food Preference and Intake in Mice
The discovery of the sweet receptor is an example of how the study of behavioral differences among inbred strains can identify genes for a trait that is as seemingly complex as sweet preference. Earlier strain surveys have been done for susceptibility to dietary obesity (21), total caloric intake (22), the preference for taste solutions (23,24,25,26,27,28, 29,30,31,32,33), and fat preference (34,35). Because these surveys were useful, larger studies are underway, with a panel of 40 of the most widely used inbred mouse strains. These strains will be assessed for physiological, metabolic, neurological, and behavioral traits. Strain surveys can be followed by QTL mapping studies designed to find genes involved in food selection, such as the overconsumption of high-carbohydrate or high-fat diets (36,37). One unexplored area of research using these methods is to focus on the genetic control of the preference for compounds found in fruits and vegetables, e.g., malic acid in apples and sinigrin in kale (38). This approach may hold clues to the uneven consumption of fruits and vegetables among members of the human population.
Beyond Single Gene Effects
The genetic locus described earlier, which influenced saccharin preference in mice, had a large effect on mouse sweet preference. It is joined by many other genes of smaller effect to determine this trait, and these remaining genes are currently unknown. How to discover genes with smaller effect sizes is one of the main challenges facing geneticists. In addition to the effects of single locus and its alleles, there are gene–gene interactions and epigenetic influences on food selection. At the moment, there is no way to estimate the number of genes that participate directly in a behavior as complex as food selection, but if we look at a simpler example, body weight, a recent genetic survey indicates that up to 6,000 genes may be capable of influencing how much a mouse weighs (D.R. Reed et al., submitted). Likewise, there is currently no way to estimate the number of gene–gene interactions for food selection. However, because they are part of a homeostatic biological system (finding and metabolizing food for energy), genes and their alleles are likely to interact with others in their particular pathway. Because little is known about gene–gene interactions and food selection, we can again turn to body weight to provide clues, and from this type of data we learn that over 30% of the genetic variance in body weight is due to gene–gene (also called epistatic) interactions (39). Finally, epigenetic influences have been studied in animal models. The word "epigenetics" has several definitions, but we use it here to mean changes in gene expression that are stable and can be transmitted during mitosis and meiosis. Epigenetics is especially relevant because genes involved in the perception of taste and smell, as well as those involved in food metabolism, are susceptible to these effects (40,41). Taken together, the number of genes, their interactions, and modifications of gene expression through experience will all be present in humans and in animal models. However by controlling the breeding and environment, these influences can be assessed more easily in animal models than would be possible when studying humans.
Animal Models of Human Food Selection
As humans, we choose among a variety of foods: apples or cake, hot dogs or tofu, butter or margarine, and there are more choices available now than ever before in our history. In the face of all the choices available, there is a superficial uniformity of human food likes and dislikes (e.g., ice cream and pizza), but if we look deeper we see that individuals differ, and not everyone has a sweet tooth (42) or prefers high-fat foods (43). The determinants of what people like and what people eat are necessarily complex, but one of the influences is the genetic makeup of the subject. As an illustration, recent work has determined that alleles in taste genes such as those for bitter perception can influence taste perception, food preference, and choice in humans (44,45,46,47). If we can understand why people differ in food selection, we have the capability to harness this information to improve human health (48). Although animals will never perfectly recapitulate the behavioral repertoire of humans, they do provide several advantages in the study of food selection, such as the ability to absolutely control the food choices available, to measure food intake accurately and for long periods of time, to control the animal's experience and environment, and to manipulate their genomes in ways that are not possible in humans. Therefore one pathway to understand human food selection and its genetic control is through the use of animal models.
Acknowledgments
Alexander A. Bachmanov, Julie A. Mennella, Ian Tarr, and Michael G. Tordoff provided helpful comments and discussion. This work was supported by a grant from the National Institutes of Health (R01 DK58797). This publication was sponsored by the National Cancer Institute (NCI) to present the talks from the "Gene–Nutrition and Gene–Physical Activity Interactions in the Etiology of Obesity" workshop held on 24–25 September 2007. The opinions or assertions contained herein are the views of the authors and are not to be considered as official or reflecting the views of the National Institutes of Health.
Footnotes
Disclosure
The author declared no conflict of interest.
- To assess the number of known human polymorphisms, a query was made of this database on 8 November 2007 (Build 128): http://www.ncbi.nlm.nih.gov/sites/entrez?db=snp.
- To assess the number of biomedical research papers published for each animal species, a query was made of this database on 9 August 2008: http://www.ncbi.nlm.nih.gov/sites/entrezPubMed.
- The number of inbred strains listed in the online catalogue at the Jackson Laboratory was counted on 8 November 2008: www.jax.org.
References
- 1.Robinson TN, Borzekowski DL, Matheson DM, Kraemer HC. Effects of fast food branding on young children's taste preferences. Arch Pediatr Adolesc Med. 2007;161:792–797. doi: 10.1001/archpedi.161.8.792. [DOI] [PubMed] [Google Scholar]
- 2.Wansink B, Painter JE, Lee YK. The office candy dish: proximity's influence on estimated and actual consumption. Int J Obes (Lond) 2006;30:871–875. doi: 10.1038/sj.ijo.0803217. [DOI] [PubMed] [Google Scholar]
- 3.Reed DR, Bachmanov AA, Beauchamp GK, Tordoff MG, Price RA. Heritable variation in food preferences and their contribution to obesity. Behav Genet. 1997;27:373–387. doi: 10.1023/a:1025692031673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams RJ. Biochemical Individuality; The Basis for the Genetotrophic Concept. New York: Wiley; 1956. [Google Scholar]
- 5.Cawley S, Bekiranov S, Ng HH, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 6.McKay RM, McKay JP, Avery L, Graff JM. C. elegans: a model for exploring the genetics of fat storage. Dev Cell. 2003;4:131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G. Polygenic control of Caenorhabditis elegans fat storage. Nat Genet. 2006;38:363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- 8.De Luca M, Yi N, Allison DB, Leips J, Ruden DM. Mapping quantitative trait loci affecting variation in Drosophila triacylglycerol storage. Obes Res. 2005;13:1596–1605. doi: 10.1038/oby.2005.196. [DOI] [PubMed] [Google Scholar]
- 9.Keeler CE. The Laboratory Mouse; Its Origin, Heredity, and Culture. Cambridge, MA: Harvard University Press; 1931. [Google Scholar]
- 10.Sclafani A. Dietary-induced overeating. Ann N Y Acad Sci. 1989;575:281–289. doi: 10.1111/j.1749-6632.1989.tb53250.x. [DOI] [PubMed] [Google Scholar]
- 11.Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 12.Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 13.Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJ. The receptors and coding logic for bitter taste. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- 14.Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 15.Kos CH. Cre/loxP system for generating tissue-specific knockout mouse models. Nutr Rev. 2004;62:243–246. doi: 10.1301/nr2004.jun243-246. [DOI] [PubMed] [Google Scholar]
- 16.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4th edn. England: Longman: Essex; 1996. [Google Scholar]
- 17.Bogue MA, Grubb SC. The Mouse Phenome Project. Genetica. 2004;122:71–74. doi: 10.1007/s10709-004-1438-4. [DOI] [PubMed] [Google Scholar]
- 18.Fuller JL. Single-locus control of saccharin preference in mice. J Hered. 1974;65:33–36. doi: 10.1093/oxfordjournals.jhered.a108452. [DOI] [PubMed] [Google Scholar]
- 19.Bachmanov AA, Reed DR, Ninomiya Y, et al. Genetic locus on mouse chromosome 4 influencing taste responses to sweeteners (Abstract) Chem Senses. 1997;22:642. [Google Scholar]
- 20.Bachmanov AA, Li X, Reed DR, et al. Sac (saccharin preference) locus: positional cloning and effects on ethanol consumption (Abstract) Alcohol Clin Exp Res. 2001;25 (Suppl):118A. [Google Scholar]
- 21.Svenson KL, Von Smith R, Magnani PA, et al. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J Appl Physiol. 2007;102:2369–2378. doi: 10.1152/japplphysiol.01077.2006. [DOI] [PubMed] [Google Scholar]
- 22.Bachmanov AA, Reed DR, Beauchamp GK, Tordoff MG. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behav Genet. 2002;32:435–443. doi: 10.1023/a:1020884312053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmanov AA, Beauchamp GK, Tordoff MG. Voluntary consumption of NaCl, KCl, CaCl2 and NH4Cl solutions by 28 mouse strains. Behav Genet. 2002;32:445–457. doi: 10.1023/a:1020832327983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tordoff MG, Bachmanov AA, Reed DR. Forty mouse strain survey of water and sodium intake. Physiol Behav. 2007;91:620–631. doi: 10.1016/j.physbeh.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tordoff MG, Bachmanov AA, Reed DR. Forty mouse strain survey of voluntary calcium intake, blood calcium, and bone mineral content. Physiol Behav. 2007;91:632–643. doi: 10.1016/j.physbeh.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis SR, Ahmed S, Dym C, Khaimova E, Kest B, Bodnar RJ. Inbred mouse strain survey of sucrose intake. Physiol Behav. 2005;85:546–556. doi: 10.1016/j.physbeh.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Lush IE. The genetics of tasting in mice. I. Sucrose octaacetate. Genet Res. 1981;38:93–95. doi: 10.1017/s0016672300020425. [DOI] [PubMed] [Google Scholar]
- 28.Lush IE. The genetics of tasting in mice. II. Strychnine. Chem Senses. 1982;7:93–98. [Google Scholar]
- 29.Lush IE. The genetics of tasting in mice. III. Quinine. Genet Res. 1984;44:151–160. doi: 10.1017/s0016672300026355. [DOI] [PubMed] [Google Scholar]
- 30.Lush IE. The genetics of tasting in mice. IV. The acetates of raffinose, galactose and blactose. Genet Res. 1986;47:117–123. doi: 10.1017/s0016672300022941. [DOI] [PubMed] [Google Scholar]
- 31.Lush IE, Holland G. The genetics of tasting in mice. V. Glycine and cycloheximide. Genet Res. 1988;52:207–212. doi: 10.1017/s0016672300027671. [DOI] [PubMed] [Google Scholar]
- 32.Lush IE. The genetics of tasting in mice. VI. Saccharin, acesulfame, dulcin and sucrose. Genet Res. 1989;53:95–99. doi: 10.1017/s0016672300027968. [DOI] [PubMed] [Google Scholar]
- 33.Lush IE. The genetics of bitterness, sweetness, and saltiness in strains of mice. In: Wysocki CJ, Kare MR, editors. Genetics of Perception and Communication. New York: Marcel Dekker; 1991. pp. 227–241. [Google Scholar]
- 34.Lewis SR, Dym C, Chai C, Singh A, Kest B, Bodnar RJ. Genetic variance contributes to ingestive processes: a survey of eleven inbred mouse strains for fat (Intralipid) intake. Physiol Behav. 2007;90:82–94. doi: 10.1016/j.physbeh.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 35.Smith BK, Andrews PK, West DB. Macronutrient diet selection in thirteen mouse strains. Am J Physiol Regul Integr Comp Physiol. 2000;278:R797–R805. doi: 10.1152/ajpregu.2000.278.4.R797. [DOI] [PubMed] [Google Scholar]
- 36.Smith Richards BK, Belton BN, Poole AC, et al. QTL analysis of self-selected macronutrient diet intake: fat, carbohydrate, and total kilocalories. Physiol Genomics. 2002;11:205–217. doi: 10.1152/physiolgenomics.00037.2002. [DOI] [PubMed] [Google Scholar]
- 37.Kumar KG, Poole AC, York B, Volaufova J, Zuberi A, Richards BK. Quantitative trait loci for carbohydrate and total energy intake on mouse chromosome 17: congenic strain confirmation and candidate gene analyses (Glo1, Glp1r) Am J Physiol Regul Integr Comp Physiol. 2007;292:R207–R216. doi: 10.1152/ajpregu.00491.2006. [DOI] [PubMed] [Google Scholar]
- 38.VanEtten C, Goitrogens . Toxic Constituents of Plant Foodstuffs. In: Liener IE, editor. Food Science and Technology. New York: Academic Press; 1969. pp. pp 103–142. [Google Scholar]
- 39.Brockmann GA, Kratzsch J, Haley CS, Renne U, Schwerin M, Karle S. Single QTL effects, epistasis, and pleiotropy account for two-thirds of the phenotypic F(2) variance of growth and obesity in DU6i × DBA/2 mice. Genome Res. 2000;10:1941–1957. doi: 10.1101/gr.gr1499r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi JK, Kim SC. Environmental effects on gene expression phenotype have regional biases in the human genome. Genetics. 2007;175:1607–1613. doi: 10.1534/genetics.106.069047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr. 2007;97:435–439. doi: 10.1017/S0007114507352392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pepino MY, Mennella JA. Factors contributing to individual differences in sucrose preference. Chem Senses. 2005 (Suppl 1):i319–i320. doi: 10.1093/chemse/bjh243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salbe AD, DelParigi A, Pratley RE, Drewnowski A, Tataranni PA. Taste preferences and body weight changes in an obesity-prone population. Am J Clin Nutr. 2004;79:372–378. doi: 10.1093/ajcn/79.3.372. [DOI] [PubMed] [Google Scholar]
- 44.Mennella JA, Pepino MY, Reed DR. Genetic and environmental determinants of bitter perception and sweet preferences. Pediatrics. 2005;115:e216–e222. doi: 10.1542/peds.2004-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandell MA, Breslin PA. Variability in a taste-receptor gene determines whether we taste toxins in food. Curr Biol. 2006;16:R792–R794. doi: 10.1016/j.cub.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 46.Hansen JL, Reed DR, Wright MJ, Martin NG, Breslin PA. Heritability and genetic covariation of sensitivity to PROP, SOA, quinine HCl, and caffeine. Chem Senses. 2006;31:403–413. doi: 10.1093/chemse/bjj044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wise PM, Hansen JL, Reed DR, Breslin PA. Twin study of the heritability of recognition thresholds for sour and salty taste. Chem Senses. 2007;32:749–754. doi: 10.1093/chemse/bjm042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka T, Reed DR, Ordovos J. Taste as the gatekeeper of personalized nutrition. In: Desiere F, Kok F, Bouwman L, editors. Personalized Nutrition: Principles and Applications. Boca Raton, FL: Taylor & Francis CRC Press; 2008. pp. pp 115–132. [Google Scholar]