SUMMARY
Enzymes that regulate their activity by modulating an equilibrium of alternate, non-additive, functionally distinct oligomeric assemblies (morpheeins) define a novel mode of allostery (Jaffe, TiBS 30:490-7, 2005). The oligomeric equilibrium for porphobilinogen synthase (PBGS) consists of high-activity octamers, low-activity hexamers, and two dimer conformations. A phylogenetically diverse allosteric site specific to hexamers is proposed as an inhibitor binding site. Inhibitor binding is predicted to draw the oligomeric equilibrium toward the low-activity hexamer. In silico docking enriched a selection from a small molecule library for compounds predicted to bind to this allosteric site. In vitro testing of selected compounds identified one compound whose inhibition mechanism is species-specific conversion of PBGS octamers to hexamers. We propose that this novel strategy for inhibitor discovery can be applied to other proteins that use the morpheein model for allosteric regulation.
INTRODUCTION
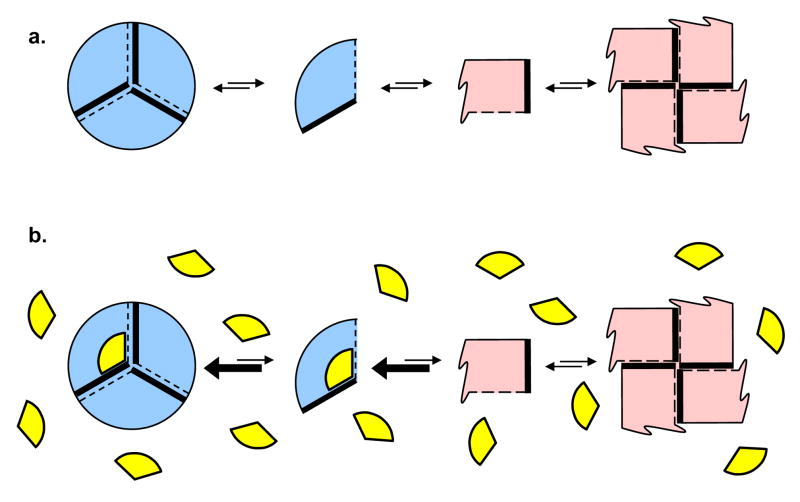
Dissociating allosteric enzymes are well established (Traut, 1994) and provide opportunities for small molecule inhibition by interference with the assembly process. If one couples dissociating enzymes with conformational flexibility (James and Tawfik, 2003), one can achieve a system wherein subunits of an oligomer of one symmetry may dissociate and then change conformation such that they can only assemble to an oligomer of different symmetry. In this case, it may be possible for small molecules to trap one oligomeric assembly and prevent equilibration to the alternate assembly. The morpheein model for allosteric regulation of protein function describes such a situation wherein proteins can exist as an ensemble of physiologically significant and functionally distinct alternate quaternary assemblies (Fig. 1a) (Jaffe, 2005). The current paper addresses the hypothesis that small molecules can act as inhibitors of such proteins by selectively binding to and stabilizing the less active assembly (Fig. 1b).
Figure 1. Proteins that can exist as an equilibrium of alternate quaternary structure assemblies (morpheeins) provide a structural foundation for allosteric regulation of protein function.
(a) The morpheein model for allosteric regulation includes oligomer dissociation, conformational change in the dissociated form, and may involve reassociation to an alternate oligomeric assembly. In the illustrated case, two alternate quaternary structure assemblies are shown (a trimer and a tetramer) and the common rule of assembly of the fundamental units is that a thick solid line must associate with a dashed line. The conformation of the dissociated form, shown as a blue or pink fundamental unit, dictates the geometry and stoichiometry of the respectively colored oligomers. The functions of the two oligomers are distinct (e.g. low activity vs. high activity), analogous to the R and T states of the traditional non-dissociating models for allosteric regulation. The fundamental unit can be monomeric or of higher order. In the case of porphobilinogen synthase, the fundamental unit is an asymmetric homo-dimer and the analogous transition is between a hexamer and an octamer. (b) In the morpheein model for allosteric regulation, a regulator molecule (depicted as a yellow wedge) binds to the structural elements on one side of this equilibrium. The yellow wedge has the appropriate geometry to bind only to the blue forms and draw the equilibrium in that direction, thus acting as an allosteric activator or inhibitor. The binding site for the yellow wedge is required to be specific for one oligomeric assembly and not the other. However, the binding site is not required to be interfacial (between subunits), as is the case for the small molecule binding site in the PBGS hexamer.
The morpheein model for allostery is distinct from the classic Monod-Wyman-Changeau and Koshland-Nemanthy-Filmer models for allostery (Koshland et al., 1966; Monod et al., 1965), both of which contain the assumption of a conserved oligomeric assembly throughout the allosteric transition. This implies a fixed stoichiometry, which need not hold true for homo-oligomeric proteins that function as morpheeins. The distinguishing feature between the morpheein model for allosteric regulation and both classic models is that the former must involve a dissociation event and a conformational change in the dissociated state; it may also involve reassembly into a functionally distinct alternate oligomer. The structurally distinct quaternary assemblies available to a morpheein present a previously unforeseen opportunity for allosteric chemical inhibition. The mode of action for the proposed quaternary structure-trapping agent is to bind to an oligomer-specific surface cavity and draw the equilibrium toward the targeted oligomeric form (Fig. 1b). Like many allosteric sites, the novel small molecule binding sites are likely to be more phylogenetically variable than enzyme active sites, allowing one to target universally essential enzymes or proteins for drug discovery.
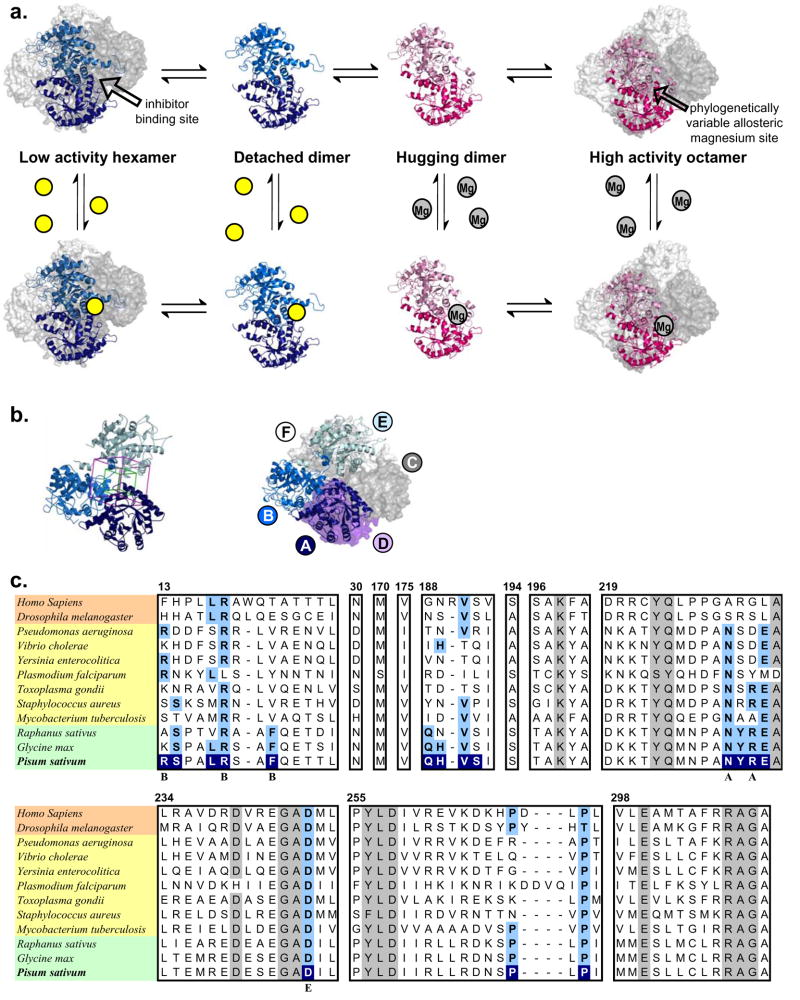
The prototype morpheein, porphobilinogen synthase (PBGS, EC 4.2.1.24, a.k.a. 5-aminolevulinate dehydratase), catalyzes a fundamental step in the biosynthesis of tetrapyrrole pigments, an activity that is essential to all organisms that carry out respiration, photosynthesis, or methanogensis. PBGS has been shown to exist in an equilibrium of high activity octamers and low activity hexamers whose interconversion is at the level of two different dimer conformations (Breinig et al., 2003; Selwood et al., 2008; Tang et al., 2006; Tang et al., 2005) (Fig. 2a). Ligand binding to the active site draws the equilibrium toward the octamer. A consequence of this equilibrium of oliogmeric assemblies is a protein concentration dependent specific activity (Kervinen et al., 2000), which we now interpret to reflect a low-activity hexamer dissociating to dimers, changing configuration, and then re-associating to an active octamer (or vice versa). Crystal structures reveal the alternate assemblies of PBGS (Fig. 2a) (Breinig et al., 2003; Frankenberg et al., 1999) and show that each monomer is comprised of an αβ-barrel domain and an extended N-terminal arm with phylogenetically variable length; the orientation of the arm with respect to the barrel is a determinant of the quaternary structure assembly. The crystallographic asymmetric units are the illustrated dimers. For both the octamer and the hexamer the active site is located in the center of the αβ-barrel. The N-terminal arm differentially participates in subunit-subunit interactions in the octameric and hexameric assemblies. It is the difference in intersubunit arm-to-barrel interactions that contributes to dramatic kinetic differenced between the octameric and hexameric assemblies (Jaffe, 2004). Phylogenetic variations in the N-terminal arm sequence result in a phylogenetic variation in the thermodynamics and kinetics of the equilibration of PBGS oligomers.
Figure 2. Porphobilinogen synthase (PBGS) is the prototype morpheein ensemble.
(a) The equilibrium of pea PBGS quaternary structure forms is shown. For most species, the asymmetric unit of the crystal structure is an asymmetric homo-dimer. The hexamer and its asymmetric unit, the detached dimer (PDB code: 1PV8) are shown in shades of blue. The octamer and its asymmetric unit, the hugging dimer (PDB code: 1GZG) are shown in shades of pink. For the octamer, the dimers assemble at a 90° rotation around a central axis; for the hexamer, the dimers assemble at a 120° rotation around a central axis. The octamer contains a phylogenetically variable binding site for an allosteric magnesium ion which binds to the arm-to-barrel interface that is unique to the octamer; the allosteric magnesium binding site is not present in the hexamer (Breinig et al., 2003; Jaffe, 2003). The hexamer contains a surface cavity not present in the octamer and is the predicted small molecule binding site. The small molecule inhibitor (depicted as yellow balls) draws the equilibrium toward the hexamer. (b) Left - The small molecule binding site in the pea PBGS hexamer model contains components from three subunits, shown as ribbons. The GLIDE docking box is superimposed at the subunit interface. The box into which docked ligands must fit is in magenta, the box in which the center of the docked ligands must fit is in green. Right – The three subunits forming the docking site are shown as ribbons in the context of the hexamer with the remaining subunits shown as surfaces, subunit labels correspond to subunit colors. (c) A multiple sequence alignment of the regions of PBGS contained within the hexamer-specific inhibitor docking site of pea PBGS is shown with highly conserved residues shaded in grey. Organisms highlighted in peach, yellow, and green are representative metazoa, microbes, and plants, respectively. Sequence conservation was determined from an alignment of 33 PBGS sequences (not shown); a residue was defined as “highly conserved” if it was present in at least 32 sequences. Numbers correspond to the pea PBGS sequence. Residues highlighted in blue of the P. sativum PBGS are within 4 Å of docked morphlock-1; these residues are highlighted in light blue where they are conserved in other sequences. Residues marked with “A”, “B”, or “E” indicate a sidechain interaction between subunits A, B, or E of pea PBGS and docked morphlock-1. A closeup of the docked structure of morphlock-1 is shown in Fig. 3a.
The physiologic relevance of the octamer-hexamer equilibrium illustrated in Fig 2a is established for PBGS from both humans and plants. In humans, the disease ALAD porphyria arises from mutations to the gene encoding PBGS (Gross et al., 1998). There are eight known human mutations associated with ALAD porphyria; all eight are associated with an increased propensity of the protein to exist as the inactive hexameric assembly (Jaffe and Stith, 2007). In plants, PBGS resides in the chloroplast (Boese et al., 1991) and is established to contain an allosteric magnesium binding site (Kervinen et al., 2000). Magnesium binding to this site facilitates the hexamer to octamer transition by stabilizing a subunit interface that is present in the octamer but not in the hexamer (Fig 2a) (Breinig et al., 2003). During the greening process in plants the resting magnesium concentration in the chloroplast is below 1 mM and the resultant low PBGS activity helps prevent the accumulation of phototoxic chlorophyll precursors. Upon exposure to light the chloroplast magnesium concentration increases to ~10 mM (Walker, 1976); appropriately, the Kd for the allosteric magnesium of the green plant Pisum sativum (pea) PBGS is 2.5 mM (Kervinen et al., 2000). The physiologic relevance of the quaternary structure equilibrium of PBGS from human pathogens has not yet been established. However, preliminary results on PBGS from several pathogens show the protein concentration dependent specific activity indicative of an equilibrium of quaternary structure assemblies under native conditions (unpublished results).
The current study was undertaken to establish whether species-selective inhibition of PBGS could be accomplished by perturbing its quaternary structure equilibrium. The PBGS from pea was selected as the inhibitor target. This well-characterized protein exhibits protein concentration dependent specific activity and is available in large quantities (Kervinen et al., 2000). A cavity on the surface of the hexameric assembly (inactive oligomer) serves as a putative small molecule binding site (Fig. 2a and b); this site is not present in the octamer (active oligomer). A quaternary-structure-perturbing inhibitor for pea PBGS would function by binding to the illustrated site in the hexamer (Fig. 2b), thereby drawing the equilibrium toward the stabilized inactive form and reducing the total activity. The sequence of PBGS from both plants and human pathogens differ from the human protein at this proposed small molecule binding site (Fig. 2c). Due to these phylogenetic sequence differences, an inhibitor selected for this site in plant or pathogen PBGS would not be expected to affect the activity of human PBGS. This is in stark contrast to an active site directed PBGS inhibitor as the active site residues are highly conserved (Jaffe, 2003).
In silico docking using the GLIDE program (Halgren et al., 2004) was utilized to identify a suite of small molecules predicted to bind to the hexamer-specific binding site of a homology model of pea PBGS. A selection of these molecules were purchased and tested in vitro for their ability to stabilize the hexameric form of pea PBGS and to inhibit enzyme activity. One potent inhibitor, which drove the pea PBGS oligomeric distribution dramatically toward the hexamer, was identified from the screen. This compound, given the name morphlock-1, did not inhibit the activity of human PBGS, nor did it alter the quaternary structure equilibrium of PBGS from humans, Drosophila melanogaster, Pseudomonas aeruginosa, or Vibrio cholerae. Thus, we have demonstrated that perturbation of a morpheein equilibrium of non-additive quaternary structure assemblies (see Fig. 1) is a viable approach for the development of species-specific drugs with novel modes of action.
RESULTS
Model Building
The model of octameric pea PBGS was published previously (Kervinen et al., 2000). The model of hexameric pea PBGS (see Methods) is comprised of three copies of an asymmetric dimer (Supplemental Material). The putative inhibitor binding site contains components from three subunits of the hexamer: one detached dimer and one subunit of an adjacent dimer (Fig. 2b). The asymmetry in each dimer results in the hexameric model containing two sequence-identical but structurally distinct inhibitor binding sites. Rotating counter-clockwise around the central axis, the detached dimers are comprised of subunits A and B, C and D, and E and F. One inhibitor binding site is at the junction of subunits A, B, and E (ABE); the other is at the junction of subunits B, A, and D (BAD). Each hexamer has three sites equivalent to the ABE site and three sites equivalent to the BAD site.
Docking
Docking studies used Life Chemical, Inc.’s compound libraries, which are comprised of small molecules (molecular weight ~500) designed to be relatively “drug-like” (e.g. adhering to Lipinski’s Rule of Five (Lipinski et al., 2001)). Two libraries, named “G-protein coupled receptor-targeted” and “Kinase-targeted” were used. For each of the proposed binding sites, ABE and BAD, a cubic region 25Å in length was defined at the interface of the three subunits (shown for ABE in Fig. 2b, purple box). This box was used by GLIDE as the search region within which each entire docked molecule must fit in order to be scored. A second concentric cubic region 14Å in length was defined and used to restrict the location of the center of each docked molecule (Fig. 2b, green box). The docking process used the default settings of GLIDE (version 3.5) Standard Precision (SP) mode, which produced a “GLIDE SCORE” for each molecule; the GLIDE SCORE is based on a proprietary modification of the CHEMSCORE algorithm (Eldridge et al., 1997) that quantitatively accounts for characteristics of the binding interaction between each docked small molecule and the protein. The GLIDE SP mode scores goodness of fit based predominantly on geometries. For each library and each binding site, we found that the GLIDE SCORES of approximately 80% of the docked compounds were similar to each other; 10% were significantly lower (better fit), and 10% were significantly higher (inferior fit). Compounds with the best SP GLIDE SCORES,~10% for each library, were then docked a second time using the Extra Precision (XP) mode of GLIDE, resulting in a set of new, quantitatively unrelated GLIDE SCORES. XP mode scoring is more rigorous as it takes into account polarity and hydrophobicity, and penalizes mismatches of hydrophobic-hydrophilic contacts or charges. Again, roughly 80% of the molecules had similar scores. Molecules with XP GLIDE SCORES in the top ~10% (about 1% of the starting library) were then analyzed further to select for predicted solubility, interactions with each of the three subunits, and variety in both chemical structure and binding interactions (see Methods). This resulted in the selection of a set of ~100 diverse, putatively soluble, small molecules for purchase and in vitro testing. Of the 100 molecules ordered from Life Chemicals, Inc., only 76 were available without requiring new syntheses. The selected compounds represented an equal distribution of fits to ABE and BAD. The purchased compounds are shown in their docked orientation in supplemental Table S1.
Initial Screening of Compounds
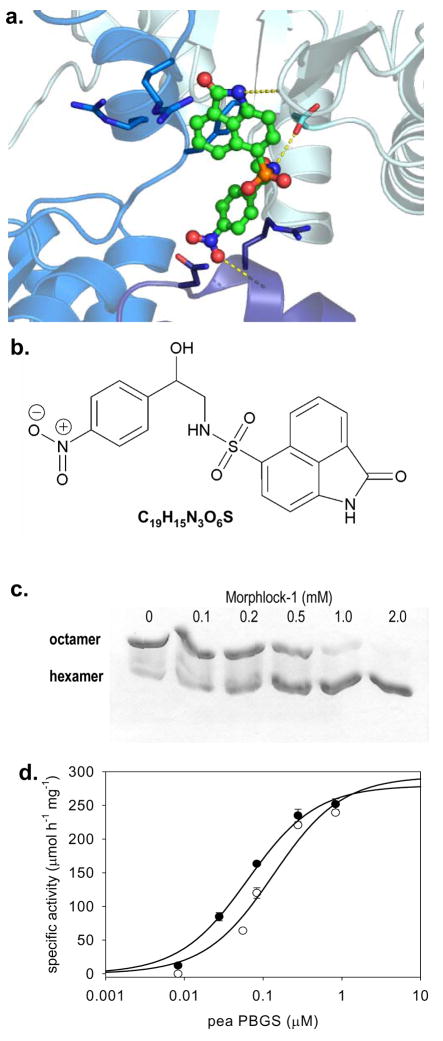
The distribution of PBGS into hexamers and octamers can be monitored by native polyacrylamide gel electrophoresis (PAGE) (Breinig et al., 2003), and this property was utilized to screen the compounds identified by GLIDE. Of the 76 purchased compounds, only morphlock-1 (Fig. 3a and 3b) was found to dramatically shift the oligomeric equilibrium to the hexamer under the screening conditions (Supplemental Fig. S1). Most significantly, the ratio of octamer to hexamer is dependent on the concentration of morphlock-1 (Fig. 3c). In these native PAGE analyses, which are at non-equilibrium conditions and use high protein concentrations (1 mg/ml, ~28 μM subunits), the observed IC50 is ~500 μM. Kinetic analysis and additional native PAGE analysis using more sensitive staining techniques show an IC50 of ~ 1 μM (see below).
Figure 3. The oligomer-trapping inhibitor, morphlock-1, stabilizes the hexameric assembly of pea PBGS.
(a) Morphlock-1 is illustrated as posed by GLIDE. Subunits A, B, and E are shown as ribbons using colors corresponding to Fig. 2b. Morphlock-1 is shown with carbon atoms in green (other atoms colored CPK) and illustrated as ball and stick. Side chains within 4.0 Å of morphlock-1 are shown as sticks, carbons are colored by chain (other atoms colored CPK). Hydrogen bonds to morphlock-1 are shown in yellow. (b) The chemical structure of morphlock-1. (c) Pea PBGS (1 mg/mL) resolves on native PAGE into its octameric and hexameric components. Pea PBGS was incubated with various concentrations of morphlock-1 (in a DMSO solution). Each concentration of morphlock-1 with a fixed amount of DMSO was incubated with protein for 30 minutes at 37 °C prior to resolution on 12.5% polyacrylamide native PhastGels. Morphlock-1 draws the pea PBGS morpheein equilibrium entirely to the hexamer in a dose dependent fashion. (d) The protein concentration-dependent pea PBGS specific activity is shown. PBGS (at varied concentrations) was incubated in the presence of DMSO (●) or 50 μM morphlock-1 in DMSO (○) for 30 minutes at 37 °C prior to assay. The final concentration of morphlock-1 in the inhibited assay was 5 μM, and the concentration of PBGS in the inhibited and control assays ranged from 0.05 – 50 μg/mL (0.014 – 1.4 μM).
Confirmation of the Chemical Identity of Morphlock-1
1H-NMR, mass and UV/Vis spectroscopic analyses of morphlock-1 were consistent with 2-oxo-1,2-dihydro-benzo(cd)indole-6-sulfonic acid[2-hydroxy-2-(4-nitro-phenyl)-ethyl]-amide (Fig. 3a). 1H-NMR (300 MHz, D2O): δ 8.89 (brs, 1H), 8.45 (d, J = 8.4 Hz, 1H), 8.01 (d, J = 2.7 Hz, 1H), 7.98 (d, J = 3.3 Hz, 1H), 7.83 (d, J = 8.4 Hz, 1H), 7.76 (d, J = 8.7 Hz, 1H), 7.24 (d, J = 9.0 Hz, 1H), 6.95 (d, J = 7.8 Hz, 1H), 5.93 (brs, 1H), 4.71 (q, J = 9.9, 1H), 3.77 (d, J = 4.2, 1H), 3.25-3.19 (m, 2H); MS (m/z): [MH]+ calculated for C19H15N3O6S, 414.07; found, 414; [MNa]+ calculated for C19H15N3O6S, 436.06; found, 436; UV/Vis: λmax 267 nm.
Kinetics of Pea PBGS Inhibition by Morphlock-1
Pea PBGS exhibits a protein concentration dependent specific activity that represents a dynamic equilibrium between high activity octamers and low activity hexamers, only the latter of which contain the putative binding site for morphlock-1 (Fig. 2). The protein concentration dependent specific activity data for pea PBGS in the absence of morphlock-1 fit well to a simple hyperbolic equation with a K(0.5) of 0.064 μM PBGS (Fig. 3d), suggesting that the protein is 50% hexamer at this concentration. At the lowest protein concentration assayed, the specific activity of pea PBGS was less than 1% of the maximal activity, suggesting that the hexamer is virtually inactive under these conditions. The dose response curve for morphlock-1 inhibition of pea PBGS (Fig. 4a) was initially carried out at 0.03 μM pea PBGS, where the hexamer is a significant component of the equilibrium mixture; the morphlock-1 concentration was varied from 0.1 μM to 0.1 mM. These inhibition data fit to a hyperbolic equation, and yield an apparent IC50 of 1.2 μM morphlock-1. The protein concentration dependent specific activity of pea PBGS predicts that the apparent IC50 of morphlock-1 will vary with protein concentration, showing less inhibition at higher protein concentration in the range of the K(0.5), as confirmed in Fig. 4b. Corroborating evidence that morphlock-1 inhibits through stabilization of the hexameric assembly is the increased K(0.5) of the protein concentration dependent activity in the presence of morphlock-1 (Fig. 3d). For this determination, the morphlock-1 concentration was set at 5 μM, which more than doubles the K(0.5) to 0.134 μM pea PBGS relative to the K(0.5) without morphlock-1 (0.064 μM).
Figure 4. Species-specific effects of morphlock-1 on PBGS.
(a)) Dose-response curves showing morphlock-1 inhibition of pea PBGS at 1 μg/ml (○) and human PBGS at 0.6 μg/ml (Δ) and 10 μg/ml (□). Proteins were incubated in the presence of 10X inhibitor for 30 minutes at 37 °C prior to assay. Concentrations on the X-axis represent the final concentration of morphlock-1 in the assay. (b) The IC50 for morphlock-1 is a function of pea PBGS concentration. IC50 values were determined as in (a) using the noted concentrations of pea PBGS. (c) Native PAGE evaluation of the effect of morphlock-1 on the quaternary structure equilibria of PBGS (~1 mg/ml) from D. melanogaster, P. aeruginosa, V. cholerae, and H. sapiens. Note that the charge/mass ratio is not the same for PBGS from different species. The position of the oligomeric equilibrium varies among PBGS from different species; thus, only one form is observed under these conditions for the PBGS from D. melanogaster, P. aeruginosa, V. cholerae. The lanes marked with an “X” contain unrelated compounds.
Species-selectivity of morphlock-1 inhibition
The putative morphlock-1 binding site is phylogenetically variable (Fig. 2c). As such, morphlock-1 would not be expected to inhibit human PBGS. The effect of morphlock-1 on the activity of human PBGS was examined; morphlock-1 does not inhibit human PBGS (Fig 4a) nor does it affect the mobility of human PBGS on a native PAGE gel (Fig. 4c). We also demonstrate that 2 mM morphlock-1 does not alter the quaternary structure equilibrium of PBGS from Drosophila melanogaster (the fruit fly), Pseudomonas aeruginosa (a common cause of nosocomial infection), or Vibrio cholera (epidemic when clean water is lacking) (Fig. 4c). Furthermore, an in silico experiment showed that docking the same Life Chemicals libraries to the comparable hexamer-trapping inhibitor-binding site on human PBGS (PDB code 1PV8) resulted in a very small overlap (3%) in the top 100 docked poses relative to the results of docking to the pea PBGS hexamer.
Morphlock-1 specifically binds to the pea PBGS hexamer
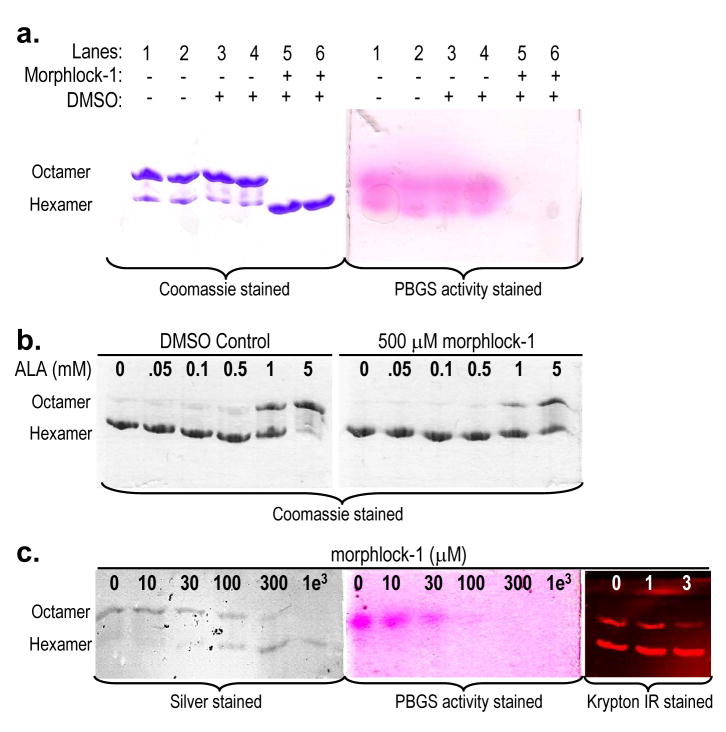
The specificity of morphlock-1 for the pea PBGS hexamer was demonstrated using native PAGE that has been stained both for protein and for PBGS activity. Using two dimensional native PAGE, it was previously established that the substrate induced stabilization of the PBGS octameric assembly can occur within the gel matrix (Jaffe et al., 1995). The substrate-mediated in-gel transition of pea PBGS hexamer to octamer is documented in Supplemental Figure S2. Thus, even though pea PBGS separates into octamer and hexamer under native PAGE conditions, incubation of the resolved gel with substrate under assay conditions causes the transition of the inactive hexamer to the active octamer; consequently both bands stain for PBGS activity. Figure 5a illustrates this phenomenon and how it is affected by morphlock-1. Pea PBGS resolves to octamer and hexamer on a native gel in the absence or presence of DMSO, and when activity stained, both bands show the bright pink complex formed between the product porphobilinogen and Ehrlich’s reagent. However, under conditions where morphlock-1 stabilizes the hexamer, no activity is observed. The Coomassie and activity stained gels (Fig. 5a) illustrate that morphlock-1 specifically binds to the hexameric assembly of pea PBGS, remains bound during the electrophoresis, and prevents the substrate mediated conversion of hexamer to octamer that allows unbound hexamer to stain for PBGS activity.
Figure 5. Morphlock-1 and substrate induced interconversion of PBGS quaternary structure assemblies.
(a) Matched Coomassie (left) and PBGS activity (right) stained native PAGE (at 1 mg/ml PBGS) illustrates specific binding of morphlock-1 (at 2 mM) to the hexameric assembly. Enzyme-bound morphlock-1 prevents the in-gel transition of inactive hexamer to active octamer (Fig. S2). (b) Hexameric pea PBGS (1 mg/mL) was incubated with DMSO (left) or 500 μM morphlock-1 (right) for 30 minutes at 37 °C. ALA was added at the indicated concentrations, and the incubation was continued for 30 minutes prior to resolution on 12.5 % polyacrylamide native PhastGels. (c) Morphlock-1 induced stabilization of the pea PBGS hexameric assembly. Morphlock-1 concentrations are listed above each lane. Protein samples for the silver and activity stained gels were at 50 μg/ml (1.4 μM subunit) and 1 mM ALA. Samples for the Krypton Infrared stained gel were at 5 μg/ml (0.14 μM subunit) and 10 mM magnesium.
Morphlock-1 inhibits the substrate-induced stabilization of the PBGS octamer
Substrate binding to a hexameric assembly of human PBGS has been shown to shift the equilibrium toward the active octamer because it stabilizes octamer-specific quaternary structure interactions (Tang et al., 2006; Tang et al., 2005). Here we demonstrate the same phenomenon for pea PBGS and show that morphlock-1 competes with this process through stabilization of the hexamer (Fig. 5b). In the absence of morphlock-1, 5 mM ALA is sufficient to support nearly total conversion to the octamer. In the presence of 2 mM morphlock-1, this same concentration of ALA only converts ~50% of the hexamer to octamer.
Correlation of inhibition studies with native gel shift studies
The interconversion of pea PBGS hexamer and octamer is a dynamic equilibrium that responds to protein concentration, substrate concentration, magnesium concentration, inhibitor concentration, and ionic strength. Under the conditions illustrated in Fig 3c, where the protein concentration is ~ 1 mg/ml (28 μM subunit), the gel shows ~90% octamer in the absence of morphlock-1 and it is easy to show that addition of morphlock-1 stabilizes the hexamer. However, 1 mg/ml is three orders of magnitude above that used for the activity assays ( Fig. 4a) where the apparent IC50 is ~1 μM. A conundrum is that native PAGE at lower protein concentration (like those used for the activity assay) shows predominantly hexamer in the absence of additives. Consequently, when a dilute protein sample starts as predominantly hexamer in the gel matrix environment, it is not possible to illustrate the hexamer-stabilizing effect of morphlock-1 upon addition to the protein. To provide such a demonstration, we take advantage of the fact that both substrate and magnesium favor formation of the octameric assembly. As shown above (Fig. 5b), there is a competition between the hexamer-stabilizing effect of morphlock-1 and the octamer-stabilizing effects of substrate and/or magnesium. Hexamer stabilization by morphlock-1 is demonstrated at lower protein concentrations (50 μg/ml using silver stain detection or 5 μg/ml using Krypton IR detection) in the presence of either substrate or magnesium (Fig. 5c). At 50 μg/ml (1.4 μM subunit) and 1 mM ALA, native PAGE shows an apparent IC50 of ~100 μM; at 5 μg/ml and 10 mM magnesium, the apparent IC50 is ~ 1 - 3 μM. The gel run at 50 μg/ml is also shown activity stained and again the activity is only observed in the octameric assembly (Fig. 5c). The fact that no activity is associated with the hexamer in Fig 5c, while activity is seen in the octamer, further supports the specific binding of morphlock-1 to the pea PBGS hexamer (see above).
DISCUSSION
Targeting alternate quaternary structure assemblies provides a new approach to drug discovery
We have sought to develop a novel paradigm for drug discovery that capitalizes on the morpheein model of allostery to target universally essential pathways in a species specific manner. While the morpheein model of allostery is only recently described (Jaffe, 2005), the advantages of targeting allosteric sites for drug discovery are well established. The possibility that a known drug target might exist as a morpheein provides new insight into how existing drugs, whose mechanism is not currently understood, might function to inhibit or activate these protein targets.
The term allosteric was introduced in 1963 by Monod et al., who realized the unlimited capacity for metabolic regulation (activation or inhibition) when nature does not require chemical similarity between the allosteric effector molecule and the substrate (Monod et al., 1963). Phylogenetic variations in allosteric binding sites allow fine tuning of metabolic regulation to the specific needs of a given species without compromising the conserved chemical specificity of an enzyme active site. These variations, in turn, allow targeting conserved pathways for the development of therapeutic agents such as antimicrobials. Significant recent effort in drug discovery has been directed toward allosteric sites, and it has been pointed out that “it is not a simple matter to find and characterize new allosteric sites” (Hardy and Wells, 2004). However, surface topologies that are specific to alternate quaternary structure assemblies form structurally defined allosteric sites. Molecules that bind to these sites may stabilize one specific assembly and draw the quaternary structure equilibrium toward that assembly. Thus, enzymes that utilize the morpheein model of allosteric regulation are predicted to contain these newly defined allosteric sites and provide new drug targets. We put PBGS forth as a prototype.
The behavior of PBGS establishes that alternate homo-oligomerization need not represent a misfolding process or the uncontrolled aggregation observed for prions or amyloid plaque formation. The morpheein model of allostery describes a conformational change in a given protein that gives rise to a reversible equilibrium between alternate physiologically relevant oligomers of fixed stoichiometries. Within the context of a recent review on proteostasis (Balch et al., 2008), which considers the equilibrium between unfolded protein chains, non-functional misfolded proteins, protein aggregates, and a folded protein ensemble, one might place all native assemblies of proteins that use a morpheein model of allosteric regulation within the context of the folded protein ensemble.
Allosteric regulation of PBGS as a new drug target
PBGS functions in the highly conserved biosynthesis of tetrapyrroles such as heme, chlorophyll, vitamin B12, siroheme, or cofactor F430 (Battersby, 2000). The phototoxic intermediates in this pathway necessitate different requirements for control in different organisms (e.g. respiration vs. photosynthesis vs. methanogenesis). There is phylogenetic variation in the presence or absence of an allosteric magnesium binding site, which has been proposed to function in activating PBGS in greening chloroplasts (Breinig et al., 2003; Prisic and Peters, 2007). This site is present in the octameric form of PBGS (Fig 2a) from plants and human bacterial pathogens (Jaffe, 2003). Treatment of octameric pea PBGS with the magnesium chelator EDTA favors formation of the hexamer; addition of magnesium to proteins containing the allosteric magnesium binding site draws the equilibrium to the octamer and increases the observed activity (Fig. 2a) (Breinig et al., 2003; Jaffe et al., 1995). The residues that bind the allosteric magnesium are not present in human PBGS nor in PBGS from other metazoa or fungi; it is established that a spatially equivalent arginine residue functions to stabilize the octameric assembly in these species of PBGS (Tang et al., 2006).
Activation of PBGS through structural stabilization of the octamer may be counteracted through structural stabilization of the hexamer. There is a mechanistic difference between disfavoring the octamer by removing the activator (here magnesium) vs. adding an inhibitor that stabilizes the hexamer. While the magnesium binding site on the octamer is not the same as the docking site used to find hexamer-stabilizing inhibitors (Fig. 2b), there is some overlap in the residues that comprise these two sites. While both qualify as allosteric sites, their existence is mutually exclusive (one is specific to the octamer while the other is specific to the hexamer), and the chemical characteristics of ligands that would bind at each of these two sites are disparate.
The emergence of multi-drug resistant organisms drives the continued need to discover or develop antibiotics that function via novel mechanisms. The current golden age of structural genomics has yielded thousands of detailed protein X-ray crystal structures. Initially, there were expectations that this wealth of structural information would lead directly to drugs custom-designed to block the active sites of, and thereby inhibit, medically relevant enzymes. With a few exceptions, this approach has not been fruitful. With an appreciation for Monod’s prescience that allosteric sites would allow the greatest flexibility in chemical specificity (Monod et al., 1963), we advance the hypothesis that surface topologies specific to one quaternary structure assembly of a morpheein equilibrium provide an opportunity for prediction of allosteric sites and present new drug binding sites. The demonstration that morphlock-1 is selective for inhibition of pea, and not human PBGS, or the tested fly or bacterial PBGS (Figs. 4a and 4c), coupled with the phylogenetic variation in the inhibitor binding site (Fig. 2c), suggests that PBGS is a viable target for the development of antimicrobial therapeutics as well as herbicides. As there is no inherent requirement for chemical similarity between different hexamer-stabilizing compounds, a cocktail of such chemically dissimilar compounds may help evade the development of drug resistance.
Approaches to discovering compounds that perturb an equilibrium of quaternary structure assemblies
Designing or discovering drugs directed at one quaternary structure assembly of a morpheein equilibrium is aided by a non-traditional approach to drug discovery. For example, standard in vitro high-throughput screening methods are designed to discover inhibitors, regardless of the mechanism of inhibition. A mechanism-blind approach is unlikely to yield species-specific inhibitors that bind to a cryptic allosteric site that is specific to one alternate quaternary assembly. It may prove possible to discover specific oligomer-stabilizing inhibitors using high throughput screening if there is an assay technique that is sensitive to changes in molecular mass, such as native PAGE or light scattering; these are not yet available as high-throughput methods. Alternatively, for a morpheein such as PBGS, where the target species show a protein-concentration-dependent specific activity, one could carry out an inhibition screen at several protein concentrations and focus on those inhibitors whose action is protein concentration dependent. One caveat is that promiscuous inhibitors, which can be discovered regardless of the search technique, would also be expected to be protein concentration dependent (Feng and Shoichet, 2006). Consequently, we have employed in silico docking methods for discovery of morphlock-1 and suggest this approach for future discovery efforts at least until the necessary experimental high throughput screening methods for oligomer-stabilizing molecules are available.
Molecular models as viable targets
One notable aspect of our in silico studies is the use of protein structure homology models to define the small molecule binding site. Although docking more often uses crystal structures, crystal structures of morpheeins can be elusive due to the proteins’ dynamic interconversion of alternate assemblies. Comparative analyses conclude that good homology models are an equally effective as crystal structures for docking studies (Kairys et al., 2006). Furthermore, the putative hexamer-trapping ligand binding site on PBGS is solvent exposed, the side chains are likely flexible, and a model, or even a crystal structure, represents only one of many plausible suites of side chain rotamer configurations (Shapovalov and Dunbrack, 2007). A complex of a morpheein-trapping inhibitor, like morphlock-1, and its stabilized oligomer is expected to assist the acquisition of high resolution crystal structures. For instance, both in our hands and those of others, pea PBGS crystals have been obtained, but crystal quality did not allow for structure solution (Youell, 2004). Efforts are underway to determine if a morphlock-1:pea PBGS complex can help solve this problem, lead to new crystal structures, and thus provide improved information to guide the future selection of potential inhibitors.
SIGNIFICANCE
The advantages of targeting allosteric sites for drug discovery are long appreciated (Monod et al., 1963) and well established, but the locations of allosteric sites are difficult to predict (Hardy and Wells, 2004). This article introduces the concept that the structural differences between morpheein assemblies can be used to predict allosteric sites. We report a hexamer-trapping, species-selective inhibitor of PBGS, which opens the door to the development of novel antimicrobial therapeutics by targeting PBGS from human pathogens. The morpheein model for allosteric regulation is not likely to be limited to PBGS. The number of proteins that behave as morpheeins may only be appreciated once it is demonstrated that other homo-oligomeric proteins can reversibly dissociate, change shape, and re-assemble differently with physiologically relevant functional consequences. We have previously proposed a number of essential enzymes as putative morpheeins, for example ribonucleotide reductase and purine nucleoside phosphorylase (Jaffe, 2005). Application of an oligomer-stabilizing approach may assist in the design or discovery of species-selective inhibitors (or activators) of these proteins. The recent literature has revealed two established drug targets for which it is possible to affect inhibition by shifting a quaternary structure equilibrium. One is tumor necrosis factor α (TNFα), for which the accepted quaternary structure assembly is a trimer (Baeyens et al., 1999), and for which one inhibitor functions by stabilizing a dimeric assembly (He et al., 2005). A second example is HIV integrase, for which small peptides affect function by shifting its oligomeric equilibrium (Hayouka et al., 2007). It is unknown if the behavior of TNFα or HIV integrase fulfill the characteristics we have set forth to define morpheeins (Jaffe, 2005). However, the discovery of the quaternary structure stabilizing inhibitors of TNFα or HIV suggests a widespread applicability for drug development based on trapping alternate quaternary structure assemblies.
EXPERIMENTAL PROCEDURES
Materials
Human (N59/C162A variant) and pea PBGS (C326A variant) were expressed and purified as previously reported (Kervinen et al., 2000; Tang et al., 2006). The programs MacroModel, glide, and QikProp, and the graphical user interface Maestro were from Schrödinger, L.L.C. (New York, NY). SigmaPlot was from Systat Software, Inc. (San Jose, CA). The candidate inhibitors were from Life Chemicals, Inc. (Burlington, ON, Canada). Krypton Infrared Protein Stain was from Thermo Scientific. All other chemicals were from Fisher (Pittsburgh, PA) or Sigma (St. Louis, MO), and were the highest purity available. Electrophoresis equipment and reagents, and chromatography equipment and resins were from GE Healthcare (Piscataway, NJ).
Preparation of Protein Structure Model Used for in silico Docking
The only crystal structure of a hexameric assembly of PBGS is that of the naturally occurring human PBGS variant F12L (PDB code 1PV8). However, its utility as a template for a model of the pea PBGS hexamer is limited because that structure contains significant regions of disorder, the overall sequence identity between human and pea PBGS is 41%, and the sequence identity in the N-terminal arm is less than 10%. There are, however, excellent high resolution structures for the octameric assembly of Pseudomonas aeruginosa PBGS (e.g. PDB code 1GZG, (Frere et al., 2002)), which has overall sequence identity of 46% with pea PBGS, and 35% sequence identity in the N-terminal arm. We previously reported merging a variety of techniques to use the information in both structures 1PV8 and 1GZG to build a model for a hexameric assembly of P. aeruginosa PBGS (Bollivar et al., 2004). A model for the hexameric assembly of pea PBGS was prepared from the model of the hexameric assembly of P. aeruginosa PBGS using standard homology modeling techniques and the program MolIDE (Canutescu and Dunbrack, 2005). This software integrates sequence alignment, threading, loop model building to accommodate insertions and deletions, and side chain optimization similar to that used for our previously published models of PBGS.
Building the Library of Compounds for Docking
Two dimensional representations of compounds available from LifeChemicals, Inc. (60,593 structures) were obtained in SD format from the vendor. Scripts from Schrödinger Inc. were used to convert these into three dimensional, energy minimized representations in a file format used by GLIDE as detailed in Supplemental Methods.
In silico Docking of Inhibitors
Glide (Halgren et al., 2004) was used for docking subsets of the virtual small molecule library to the pea PBGS hexamer homology model. Compounds were selected for testing based on the following five criteria. 1) Each selected molecule must make van der Waals contacts or hydrogen bonds with all three of the subunits. 2) Each selected molecule must have a predicted solubility (Log S) estimate of at least -6 (calculated using QikProp (Jorgensen and Duffy, 2000)). 3) The set of selected molecules must contain a broad sampling of dissimilar structures. 4) The set of selected molecules must occupy a variety of predicted docking locations within the binding site (Fig 2b). 5) The final criterion was cost, limiting our initial purchase to ~100 molecules.
Preparation of the Candidate Inhibitors
Solutions were prepared by the addition of 1 mL of dimethylsulfoxide (DMSO) to each vial purchased at 10 μmoles to yield a calculated final concentration of 10 mM. Fourteen compounds were insoluble in DMSO at this concentration and were not analyzed further. Compounds were stored in the dark at room temperature.
Dialysis Preparation of PBGS Oligomers
Samples were dialyzed prior to native gel electrophoreses experiments. To produce octameric pea PBGS, 50 μL of PBGS (3 mg/mL) was dialyzed against 2 L of 100 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane hydrochloride (BTP-HCl) pH 8.5. To produce hexameric pea PBGS, 50 μL of PBGS (3 mg/mL) was dialyzed against 2 L of 10 mM BTP pH 8.5, 1 mM ethylenediamine tetra-acetic acid (EDTA). All dialyses were performed overnight at 4 °C.
Native Gel Electrophoresis
Electrophoresis was performed using a PhastSystem with PhastGel native buffer strips (880 mM L-alanine and 250 mM Tris pH 8.8, made of 3% Agarose IEF). Using a 6-lane sample applicator, 4 μl of each sample were loaded onto a homogeneous 12.5% polyacrylamide gel (12.5% total acrylamide in the separation zone; buffer of 112 mM acetate and 112 mM Tris pH 6.5. The two dimensional native PAGE experiment (Fig. S2) is detailed in Supplemental Methods. After separation, gels were developed on the PhastSystem using Coomassie Blue, or stained using silver stain, Krypton Infrared stain, or the PBGS activity stain as detailed in Supplemental Methods.
PBGS Activity Assays
The activity assay was based on the measurement of the porphobilinogen formed from δ-aminolevulinic acid (ALA) by PBGS. For human PBGS, the standard assay contained 100 mM BTP-HCl pH 7.3, 10 mM β-mercaptoethanol (β-ME), 10 μM ZnCl2 and PBGS in a volume of 900 μL. For pea PBGS, the standard assay contained 100 mM BTP-HCl pH 8.5, 10 mM MgCl2 and an appropriate concentration of PBGS in a volume of 900 μL. Reactions were initiated by the addition of 100 μL of 100 mM ALA-HCl to a final concentration of 10 mM, yielding a final assay volume of 1 mL. The assays were allowed to proceed for various fixed lengths of time at 37 °C prior to termination by the addition of 500 μL of STOP reagent (20% trichloroacetic acid). For human PBGS, STOP reagent also contained 100 mM HgCl2, which precipitates as a complex with β-ME to prevent β-ME interference with subsequent steps. The stopped reactions were vortexed vigorously and centrifuged for 3 min. at 5000 rpm in a Fisher Centrific benchtop centrifuge. Porphobilinogen was quantified by formation of the pink-colored porphobilinogen:ρ -dimethylaminobenzaldehyde complex upon reaction with Ehrlich’s reagent. 800 μL of assay solution was added to 800 μL of Ehrlich’s reagent, and the color was allowed to develop for 8 min. prior to measuring the absorbance at 555 nm (ε555 = 60,200 M−1cm−1) utilizing a Varian Cary UV-visible spectrophotometer equipped with a quartz dip probe. When necessary, dilutions were performed as previously described (Tang et al., 2006).
For the inhibition studies, human and pea PBGS were pre-incubated with either the inhibitor or DMSO for 30 minutes prior to assay. The enzyme was diluted to a concentration 11.1-fold greater than the desired final assay concentration using the corresponding assay buffer as the diluent. For each reaction (final volume of 1 mL), 90 μL of diluted enzyme was placed in a test tube and 10 μL of inhibitor or 10 μL of DMSO was added. Following pre-incubation, 800 μL of the appropriate assay buffer was added, and the samples were equilibrated at 37 °C for 15 min. prior to the initiation of the reaction with 100 μL of 100 mM ALA.
Data fitting
All kinetic data were fit using the program SigmaPlot®. The protein concentration dependence of pea PBGS was described by the hyperbolic function: v = (Vmax[PBGS])/(K(0.5) + [PBGS]), where v is the observed rate, Vmax is the maximum rate, [PBGS] is the subunit concentration of PBGS, and K(0.5) is the midpoint of the protein concentration dependence curve. The inhibition of pea PBGS was described by the hyperbolic function: vrel = (Vmax[I])/(IC50 + [I]), where vrel is the relative rate observed, Vmax is the maximum rate, [I] is the concentration of inhibitor, and IC50 is the concentration of inhibitor at which the rate is reduced by 50%.
Supplementary Material
1: Coordinates for the model of the hexameric assembly of pea PBGS
2: Table S1 – The Life Chemicals identifier for each of the 76 compounds and the poses of their structures that led to their selection
3: Figure S1 – Native PAGE gel shift evaluation of compounds purchased from Life Chemicals
4: Figure S2 – Two dimensional native PAGE showing substrate mediated in-gel hexamer to octamer transition
5: Supplemental Methods
Acknowledgments
This publication was supported by NIH Grants CA006927 (FCCC), CA-009035-31 (ICR), AI063324 (EKJ), and GM31186 (GDM), and by an appropriation from the Commonwealth of Pennsylvania. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute, the National Institute for Allergy and Infectious Disease, nor the National Institute of Environmental Health Sciences of the National Institutes of Health. This project was funded, in part, under a grant with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. We acknowledge the contributions of the FCCC Organic Synthesis and Molecular Modeling core facilities, in particular the expertise of Roland Dunbrack and Adrian Canutescu, the authors of MolIDE. The authors thank David Bollivar and Jeffrey Peterson for helpful discussions and John Taylor for use of the Li-Cor Odyssey scanner.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baeyens KJ, De Bondt HL, Raeymaekers A, Fiers W, De Ranter CJ. The structure of mouse tumour-necrosis factor at 1.4 A resolution: towards modulation of its selectivity and trimerization. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):772–778. doi: 10.1107/s0907444998018435. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Battersby AR. Tetrapyrroles: the pigments of life. Nat Prod Rep. 2000;17(6):507–526. doi: 10.1039/b002635m. [DOI] [PubMed] [Google Scholar]
- Boese QF, Spano AJ, Li JM, Timko MP. Aminolevulinic acid dehydratase in pea (Pisum sativum L.). Identification of an unusual metal-binding domain in the plant enzyme. J Biol Chem. 1991;266(26):17060–17066. [PubMed] [Google Scholar]
- Bollivar DW, Clauson C, Lighthall R, Forbes S, Kokona B, Fairman R, Kundrat L, Jaffe EK. Rhodobacter capsulatus porphobilinogen synthase, a high activity metal ion independent hexamer. BMC Biochemistry. 2004;5:17. doi: 10.1186/1471-2091-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breinig S, Kervinen J, Stith L, Wasson AS, Fairman R, Wlodawer A, Zdanov A, Jaffe EK. Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase. Nature Structural Biology. 2003;10(9):757–763. doi: 10.1038/nsb963. [DOI] [PubMed] [Google Scholar]
- Canutescu AA, Dunbrack RL., Jr MollDE: a homology modeling framework you can click with. Bioinformatics. 2005;21(12):2914–2916. doi: 10.1093/bioinformatics/bti438. [DOI] [PubMed] [Google Scholar]
- Eldridge M, Murray CW, Auton TA, Paolini GV, Lee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput-Aided Mol Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- Feng BY, Shoichet BK. A detergent-based assay for the detection of promiscuous inhibitors. Nat Protoc. 2006;1(2):550–553. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg N, Erskine PT, Cooper JB, Shoolingin-Jordan PM, Jahn D, Heinz DW. High resolution crystal structure of a Mg2+-dependent porphobilinogen synthase. J Mol Biol. 1999;289(3):591–602. doi: 10.1006/jmbi.1999.2808. [DOI] [PubMed] [Google Scholar]
- Frere F, Schubert WD, Stauffer F, Frankenberg N, Neier R, Jahn D, Heinz DW. Structure of porphobilinogen synthase from Pseudomonas aeruginosa in complex with 5-fluorolevulinic acid suggests a double Schiff base mechanism. J Mol Biol. 2002;320(2):237–247. doi: 10.1016/S0022-2836(02)00472-2. [DOI] [PubMed] [Google Scholar]
- Gross U, Sassa S, Jacob K, Deybach JC, Nordmann Y, Frank M, Doss MO. 5-Aminolevulinic acid dehydratase deficiency porphyria: a twenty-year clinical and biochemical follow-up. Clin Chem. 1998;44(9):1892–1896. [PubMed] [Google Scholar]
- Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Wells JA. Searching for new allosteric sites in enzymes. Curr Opin Struct Biol. 2004;14(6):706–715. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Hayouka Z, et al. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc Natl Acad Sci U S A. 2007;104(20):8316–8321. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He MM, et al. Small-molecule inhibition of TNF-alpha. Science. 2005;310(5750):1022–1025. doi: 10.1126/science.1116304. [DOI] [PubMed] [Google Scholar]
- Jaffe EK. An unusual phylogenetic variation in the metal ion binding sites of porphobilinogen synthase. Chem Biol. 2003;10(1):25–34. doi: 10.1016/s1074-5521(02)00296-x. [DOI] [PubMed] [Google Scholar]
- Jaffe EK. The porphobilinogen synthase catalyzed reaction mechanism. Bioorganic Chemistry. 2004;32(5):316–325. doi: 10.1016/j.bioorg.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Jaffe EK. Morpheeins--a new structural paradigm for allosteric regulation. Trends in Biochemical Sciences. 2005;30(9):490–497. doi: 10.1016/j.tibs.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Jaffe EK, Ali S, Mitchell LW, Taylor KM, Volin M, Markham GD. Characterization of the role of the stimulatory magnesium of Escherichia coli porphobilinogen synthase. Biochemistry. 1995;34(1):244–251. doi: 10.1021/bi00001a029. [DOI] [PubMed] [Google Scholar]
- Jaffe EK, Stith L. ALAD porphyria is a conformational disease. Am J Hum Genet. 2007;80(2):329–337. doi: 10.1086/511444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LC, Tawfik DS. Conformational diversity and protein evolution--a 60-year-old hypothesis revisited. Trends Biochem Sci. 2003;28(7):361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Duffy EM. Prediction of drug solubility from Monte Carlo simulations. Bioorg Med Chem Lett. 2000;10(11):1155–1158. doi: 10.1016/s0960-894x(00)00172-4. [DOI] [PubMed] [Google Scholar]
- Kairys V, Fernandes MX, Gilson MK. Screening drug-like compounds by docking to homology models: A systematic study. Journal of Chemical Information and Modeling. 2006;46(1):365–379. doi: 10.1021/ci050238c. [DOI] [PubMed] [Google Scholar]
- Kervinen J, Dunbrack RL, Jr, Litwin S, Martins J, Scarrow RC, Volin M, Yeung AT, Yoon E, Jaffe EK. Porphobilinogen synthase from pea: expression from an artificial gene, kinetic characterization, and novel implications for subunit interactions. Biochemistry. 2000;39(30):9018–9029. doi: 10.1021/bi000620c. [DOI] [PubMed] [Google Scholar]
- Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5(1):365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: a Plausible Model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Prisic S, Peters RJ. Synergistic substrate inhibition of ent-copalyl diphosphate synthase: a potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol. 2007;144(1):445–454. doi: 10.1104/pp.106.095208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwood T, Tang L, Lawrence SH, Anokhina Y, Jaffe EK. Kinetics and thermodynamics of the interchange of the morpheein forms of human porphobilinogen synthase. Biochemistry. 2008;47(10):3245–3257. doi: 10.1021/bi702113z. [DOI] [PubMed] [Google Scholar]
- Shapovalov MV, Dunbrack RL., Jr Statistical and conformational analysis of the electron density of protein side chains. Proteins. 2007;66(2):279–303. doi: 10.1002/prot.21150. [DOI] [PubMed] [Google Scholar]
- Tang L, Breinig S, Stith L, Mischel A, Tannir J, Kokona B, Fairman R, Jaffe EK. Single amino acid mutations alter the distribution of human porphobilinogen synthase quaternary structure isoforms (morpheeins) Journal of Biological Chemistry. 2006;281(10):6682–6690. doi: 10.1074/jbc.M511134200. [DOI] [PubMed] [Google Scholar]
- Tang L, Stith L, Jaffe EK. Substrate-induced interconversion of protein quaternary structure isoforms. Journal of Biological Chemistry. 2005;280(16):15786–15793. doi: 10.1074/jbc.M500218200. [DOI] [PubMed] [Google Scholar]
- Traut TW. Dissociation of enzyme oligomers: a mechanism for allosteric regulation. Crit Rev Biochem Mol Biol. 1994;29(2):125–163. doi: 10.3109/10409239409086799. [DOI] [PubMed] [Google Scholar]
- Walker DA. Regulatory mechanism in photosynthetic carbon metabolism. Curr Top Cell Regul. 1976;11:203–241. doi: 10.1016/b978-0-12-152811-9.50013-4. [DOI] [PubMed] [Google Scholar]
- Youell JH. Chemistry, Biochemistry (0487) Southampton (United Kingdom): University of Southampton; 2004. Studies on recombinantPisum sativum and human 5-aminolaevulinic acid dehydratases; p. 692. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1: Coordinates for the model of the hexameric assembly of pea PBGS
2: Table S1 – The Life Chemicals identifier for each of the 76 compounds and the poses of their structures that led to their selection
3: Figure S1 – Native PAGE gel shift evaluation of compounds purchased from Life Chemicals
4: Figure S2 – Two dimensional native PAGE showing substrate mediated in-gel hexamer to octamer transition
5: Supplemental Methods