SUMMARY
Abasic (AP) sites are among the most abundant DNA lesions formed in human cells and they present a strong block to replication. DNA polymerase ι (Polι) is one of the few DNA Pols that does not follow the A-rule opposite an abasic site. We present here three structures of human Polι in complex with DNAs containing an abasic lesion and dGTP, dTTP or dATP as the incoming nucleotide. The structures reveal a mechanism of translesion synthesis across an abasic lesion that differs from that in other Pols. Both the abasic lesion and the incoming dNTPs are intrahelical and are closely apposed across a constricted active site cleft. The dNTPs partake in distinct networks of hydrogen bonds in the “void” opposite the lesion. These different patterns of hydrogen bonds, as well as stacking interactions, may underlie Polι’s small preference for insertion of dGTP over other nucleotides opposite this common lesion.
INTRODUCTION
Abasic sites are among the most abundant DNA lesions in humans, with up to 10,000 purine bases lost per day in each cell due to cleavage of the N-glycosidic bond (Lindahl, 1982; Lindahl and Nyberg, 1972). Hydrolysis of the N-glycosidic bond occurs at a high rate spontaneously, and oxidative stress and exposure to ionizing radiation and alkylating agents further increase the incidence of base loss. Although abasic lesions can be removed by the base excision repair and nucleotide excision repair pathways (Seeberg et al., 1995; Wallace, 1997), lesions that escape repair pose a problem for cellular processes such as transcription and replication that depend on an instructional template. Indeed, it has been estimated that each human liver cell contains ~50,000 apurinic/apyrimidinic (AP) sites under normal conditions (Nakamura and Swenberg, 1999). Abasic lesions block replication, and are also highly mutagenic because when replicative DNA polymerases (Pols) do manage (inefficiently) to insert a nucleotide opposite the lesion they prefer to insert an A, in a phenomenon known as the A-rule (Avkin et al., 2002; Boiteux and Laval, 1982; Lawrence et al., 1990; Loeb and Preston, 1986; Mozzherin et al., 1997; Pages et al., 2008; Sagher and Strauss, 1983; Shibutani et al., 1997; Strauss, 2002). The mechanistic basis for the A-rule remains under debate, despite the numerous biochemical and structural studies (Avkin et al., 2002; Boiteux and Laval, 1982; Chen et al., 2008; Cuniasse et al., 1990; Cuniasse et al., 1987; Fiala et al., 2007; Gelfand et al., 1998; Hogg et al., 2004; Ling et al., 2004; Loeb and Preston, 1986; Matray and Kool, 1999; Mozzherin et al., 1997; Randall et al., 1987; Reineks and Berdis, 2004; Sagher and Strauss, 1983; Shibutani et al., 1997; Strauss, 2002; Zahn et al., 2007).
Human Polι is one of the few DNA Pols that does not obey the A-rule for inserting nucleotides opposite the abasic site. Polι is a member of the Y-family DNA Pols that possess the ability to replicate through DNA lesions (Prakash et al., 2005). In addition to Polι, humans have three other Y-family Pols - Polη, Polκ and Rev1 – each with a unique DNA damage bypass and fidelity profile (Prakash et al., 2005). Polι is an unusual polymerase in that it inserts nucleotides opposite template purines with a much higher efficiency and fidelity than opposite template pyrimidines. And, even for the template purines, Polι exhibits a higher efficiency and fidelity opposite template A than opposite template G. Polι is highly inefficient at incorporating the correct nucleotide opposite template C and T, and opposite template T, it misincorporates a G ~ 10 times better than it incorporates the correct nucleotide A (Haracska et al., 2001; Johnson et al., 2000; Tissier et al., 2000; Washington et al., 2004; Zhang et al., 2000). The ternary crystal structure of Polι bound to template A and an incoming dTTP, shows that the templating A adopts a syn conformation and forms a Hoogsteen base pair with the incoming T, which remains in the anti conformation (Nair et al., 2006b; Nair et al., 2004). Similarly, in the structure of Polι paired with a template G and an incoming dCTP, a G.C+ Hoogsteen base pair is formed in the polymerase active site (Nair et al., 2005a; Nair et al., 2006b). Hoogsteen base pairing provides an elegant basis for the much higher efficiency and fidelity of nucleotide incorporation opposite template purines than opposite pyrimidines because only the templates A and G have a Hoogsteen edge by which they can establish two hydrogen bonds with the correct incoming pyrimidine nucleotide. Because of the lack of a Hoogsteen edge, there are few hydrogen bonding possibilities opposite template pyrimidines. The ability to form Hoogsteen base pairs allows Polι to promote replication through adducts such as 1, N6-ethenodeoxyadenosine that disrupt the Watson-Crick (W-C) edge but not the Hoogsteen edge of the templating purine (Nair et al., 2006a). Curiously, Polι can also incorporate nucleotides opposite an abasic lesion, albeit with reduced efficiency. Although Polι can incorporate any of the four nucleotides opposite the abasic site, it exhibits a small preference for the insertion of G followed by T, A, and then a C (Johnson et al., 2000).
To gain insights into the mechanism(s) by which Polι inserts nucleotides opposite abasic lesions, we have determined structures of Polι in complex with DNAs containing an abasic lesion at the templating position and with dGTP, dTTP or dATP as the incoming nucleotide. Together, the structures show that the Polι active site facilitates the entry of dNTPs despite the lack of an instructional templating base. Polι is able to stabilize the incoming nucleotide opposite the abasic lesion without any significant change in conformation of either the protein, the DNA or the nucleotide. Both the abasic lesion and the incoming dNTPs are intrahelical, and, the dNTPs partake in distinct networks of hydrogen bonds in the “void” opposite the lesion. We suggest that these different patterns of hydrogen bonds, as well as stacking interactions, underlie Polι’s preference for insertion of dGTP over dTTP/dATP opposite the abasic lesion.
RESULTS
Structure Determination
We crystallized Polι with a “double-ended” 18-nt template-primer designed to present an abasic lesion in the active sites of two symmetrically bound Polι molecules (see Methods). We have used a similar strategy previously to crystallize Polι with undamaged and damaged DNAs (Nair et al., 2005a; Nair et al., 2006a; Nair et al., 2006b; Nair et al., 2004). Cocrystals with incoming dGTP, dATP and dTTP were obtained under identical conditions (from PEG solutions), belonging to space group P6522 with cell dimensions of a = 98 Å, b = 98 Å, c = 203Å and α =β=90°, γ=120°. We were unable to obtain suitable crystals of Polι with an incoming dCTP. The structures were solved by molecular replacement using the structure of the PolιA.dTTP (with dA at the templating position and dTTP as the incoming nucleotide) complex as a search model (Nair et al., 2006b). The PolιAbasic.dGTP, PolιAbasic.dTTP and PolιAbasic.dATP complexes were refined to resolutions of 2.1 Å, 2.1 Å and 2.2 Å, respectively. The refined models contain Polι residues 25–370, 379–394 and 404–414 (the first 24 and the last 6 residues are disordered, and residues 371–378 and 395–403 comprising two loops in the PAD have poor density), DNA nucleotides 4–11 (three of the four unpaired template nucleotides at the 5′ end are disordered), incoming dNTP, two Mg2+ ions, and water molecules (Fig. 1). The PolιAbasic.dGTP (Rfree of 25.8%; Rcrys of 22.0%), PolιAbasic.dTTP (Rfree of 24.7%; Rcrys of 22.0%) and PolιAbasic.dATP (Rfree of 25.5%; Rcrys of 21.6%) complexes contain 224, 213 and 181 water molecules, respectively. All three structures have good stereochemistry, with ~86% of the residues in the most favored regions of the Ramachandran plot and <1% in the disallowed regions.
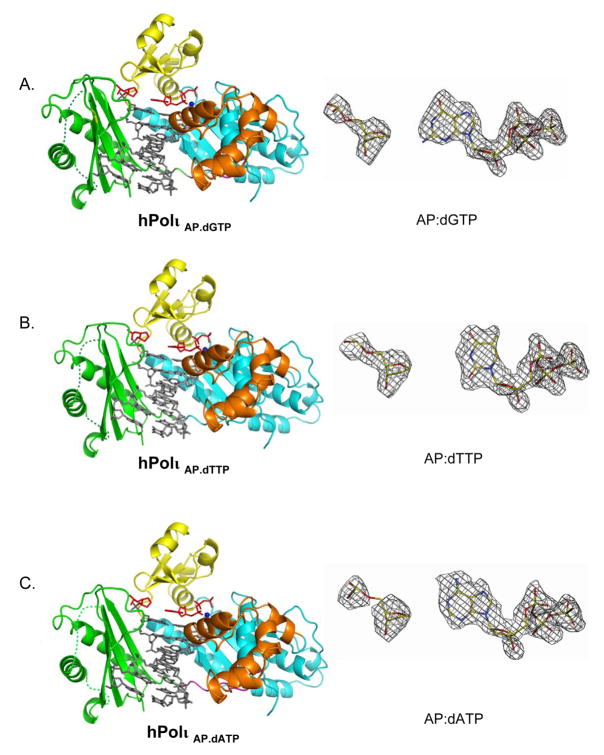
Figure 1.
Polι Abasic.dNTP ternary complexes and corresponding simulated annealed omit maps of the incipient “base pairs”. A) Polι Abasic.dGTP ternary complex (left), shown for one of the two Polι molecules bound to a double-ended template primer. The palm, fingers, thumb domain and the PAD in the Polι molecule are shown in cyan, yellow, orange and green respectively. DNA is shown in gray, abasic lesion and incoming dGTP are in red, and the putative Mg2+ ions are displayed as dark blue spheres. The simulated annealing Fo-Fc omit map (contoured at 3.0σ) for the abasic lesion and incoming dGTP is shown on right. B) Polι Abasic.dTTP ternary complex and corresponding simulated annealing Fo-Fc omit map. C) Polι Abasic.dATP ternary complex and corresponding simulated annealing Fo-Fc omit map.
Overall Arrangement
In all three ternary complexes, a Polι molecule binds to each end of the double-ended “abasic” template primer. The two molecules are related by a crystallographic two-fold axis and thus make identical contacts with the template-primer. Polι has the familiar right-handed architecture with palm (residues 25–37, 99–224), fingers (38–98), and thumb (225–288) domains, and the PAD (polymerase associated domain; residues 298–414) unique to Y-family polymerases (Fig. 1). The four domains occupy identical positions with respect to the template-primer in all three complexes (Fig. 1). The palm forms the floor of the DNA binding cavity and carries the active site residues (Asp34, Asp126 and Glu127) that catalyze the nucleotidyl transfer reaction, while the fingers domain lies over the abasic lesion and the incoming nucleotide. The thumb and the PAD are connected by a long linker that spans the width of the DNA. The thumb skims the minor groove surface on one side the template-primer, while the PAD docks in the major groove on the other side. The interface between the two Polι molecules is relatively small, with ~700 Å2 of surface area buried between the thumb domain of one polymerase and the PAD of the other. The majority of Polι-DNA interactions are mediated by the PAD, wherein the main chain amides on “outer” β-strands of the PAD β-sheet make a series of hydrogen bonds with the template and primer strands.
The Polι Abasic.dGTP ternary complex
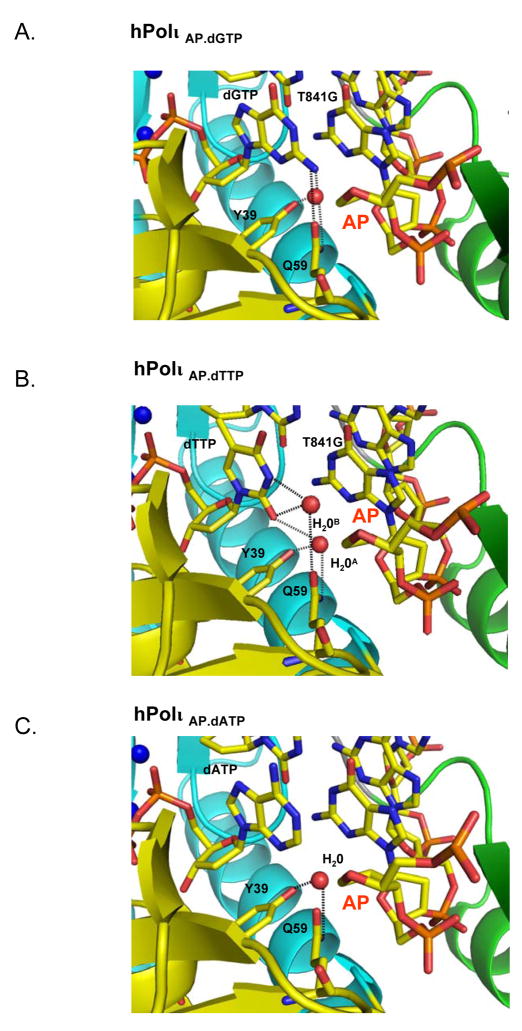
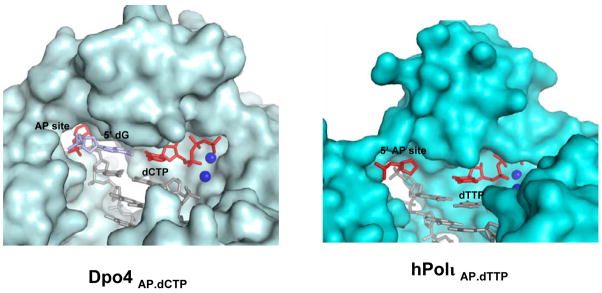
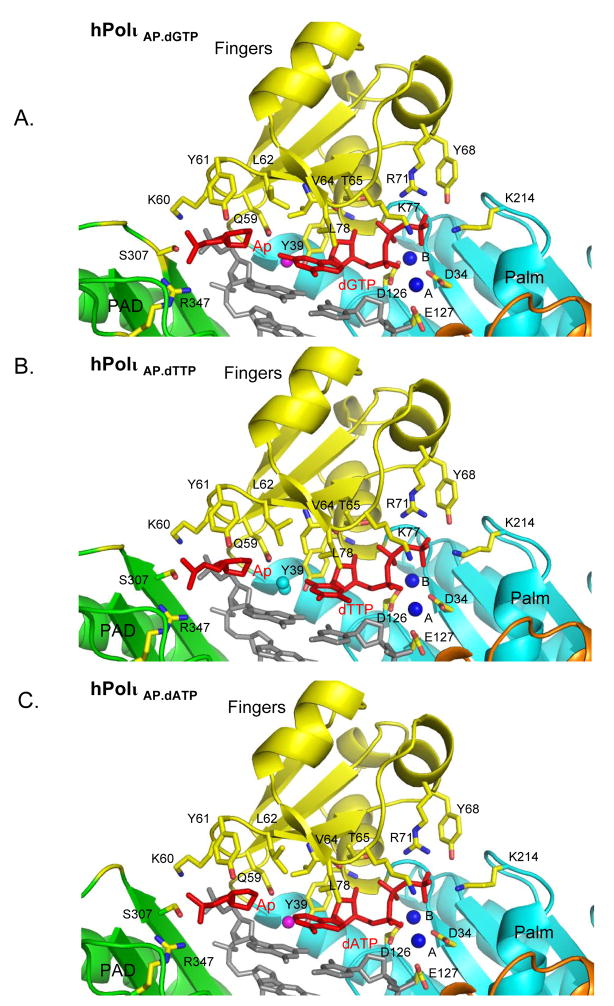
The abasic lesion does not loop or bulge out of the DNA helix (Figs. 1–3). This is in contrast to an archeael homolog of human Polκ, namely Dpo4, structures of which have been reported in complex with abasic lesions (Ling et al., 2004). In Dpo4, the abasic site loops out of the DNA helix and the incoming dNTP pairs with a base 5′ to the lesion (Fig. 4). By contrast, in Polι, the abasic sugar is accommodated intrahelically in a small hydrophobic pocket formed by the aliphatic portions of Lys60 and Gln59 side chains, the aromatic ring of Tyr61, and the non-polar side chain of Leu62 (Figs. 2 & 4). The lesion is further locked in place by a salt-link between Lys60 and a phosphate on the 3′ side of the lesion and a hydrogen bond between Ser307 (on the PAD) and the phosphate of the lesion (Fig. 2). Incoming dGTP sits opposite the abasic lesion, and not opposite the 5′ base as in Dpo4. The dGTP sugar stacks over the aromatic ring of Tyr39, and a hydrogen bond is formed between its 3′-OH and the main chain amide of Tyr39. The dGTP triphosphate moiety interleaves between the palm and fingers domains, and makes hydrogen bonds with Thr65, Tyr68, Arg71 and Lys77 from the fingers domain and Lys214 from the palm domain (Fig. 2A). The triphosphate moiety and the catalytic residues Asp34, Asp126 and Glu127 coordinate two Mg2+ ions in the active site. Based on this geometry, Polι is well poised for catalysis with the putative 3′oxygen (at the primer terminus) located ~3.2Å from the dGTP α-phosphate and aligned more or less linearly with respect to the scissile P α–O3′ bond (Fig. 2A). In all, these interactions to the dGTP sugar and triphosphate moiety are similar to those observed in previous Pol ι structures and they serve to position dGTP in the active site (Nair et al., 2005a; Nair et al., 2006b; Nair et al., 2004). The dGTP base extends towards the abasic lesion. Strikingly, the distance between the C2′ atom on the lesion sugar and the N2 amino group of dGTP base is only ~3.4Å. The interplanar distance between the dGTP base and the adjacent base pair (Guanine (template):Cytosine (primer)) at the primer terminus is ~3.4Å, characteristic of B-DNA, with ~304 Å2 of surface area buried at this interface. The intrahelical position of the base is further stabilized by hydrogen bonds in the minor groove (Fig. 3A). That is, the N2 amino group of dGTP makes a direct hydrogen bond with OE1 of Gln59 (2.8Å) on the fingers domain, and a single water molecule mediates hydrogen bonds between N2 (3.5Å) of dGTP, and OH of Tyr39 (2.7Å) and NE2 of Gln59 (3.1Å) of Polι (Fig. 3A).
Figure 3.
Hydrogen-bonding networks between dNTP base and polymerase in A) Polι Abasic.dGTP ternary complex, B) Pol ι Abasic.dTTP ternary complex, and C) Polι Abasic.dATP ternary complex. The relevant DNA and protein residues are colored according to element, the water molecules are shows as red spheres and the hydrogen bonds are displayed as dashed lines.
Figure 4.
Comparison of Dpo4Abasic.dCTP and Polι Abasic.dTTP ternary complexes. The protein surface is displayed in light blue (Dpo4) or cyan (Polι), DNA is shown in stick representation, and putative Mg2+ (Polι) or Ca2+ (Dpo4) ions are shown as dark blue spheres. The abasic site and the incoming dTTP (Polι) or dCTP (Dpo4) are colored in red. The nucleotide 5′ to the abasic site in the Dpo4 complex is colored in skyblue. The abasic site is extrahelical in the Dpo4 complex (Ling et al., 2004) but remains intrahelical in the Polι complex(es).
Figure 2.
Close-up view of the active site region in A) Polι Abasic.dGTP ternary complex, B) Polι Abasic.dTTP ternary complex, and C) Polι Abasic.dATP ternary complex. The coloring scheme is the same as in Figure 1. Highlighted and labeled are the catalytic residues (D34, D126 and E127) and some of the residues apposed close to the abasic lesion (Q59, K60, Y61, L62, S307 and R347) and incoming dNTP (Y39, V64, T65, K77, L78, Y68, R71 and K214). The water molecules that participate in forming water-mediated interactions are displayed as magenta spheres. The putative Mg2+ ions (A and B) are shown as dark blue spheres
To evaluate the contribution that hydrogen bonding by the N2 amino group of dGTP makes to its incorporation opposite the abasic lesion, we compared the catalytic efficiencies (kcat/Km) of dGTP vs. dITP incorporation using steady-state kinetic analyses (Johnson et al., 2000; Nair et al. 2006a). Since dITP lacks the N2 amino group, any reduction in the efficiency of dITP incorporation would reflect the contribution that hydrogen bonding by the N2 amino group makes to dGTP incorporation opposite the abasic lesion. Since we observe an ~ 15-fold reduction in the efficiency of dITP incorporation opposite the abasic lesion (Table 2), we infer that hydrogen bonding of the N2 amino group with the Gln59 and Tyr39 residues makes a positive contribution to dGTP incorporation opposite the abasic lesion.
TABLE 2.
Catalytic efficiency of dGTP vs. dITP incorporation opposite an abasic lesion by Polι
| Incoming nucleotide | kcat (min−1) | Km (μM) | kcat/Km | Efficiency relative to dGTP insertion |
|---|---|---|---|---|
| dGTP | 6.15±0.16 | 22.1±2.84 | 0.28 | - |
| dITP | 1.95±0.11 | 112±22.3 | 0.017 | 0.06 (16x↓) |
The Polι Abasic.dTTP ternary complex
In the Polι Abasic.dTTP complex, Polι adopts a nearly identical conformation to that observed in the Polι Abasic.dGTP complex (Fig. 1). The polymerases superimpose with an rmsd of only 0.2 Å (for 728 Cα atoms). The DNA configuration is also very similar, with an average rise of 3.2 Å and a helical twist of 30.9°/30.7° in the two complexes. Interactions with the dTTP triphosphate moiety are similar to those observed in the Polι Abasic.dGTP complex (Fig. 2), and the C1′-C1′ distance across the Abasic:dTTP pair is 9.1Å, as compared to 9.0Å in Abasic:dGTP pair. However, because of its smaller size, the dTTP base does not fill the space opposite the lesion to the same extent as dGTP. For example, the distance between the lesion sugar C2′ and the dTTP O2 atoms is 6.5Å, as compared to 3.4Å between the C2′ and N2 atoms in the Polι Abasic.dGTP complex. Accordingly, there are fewer stacking interactions with the adjacent base pair at the primer terminus, with the dTTP thymine stacking primarily over the cytosine on the primer strand (Fig. 2B). The amount of surface area buried at this interface is ~264Å2, as compared to ~304Å2 in the Polι Abasic.dGTP complex. Also, unlike the Polι Abasic.dGTP complex, there is no direct hydrogen bond between the dTTP base and the polymerase (Fig 3B). Most of the interactions in the minor groove derive from two water molecules, one of which occupies a position nearly identical to that in the Polι Abasic.dGTP complex and mediates hydrogen bonds between O2 of dTTP base (2.9Å), OH of Tyr39 (2.8Å) and NE2 of Gln59 (3.2Å). The second water molecule is located ~4Å from the first, coincides with the N2 amino group of dGTP in the Polι Abasic.dGTP complex, and mediates hydrogen bonds between N3 (2.8Å) and O2 (3.1Å) of dTTP with OE1 of Gln59 (3.2Å) (Fig. 3B).
Compared to the undamaged Polι A.dTTP complex, the configuration of Polι remains invariant, superimposing with an rmsd of 0.4Å (728 Cα atoms) (Fig. S1). The conformations of DNAs (the duplex portions) and incoming dTTPs are also essentially the same, superimposing with rmsds of 0.15Å (28 P atoms) and 0.2Å (all atoms), respectively. The most significant difference between the undamaged Polι A.dTTP and damaged Polι Abasic.dTTP complexes is a shift in the abasic sugar towards the PAD (away from the incoming nucleotide) by ~1Å, which eases the entry of the two water molecules that mediate a network of hydrogen bonds in the minor groove (Fig. 3B).
The Polι Abasic.dATP ternary complex
The structure is close to that described above for the Polι Abasic.dGTP ternary complex (Fig. 1). For example, the C1′-C1′ distance across the Abasic:dATP pair is 8.5Å, and the distance between the lesion C2′ and dATP C2 atoms is only 4.5Å. In the Polι Abasic.dGTP complex, the N2 amino group of dGTP base overlaps with the guanine 3′ to the lesion (on the template strand). Since the dATP base lacks an N2 amino group it stacks primarily over the primer cytosine (Fig. 2C). As such, the amount of surface area buried at this interface is less than in the Polι Abasic.dGTP complex; 274Å2 as compared to ~304Å2. Also, there are fewer interactions in the minor groove (Fig. 3C). For example, there is no direct or water mediated hydrogen bond between the dATP base and the polymerase. Rather, a single water molecule, at the same position as in Polι Abasic.dGTP complex, makes hydrogen bonds with OH of Tyr39 (2.8Å) and NE2 of Gln59 (3.2Å) (Fig. 3C).
Discussion
The non-instructional nature of abasic lesions makes them potentially cytotoxic and highly mutagenic. Almost all high fidelity DNA Pols prefer to incorporate an A opposite the lesion, but the mechanistic basis for this preference remains uncertain, with suggestions ranging from better stacking capabilities of dATP to better shape complementarities opposite the lesion (Chen et al., 2008; Matray and Kool, 1999; Reineks and Berdis, 2004; Zahn et al., 2007). Polι, a member of Y-family, is one of the few DNA Pols that does not obey the A-rule. Polι can insert all four nucleotides opposite an abasic lesion, but with a small preference for a G (Johnson et al., 2000).
We present here ternary complex structures of human Polι with three of the four possible nucleotides opposite an abasic lesion (Figs. 1 and S2). From the structures, the ability of Polι to insert all four nucleotides derives from the same structural feature that makes it conducive to Hoogsteen base pairing, namely a constricted active site that reduces the C1′-C1′ distance across the sugars from ~10.5Å in most DNA polymerases to <9Å in Polι (Nair et al., 2005a; Nair et al., 2006a; Nair et al., 2006b; Nair et al., 2004). The abasic lesion sugar is entrenched in a cavity lined by residues from the fingers domain, including the side chain of Leu62 and the aliphatic portions of Gln59 and Lys60. Lys60 also makes hydrogen bonds with the phosphate 3′ to the abasic lesion. Interestingly, Leu62 and Lys60 are unique to Polι, while Gln59 is present only in Polι and Polη. On the other side of the active site, the dNTP sugar is fixed by packing against the aromatic ring of Tyr39, as well as by a hydrogen bond between its 3′ OH and the main chain amide of the tyrosine. Consequently, the C1′-C1′ distance between the sugars shrinks to 8.5–9.1Å in complexes with dGTP, dTTP and dATP. Because of this shortened C1′-C1′ distance the dNTP bases almost fill the void opposite the abasic lesions, with the dGTP/dATP bases approaching within van der Waals distance (~3.4–4.5Å) of the lesion sugar. In all, this broad shape complementarity may underlie Polι’s ability to insert all four nucleotides opposite an abasic lesion.
Polι inserts nucleotides opposite an abasic lesion in the order of preference G > T > A > C. Overall, the differences in catalytic efficiencies for the incorporation of different nucleotides are relatively small, with G, T, and A incorporated ~ 3-, 2.2- and 1.5-times more efficiently than a C, respectively (Johnson et al., 2000). Despite these small differences, it is noteworthy that dGTP makes the most base stacking interactions, with ~304 Å2 of surface area buried with the adjacent base-pair, as compared to ~264Å2 for dTTP and 274Å2 for dATP. The dGTP base also makes a direct hydrogen bond with Polι, while the dATP base does not partake in direct or water mediated hydrogen bonds with the polymerase. In some respect, given that dGTP partakes in most base stacking interactions and makes a direct hydrogen bond with Polι it is surprising that it is not inserted with greater catalytic efficiency. The N2 amino group of dGTP could contribute both positively and negatively to selectivity, wherein the positive contribution would come from van der Waals contacts with the base 3′ the lesion and a direct hydrogen bond to the polymerase, while a negative contribution may derive from electrostatic repulsion between the partial negative charges on the N atom and the O2 (T/C) or N(G) atoms on the adjacent bases. To evaluate the contribution of the N2 amino group to dGTP incorporation opposite the abasic lesion, we compared the catalytic efficiencies (kcat/Km) of dGTP vs. dITP incorporation using steady-state kinetic analyses (Table II). We observe an ~ 15-fold reduction in the efficiency of dITP incorporation opposite the abasic lesion, from which we infer that the N2 amino group contributes (on balance) positively to dGTP incorporation opposite the abasic lesion.
In all three Polι structures the abasic lesion and the dNTPs are intrahelical. This is in stark contrast to the structure of Dpo4 with an abasic site (Ab-1), where the lesion is looped out towards the solvent in the gap between the fingers domain and the PAD, and the base 5′ to the lesion swings in to pair with the incoming nucleotide (Fig. 4) (Ling et al., 2004). In Polι, an abasic site is unable to loop out for two reasons. First, the abasic sugar is firmly entrenched in a hydrophobic cavity delineated by Gln59, Lys60 and Leu62 (described above), where Lys60 and Leu62 are unique to Polι. Second, an abasic lesion in an extrahelical position would sterically clash with Lys60 and Glu97 from the fingers domain and Ser307 from the PAD (Fig. 2). The gap between the fingers domain and the PAD is constricted over this region and partially blocked by Lys60 and Ser307, which extend across the space to make hydrogen bonds with the sugar-phosphate of the template strand (Figs. 2 & 4). In Dpo4, the extrahelical position of the abasic lesion gives rise to -1 frameshift mutations during DNA synthesis (Ling et al., 2004). In contrast, DNA synthesis across an abasic lesion by Polι will lend to substitution mutations that are less deleterious than the frameshift mutations generated by Dpo4 (Ling et al., 2004).
We show here that Polι inserts nucleotides opposite an abasic lesion without instruction from the template strand. The constricted active site cleft of Polι helps to stabilize an incoming nucleotide opposite the lesion without any significant change in conformation of the protein, the DNA or the nucleotide. Whereas replicative Pols insert an A opposite an abasic lesion, Polι has a small preference for a G, derived from a combination of base stacking and specific interactions with the polymerase. Interestingly, the replicative Pol from bacteriophage T4 obeys the A-rule, but it can incorporate a non-natural 5-nitro-1-indolyl-2′-deoxyriboside-5′-triphosphate (5-NITP) nucleotide with 1000-fold greater efficiency than dATP (Reineks and Berdis, 2004). From the structure of a related Pol from bacteriophage RB69 with 5-NITP (Zahn et al., 2007), stacking interactions with the base 3′ to the templating lesion appear to be the primary determinant of this preference. The most dramatic example of abasic lesion bypass is provided by Rev1, which incorporates a C opposite the lesion with much greater catalytic efficiency than A, G or T (Haracska et al., 2002; Nelson et al., 1996). The structure of Rev1 reveals a novel protein-template directed mechanism of DNA synthesis, wherein an arginine makes specific hydrogen bonds with the WC edge of incoming dCTP (Nair et al., 2005b). The pattern of hydrogen bonding is such that substitution by any other incoming nucleotide would lead to loss of hydrogen bonds, and unfavorable electrostatic and steric intrusion. Polι and Rev1 have thus evolved somewhat different strategies (and nucleotide preferences) to cope with a non-instructional template, but both polymerases participate directly in specifying incoming dGTP or dCTP, respectively.
EXPERIMENTAL PROCEDURES
Crystallization
The GST-Polι (residues 1–420) fusion protein was expressed and purified as described previously (Johnson et al., 2000). A single self-complementary oligonucleotide (18-nt) containing the abasic site (tetrahydrofuran analogue) at the fourth position from the 5′ end, and with a dideoxycytosine at the 3′ end (5′-TCTApGGGTCCTAGGACCCdd-3′) was synthesized, purified, desalted and lyophilized. Prior to crystallization, the oligonucleotide was annealed with itself to give a “double-ended” template-primer with two replicative ends. For crystallization, Polι and DNA were mixed in the ratio of 1:1.2, followed by the addition of incoming nucleotide (dGTP, dTTP or dATP) and MgCl2 to final concentrations of 20 mM and 10 mM respectively. All three ternary complexes crystallized under similar conditions, namely from solutions containing 10–15% PEG 5000 MME and 0.2 –0.4M (NH4)2SO4 in 0.1 M MES buffer (pH=6.5). The cocrystals belong to space group P6522 with cell dimensions of a = 98 Å, b = 98 Å, c = 203Å, α =β =90°, and γ=120°. For data collection, the cocrystals were cryoprotected by soaks for 5 minutes in mother liquor solutions containing 5%, 10% and 15% glycerol, respectively, and then flash frozen in liquid nitrogen.
Structure determination and refinement
X-ray data on cryocooled cocrystals were measured at the Advanced Photon Source (APS, beamline 17ID) (Table 1). The data were indexed and integrated using DENZO and reduced using SCALEPACK (Otwinowski and Minor, 1997). The structure was determined by molecular replacement (MR), using the earlier Polι A.dTTP complex as a search model (Nair et al., 2006b; Nair et al., 2004). The program AmoRe gave a unique MR solution (Navaza, 1994), which was then subjected to rigid-body refinement using CNS (Brunger et al., 1998). Electron density maps showed the clear presence of incoming nucleotide as well as clear density for the abasic lesion. Following further iterative rounds of refinement with CNS, model building with program O (Jones et al., 1991), and water picking, the Rfree lowered to around 28 % for all three complexes. The presence and conformation of the abasic site was confirmed with simulated annealing omit maps (Fig. 1). At this point, the refinement was continued with REFMAC (Murshudov et al., 1997), which allows for the anisotropic motion of rigid bodies, described as TLS parameters (Winn et al., 2003). Briefly, TLS refinement was performed with the Polι palm, fingers and thumb domains, and the PAD defined as four rigid units, and the DNA, incoming nucleotide, and a Mg2+ ion defined together as a fifth rigid unit. TLS refinement with REFMAC (using the same test set of reflections used to calculate Rfree in CNS) dropped the Rfree and Rcrys at the end of refinement to about 25% and 22%, respectively (Table 1). The final models for all three complexes include Polι residues 25–414 (there is no density for the first 24 residues), DNA nucleotides 4–11 (electron density for three of the four unpaired template nucleotides at the 5′ end was not visible), incoming nucleotide, two Mg2+ ions, and a number of water molecules (Table 1). The models have good stereochemistry, as shown by PROCHECK (Laskowski et al., 1993), with about 86% of the residues in the most favored regions of the Ramachandran plot and <1% in the disallowed regions.
Table 1.
Data collection, phasing and refinement statistics
| Data Collection | Polι Abasic.dGTP | Polι Abasic.dTTP | Polι Abasic.dATP |
|---|---|---|---|
| Resolution (Å) | 2.08 (2.15–2.08) | 2.09 (2.18–2.09) | 2.2 (2.28–2.2) |
| No. of Measured | 348022 | 348373 | 304275 |
| No. of Unique | 35030 (3450) | 34554 (3769) | 30070 (2952) |
| Data Coverage (%) | 99 (100) | 99 (100) | 99.5 (100) |
| Rmerge (%)a,b | 4.9 (24.4) | 5.5 (19.7) | 5.8 (42.8) |
| I/σ | 32.6 (9.9) | 33.3 (9.9) | 37.6 (37.6) |
| Redundancy | 9.9 (10.5) | 10.5 (9.5) | 10.1 (10.3) |
|
| |||
| Resolution Range | 20–2.08 | 20–2.09 | 20–2.2 |
| Reflections | 33090 | 33416 | 29220 |
| Rcryst (%)c | 22.0 | 22.0 | 21.6 |
| Rfree (%)d | 25.8 | 24.7 | 25.5 |
| Protein | 2877 | 2877 | 2877 |
| DNA | 312 | 312 | 322 |
| dNTP | 31 | 29 | 30 |
| Mg2+ ion | 2 | 2 | 2 |
| Water | 224 | 213 | 181 |
| B Factors (Å2) | |||
| Protein | 48.9 | 45.6 | 48.4 |
| DNA | 47.4 | 44.0 | 45.2 |
| dNTP | 34.2 | 36.0 | 43.5 |
| Mg2+ ion B | 14.9 | 20.6 | 32.1 |
| Mg2+ ion A | 74.9 | 70.4 | 64.5 |
| Water | 40.6 | 38.8 | 40.7 |
| R.m.s deviations | |||
| Bond lengths (Å) | .009 | .006 | .008 |
| Bond angles (°) | 1.69 | 1.43 | 1.73 |
Values for outermost shells are given in parentheses
Rmerge =Σ|I-<I>|/ΣI, where I is the integrated intensity of a given reflection.
Rcryst = Σ||Fobserved| – |Fcalculated||/Σ|Fobserved|
For Rfree calculations 8% of data excluded from refinement was used.
Acknowledgments
We thank the staff at the Advanced Photon Source (beamline 17-ID) for facilitating X-ray data collection. This work was supported by grant CA115856 from the U. S. National Institutes of Health (S.P. and A. K. A.). We thank Anshu Bhatnagar for kinetic studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc Natl Acad Sci U S A. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S, Laval J. Coding properties of poly(deoxycytidylic acid) templates containing uracil or apyrimidinic sites: in vitro modulation of mutagenesis by deoxyribonucleic acid repair enzymes. Biochemistry. 1982;21:6746–6751. doi: 10.1021/bi00269a020. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve R, Jiang W, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A software suite for macromolecular structure determination. Acta Cryst . 1998;D54:905. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chen J, Dupradeau FY, Case DA, Turner CJ, Stubbe J. DNA oligonucleotides with A, T, G or C opposite an abasic site: structure and dynamics. Nucleic Acids Res. 2008;36:253–262. doi: 10.1093/nar/gkm622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuniasse P, Fazakerley GV, Guschlbauer W, Kaplan BE, Sowers LC. The abasic site as a challenge to DNA polymerase. A nuclear magnetic resonance study of G, C and T opposite a model abasic site. J Mol Biol. 1990;213:303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- Cuniasse P, Sowers LC, Eritja R, Kaplan B, Goodman MF, Cognet JA, LeBret M, Guschlbauer W, Fazakerley GV. An abasic site in DNA. Solution conformation determined by proton NMR and molecular mechanics calculations. Nucleic Acids Res. 1987;15:8003–8022. doi: 10.1093/nar/15.19.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala KA, Hypes CD, Suo Z. Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. J Biol Chem. 2007;282:8188–8198. doi: 10.1074/jbc.M610718200. [DOI] [PubMed] [Google Scholar]
- Gelfand CA, Plum GE, Grollman AP, Johnson F, Breslauer KJ. Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry. 1998;37:7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc Natl Acad Sci U S A. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem. 2002;277:15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- Hogg M, Wallace SS, Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. Embo J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- Jones AT, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;A47:110–119. [Google Scholar]
- Lawrence CW, Borden A, Banerjee SK, LeClerc JE. Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res. 1990;18:2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Matray TJ, Kool ET. A specific partner for abasic damage in DNA. Nature. 1999;399:704–708. doi: 10.1038/21453. [DOI] [PubMed] [Google Scholar]
- Mozzherin DJ, Shibutani S, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase delta. Proc Natl Acad Sci U S A. 1997;94:6126–6131. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA Polymerase iota Incorporates dCTP Opposite Template G via a G.C+ Hoogsteen Base Pair. Structure (Camb) 2005a;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005b;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1, N6-ethenodeoxyadenosine lesion by human DNA polymerase iota. Nat Struct Mol Biol. 2006a;13:619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site. Structure. 2006b;14:749–755. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- Navaza J. AMoRe: an automated package for molecular replacement. Acta Cryst. 1994;50:157–163. [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pages V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc Natl Acad Sci U S A. 2008;105:1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic Translesion Synthesis DNA Polymerases: Specificity of Structure and Function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Randall SK, Eritja R, Kaplan BE, Petruska J, Goodman MF. Nucleotide insertion kinetics opposite abasic lesions in DNA. J Biol Chem. 1987;262:6864–6870. [PubMed] [Google Scholar]
- Reineks EZ, Berdis AJ. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- Sagher D, Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the “A rule”. J Biol Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- Strauss BS. The “A” rule revisited: polymerases as determinants of mutational specificity. DNA Repair (Amst) 2002;1:125–135. doi: 10.1016/s1568-7864(01)00014-3. [DOI] [PubMed] [Google Scholar]
- Tissier A, McDonald JP, Frank EG, Woodgate R. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- Wallace SS, editor. Oxidative damage to DNA and its repair. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Washington MT, Johnson RE, Prakash L, Prakash S. Human DNA polymerase iota utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol Cell Biol. 2004;24:936–943. doi: 10.1128/MCB.24.2.936-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- Zahn KE, Belrhali H, Wallace SS, Doublie S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Wang Z. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase iota. Mol Cell Biol. 2000;20:7099–7108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]