Abstract
Proliferating cell nuclear antigen (PCNA) is a homotrimeric protein that functions as a sliding clamp during DNA replication. Several mutant forms of PCNA that block translesion DNA synthesis have been identified in genetic studies in yeast. One such mutant protein (encoded by the rev6-1 allele) is a glycine to serine substitution at residue 178, located at the subunit interface of PCNA. To better understand how this substitution interferes with translesion synthesis, we have determined the X-ray crystal structure of the G178S PCNA mutant protein. This substitution has little effect on the structure of the domain in which the substitution occurs. Instead, significant, local structural changes are observed in the adjacent subunit. The most notable difference between mutant and wild-type structures is in a single, extended loop (comprising amino acid residues 105-110), which we call loop J. In the mutant protein structure, loop J adopts a very different conformation in which the atoms of the protein backbone have moved by as much as 6.5 Å from their positions in the wild-type structure. To better understand the functional consequences of this structural change, we have examined the ability of this mutant protein to stimulate nucleotide incorporation by DNA polymerase eta (pol η). Steady state kinetic studies show that while wild-type PCNA stimulates incorporation by pol η opposite an abasic site, the mutant PCNA protein actually inhibits incorporation opposite this DNA lesion. These results show that the position of loop J in PCNA plays an essential role in facilitating translesion synthesis.
DNA damage in the template strand blocks replication by classical DNA polymerases, which are involved in normal DNA replication and repair. In order to overcome these replication blocks, cells employ several non-classical DNA polymerases that are capable of replicating through template DNA lesions in a process called translesion DNA synthesis (1-3). One such enzyme is eukaryotic DNA polymerase eta (pol η), which is a 71-kDa monomeric protein encoded by the RAD30 gene in yeast (4). Pol η functions in the replication of a few types of DNA lesions, including thymine dimers (4-6) and 8-oxoguanines (7,8). Deletion of the RAD30 gene in yeast leads to an increase in ultraviolet (UV) radiation-induced mutagenesis (9-11), and in humans, inactivation of pol η is responsible for the variant form of xeroderma pigmentosum (XPV) (12,13), which results in greater cancer susceptibility. Another non-classical DNA polymerase in eukaryotes is DNA polymerase zeta (pol ζ), which is comprised of a 173-kDa catalytic subunit and a 29-kDa accessory subunit encoded in yeast by the REV3 and REV7 genes, respectively (14,15). Pol ζ functions in the error-prone replication of a wide range of DNA lesions, and disruptions of the REV3 and REV7 genes result in a drastic decrease in the frequency of DNA damage-induced mutations in yeast (16,17). Moreover, expression of anti-sense RNA to pol ζ leads to a reduction in the frequency of UV radiation-induced mutations in human cells (18).
A key factor in translesion synthesis is proliferation cell nuclear antigen (PCNA). PCNA, encoded in yeast by the POL30 gene, is a ring-shaped, homotrimeric protein that acts as a sliding clamp for classical DNA polymerases (19,20). Many protein factors involved in DNA replication and repair interact with PCNA via their PCNA interacting peptide (PIP) motifs that bind along the inter-domain connector loop of PCNA (21). Pol η binds to PCNA in this manner, and this interaction is necessary for pol η function in vivo (22,23). Moreover, this interaction stimulates the enzymatic activity of pol η in vitro (22). Pol ζ, although lacking a PIP motif, also interacts with PCNA, and its enzymatic activity is stimulated by PCNA (24).
Several PCNA mutant proteins in yeast have been identified that interfere with translesion synthesis in vivo (25-27). One of these is encoded by the pol30-178 allele (formerly called the rev6-1 allele); it encodes a mutant form of PCNA in which Gly-178 is substituted with a serine (25). This amino acid substitution is at the subunit interface of PCNA, and genetic studies have shown that translesion synthesis by both pol η and pol ζ is completely blocked in cells expressing only this mutant form of PCNA (25). All other aspects of DNA replication and repair appear to occur normally in cells expressing this PCNA mutant protein (25). Another PCNA mutant protein that blocks translesion synthesis, but supports normal cell growth is encoded by the pol30-113 allele (26). In this mutant protein, Glu-113 is substituted with a glycine. Interestingly, Glu-113 is also located at the subunit interface of PCNA directly opposite from Gly-178 on the neighboring subunit. Based on the fact that these mutant proteins block translesion synthesis without interfering with normal DNA replication, the structural changes resulting from these amino acid substitutions are likely subtle.
To understand the structural and mechanistic basis of the defect in the PCNA G178S mutant protein's ability to support translesion synthesis, we determined the X-ray crystal structure of this mutant protein to a resolution of 2.9 Å. We find that the substituted serine side chain forms a new hydrogen bond with the backbone carbonyl of Glu-113 on the adjacent subunit. This contact results in an extended loop on the adjacent subunit (comprising amino acid residues 105-110, which we refer to as loop J) changing its conformation and moving it into an aberrant position that deviates as much as 6.5 Å from its position in the wild-type structure. We have examined the biochemical relevance of this structure by carrying out steady state kinetic studies with a model non-classical polymerase, pol η, in the presence of the wild-type and mutant PCNA proteins. We find that while wild-type PCNA stimulates incorporation by pol η opposite an abasic site, the mutant PCNA protein inhibits incorporation opposite this DNA lesion. These findings suggest that the proper conformation of loop J is essential for translesion DNA synthesis.
EXPERIMENTAL PROCEDURES
Protein expression and purification
Wild-type and mutant PCNA proteins from Saccharomyces cerevisiae were over-expressed in E. coli Rosetta-2 (DE3) cells harboring pET-11a vectors into which wild-type or mutant PCNA genes were cloned. PCNA proteins were N-terminally FLAG tagged for rapid purification. Transformed cells were grown at 37°C in Overnight Express Instant TB Medium (Novagen) for 12 hrs. Cells were lysed in 50mM TrisCl, pH 7.5, 150mM NaCl, 1mM PMSF, 1mg/ml lysozyme, with a Complete Mini Protease Inhibitor Cocktail tablet (Roche). Following ultracentrifugation, the crude extract was loaded onto a 15 ml resin bed of Anti-FLAG M2 Affinity Gel (Sigma) and purified as instructed. The eluted PCNA protein was then run on a Superdex G-75 column equilibrated with 20mM TrisCl, pH 7.5, 1mM DTT, and 250mM NaCl. Yeast pol η and RFC were over-expressed and purified as previously described (28,29). All purified proteins were stored in aliquots at −80 °C.
Crystallization of G178S PCNA
Crystallization of the G178S PCNA mutant protein was performed manually using the hanging drop method with 4 μl drops. To identify ideal crystallization conditions, an initial screen utilized conditions similar to those which produce wild type PCNA crystals (19). Optimally diffracting crystals were generated by combining an equal volume of protein (30 mg/ml) with a reservoir solution containing 2.06 M ammonium sulfate and 0.1 M sodium citrate, pH 5.8. Cubic crystals formed within 16 h at 18°C.
Data collection and structural determination
G178S PCNA protein crystals were presoaked in a mother liquor containing 10% (v/v) glycerol prior to being flash frozen at 100 K. Mounted crystals were subsequently used for data collection at 100 K at the 4.2.2 synchrotron beamline at the Advanced Light Source in the Ernest Orlando Lawrence Berkeley National Laboratory. The data were collected with a crystal to detector distance of 150 mm. The data were analyzed and scaled using d*trek (30) to a resolution of 2.9 Å and the space group was determined to be P213, which is the same space group into which the wild-type PCNA protein crystallizes (19).
We carried out molecular replacement using the known structure of wild type PCNA (1PLQ) and the PHASER program (31) to produce a model with the P213 space group. To remove any structural bias, simulated annealing was performed using the PHENIX program (32) prior to any refinement. Further structural refinement was executed using PHENIX and REFMAC5 from the CCP4 package (31). Model building was carried out using Coot and O (34). The coordinates have been deposited in the PDB with ID number 3F1W.
DNA substrates
For DNA polymerase activity assays, a synthetic 68-mer oligodeoxynucleotide with the sequence 5′-Biotin-GAC GGC ATT GGA TCG ACC TCX AGT TGG TTG GAC GGG TGC GAG GCT GGC TAC CTG CGA TGA GGA CTA GC-Biotin, was used as the template strand where X is an abasic site or a non-damaged guanine. For the running start abasic bypass assays, a synthetic 31-mer oligodeoxynucleotide with the sequence 5′- TCG CAG GTA GCC AGC CTC GCA CCC GTC CAA C was used as a primer. For the steady state kinetic studies, a synthetic 31-mer oligodeoxynucleotide with the sequence 5′-GGT AGC CAG CCT CGC ACC CGT CCA ACC AAC T, was used as a primer. Primer strands were 5′-32P-end-labeled using T4 polynucleotide kinase (New England Biolabs) and (32P-γ)ATP (PerkinElmer). Template strands and labeled primer strands (1 μM each) were annealed in 25 mM Tris–Cl, pH 7.5, 100 mM NaCl by heating to 90°C for 2 min and slow cooling to 22°C over several hours. Labeled and annealed DNA substrates were stored at 4°C for up to 2 weeks.
Polymerase activity assays
All experiments were carried out in 40 mM TrisCl pH 7.5, 8mM MgCl2, 150mM NaCl, 1mM DTT, and 100 μg/ml bovine serum albumin. Reactions also contained a 10-fold molar excess of streptavidin over DNA to block the ends of the DNA to prevent PCNA dissociation. Wild-type or mutant PCNA proteins were loaded onto the DNA substrates by incubating 90 nM PCNA (trimer concentration), 20 nM DNA, 25 nM RFC, and 500 μM ATP for 5 minutes at 22°C. Reactions also contained various concentrations of dGTP (0 to 100 μM for the abasic site template) or dCTP (0 to 5 μM for the non-damaged template). Reactions were initiated by adding 1 nM pol η and were quenched after 10 min by the addition of 10 volumes of formamide loading buffer (80% deionized formamide; 10 mM EDTA, pH 8.0; 1 mg/ml xylene cyanol; 1 mg/ml bromophenol blue). Extended primers (the product) and unextended primers (the substrate) were separated on a 15% polyacrylamide sequencing gel containing 8 M urea. The intensities of the labeled gel bands were determined using the InstantImager (Packard). The rate of product formation was graphed as a function of dNTP concentration, and the Vmax and Km values were obtained from the best fit of the data to the Michaelis–Menten equation using SigmaPlot 10.0.
The running start abasic site bypass assay was performed under identical conditions as the steady state kinetic studies except that 20 μM of each dNTP was used. Reactions were quenched after 5, 10, and 15 min by the addition of 10 volumes of formamide loading buffer, and reaction products were visualized on a 15% polyacrylamide sequencing gel containing 8 M urea.
RESULTS
Several mutant forms of yeast PCNA that are unable to support translesion DNA synthesis in vivo have been identified in genetic screens (25-27). One of these is the PCNA G178S mutant protein. Yeast strains expressing this mutant form of PCNA have the same phenotype as strains lacking both pol η and pol ζ (25). To better understand how this mutant form of PCNA blocks translesion synthesis, we have determined the X-ray crystal structure of this mutant protein and have carried out functional studies of pol η in the presence of this mutant protein.
Overview of the PCNA G178S protein structure
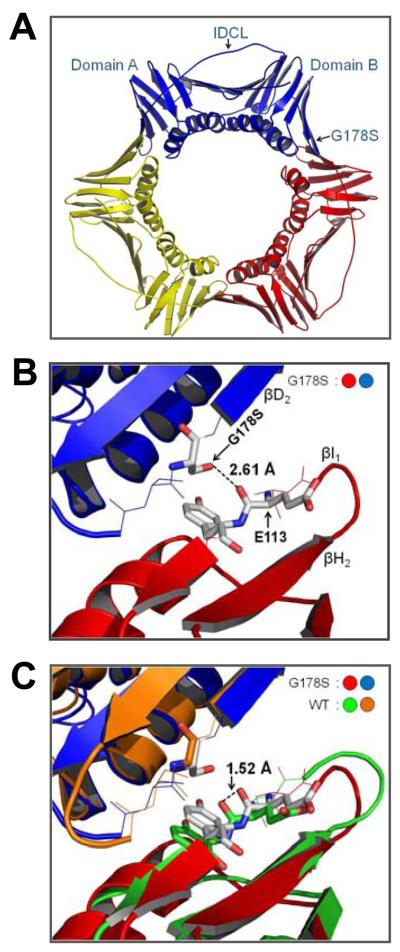
We have determined the X-ray crystal structure of the PCNA G178S mutant protein to a resolution of 2.9 Å (Table 1). Fig. 1A shows an overview of the structure of the PCNA G178S mutant protein trimer. Each subunit is comprised of two domains arranged in the trimeric ring in a head-to-tail fashion with domain A (residues 1-118) of one subunit interacting with domain B (residues 135-240) of its neighbor. Within each subunit, domains A and B are linked by the inter-domain connector loop (residues 119-134). No large-scale differences in the structure of the PCNA G178S mutant protein and the wild type PCNA protein are detectible.
Table 1.
Data collection and refinement statistics.
| (A) Data collection statistics | ||
| Resolution (Å) | 29.8– 2.9 | |
| Wavelength (Å) | 1.072 | |
| Space Group | P213 | |
| Cell (Å) | a = 123.13 b = 123.13; c = 123.13 | |
| Completeness (%) (last shell) | 100 (3.0-2.9:96.8) | |
| Redundancy | 10.79 (11.01) | |
| I/σI | 14.8 (2.50) | |
| Rmerge (%)b | 7.3 (60.5) | |
| (B) Refinement statistics | ||
| Resolution range (Å) | 19.9-2.9 | |
| R (%)c | 22.6 | |
| Rfree (%)d | 25.4 | |
| rms bonds (Å) | 0.009 | |
| rms angles (°) | 1.21 | |
| Number of water molecules | 0 | |
| Number of protein atoms | 1981 (254 AA residues) | |
| Ramachandran analysis (%) | ||
| Most favored | 89.4 | |
| Allowed | 10.6 | |
| PDB ID | 3F1W | |
Values in parentheses relate to the highest resolution shell.
Rmerge = Σ|I| - 〈I〉/ΣI, where I is the observed intensity, and 〈I〉 is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
R = Σ∥Fo| - |Fc∥/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
Rfree is defined by Brunger (38).
Fig. 1.
Structure of the PCNA G178S Mutant Protein. (A) The trimeric form of the protein is shown with monomeric subunits in red, yellow, and blue. The inter-domain connecting loop (IDCL), domains A and B, and the G178S substitution are indicated on one of the subunits. (B) Close up, side view of the subunit interface with the Ser-178 substitution of the blue monomer and Tyr-114 and Glu-113 of the red monomer shown in stick format. The distance between the hydroxyl of Ser-178 and the backbone carbonyl of Glu-113 is indicated. (C) The superimposition of the structures of the G178S PCNA mutant protein and wild type PCNA (1PLQ). The distance between backbone carbonyl of Glu-113 in the wild type and mutant PCNA structures is indicated.
Fig. 1B shows a closer view of the subunit interface of the PCNA mutant protein. The G178S substitution is located in β strand D2 of domain B of the upper subunit. The side chain hydroxyl group of the substituted serine forms a new hydrogen bond with the backbone carbonyl of Glu-113 on the adjacent subunit. The two oxygen atoms are 2.6 Å apart, which is typical for a hydroxyl group-carbonyl group hydrogen bond. Glu-113 is in β strand I1, which forms an anti-parallel β sheet with strand H1. In the mutant protein structure, the new hydrogen bond between Ser-178 and Glu-113 alters the trajectory of this anti-parallel β sheet. This is most clearly seen by superimposition of the structures of the wild-type and mutant PCNA proteins (Figs. 1C and 2A).
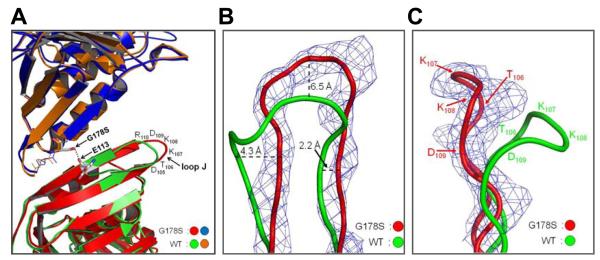
Fig. 2.
Conformation of loop J in the wild type and mutant structure. (A) Superimposition of the wild type and G178S PCNA mutant protein structures is shown with the Ser-178 substitution and Glu-113 represented in stick format and the hydrogen bond between them shown as black dots. The amino acid residues of loop J are indicated. (B) Close up view of loop J showing the electron density (level=2.0) for the G178S PCNA mutant protein and the backbone of the wild type and mutant proteins in ribbon representation. The distances between the wild type and mutant protein backbone are specified. (C) Side view of loop J with the position of the amino acid residues indicated.
Loop J in the wild-type and mutant protein structures
Altering the trajectory of the H1 and I1 anti-parallel β sheet results in structural changes in the extended loop between strands H1 and I1 (residues 105-110), which we refer to as loop J (Fig. 2A). In the mutant protein structure, this loop adopts a very different conformation in which the atoms of the protein backbone have moved significantly from their positions in the wild-type structure. For example, the α carbon of Lys-107 has moved 6.5 Å from its position in the wild-type protein structure. Fig. 2B,C shows the protein backbone and electron density of loop J in its aberrant conformation in the mutant protein structure overlaid with the protein backbone of loop J in its usual conformation in the wild-type protein structure. Based on the clear electron density of loop J in both the wild-type and mutant protein structures, it is important to note that loop J is not merely becoming more flexible and disordered in the mutant protein structure. Instead this loop has shifted from one stable configuration in the wild-type structure to another stable configuration in the mutant protein structure.
It should also be pointed out that the shift in loop J in this structure is not the result of crystal packing. The space group and unit cell dimensions of our crystals are the same as those of reported for the wild-type protein (19), so observed structural differences cannot be attributed to crystal packing. Moreover, in the crystal lattice, loop J does not contact any proteins from neighboring asymmetric units; rather, it sticks out into solvent-filled spaces of the crystal lattice (see Supplemental Fig. S1).
Comparison of the two PCNA domains
To gain insight into global structural changes occurring to PCNA as a result of the G178S substitution beyond the effected loop J, we superimposed the protein backbone of the structures of the wild-type and mutant subunits (Fig. 3). Domain B which contains the G178S substitution is structurally identical to the wild-type form of PCNA. By contrast, domain A is affected by the G178S mutation on the adjacent monomer. To quantify the degree of structural differences between the wild-type and mutant forms of PCNA, we calculated RMSD values between the backbone of the wild type and mutant forms of PCNA for each domain independently. The RMSD value for the domain which contains the G178S mutation (domain B) is 0.50 Å. The RMSD value for the domain without the mutation (domain A) is three times as large at 1.3 Å. Thus, these results show that the G178S mutation is affecting the structure of PCNA by acting in trans to alter the structure of the neighboring monomer's domain A.
Fig. 3.
Superimposition of the PCNA monomer backbone of wild type and mutant PCNA proteins. The monomeric subunit is lying on its side with the inter-domain connector loop in the back to allow the separate domains to be easily viewed. The adjacent mutant monomeric subunit is shown in blue with the G178S substitution indicated. The G178S substitution, the site of mono-ubiquitination (Lys-164), and loop J are indicated. Domains A and B of the monomeric subunit are separated by a dashed line and the RMSD values were independently determined for each domain.
In summary, the only notable structural difference between the wild-type and mutant forms of PCNA is in domain A with the largest change being in the position of loop J. Thus it is highly likely that the aberrant conformation of loop J is responsible for the effect of this mutation on translesion DNA synthesis.
Impact of the mutant PCNA protein on the activity of pol η
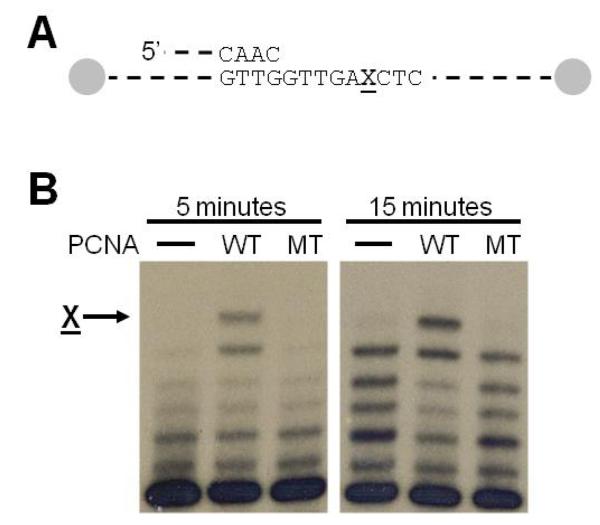
Genetic studies have shown that this mutant protein blocks translesion DNA synthesis (25). To better understand how the structural changes described here interfere with translesion DNA synthesis, we have directly measured the enzymatic activity of the non-classical DNA polymerase pol η in the presence and absence of wild-type and mutant PCNA protein. Wild-type PCNA enhances the ability of pol η to incorporate nucleotides opposite an abasic site (22). Fig. 4 shows the incorporation of pol η opposite an abasic site in a running start assay in the absence of PCNA and in the presence of either the wild-type or mutant form of PCNA. The presence of wild-type PCNA stimulates the ability of pol η to incorporate a nucleotide opposite the abasic site compared to pol η alone. By contrast, the mutant protein appears to have no ability to stimulate incorporation opposite the abasic site.
Fig. 4.
Running start experiment with pol η on an abasic site. (A) Schematic diagram of the 31/68-mer substrate used in the running start assays with the ends of the template strand containing biotinstreptavidin blocks. The X indicates the location of the abasic site (B) Autoradiograph of the synthesis products after five or fifteen minutes following the addition of pol η. The arrow indicates incorporation opposite the abasic site. Lanes labeled WT contain the wild-type PCNA protein, and lanes labeled MT contain the G178S mutant PCNA protein.
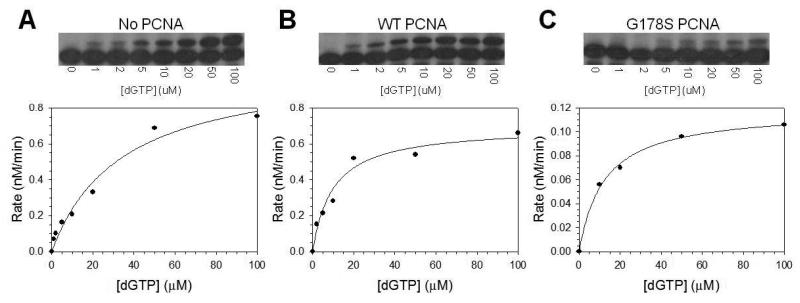
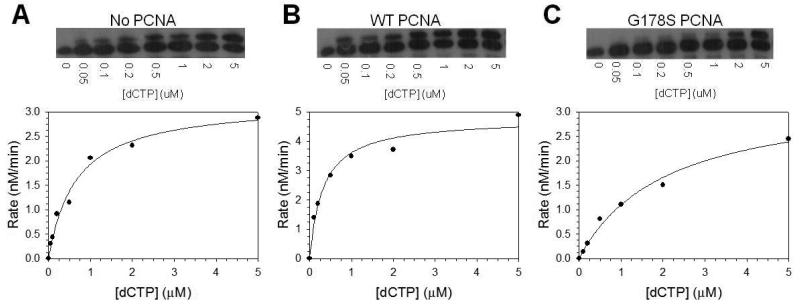
To quantify the effects of wild-type and mutant PCNA proteins on the enzymatic activity of pol η, we measured the kinetics of dGTP (the preferred incoming dNTP) incorporation opposite the abasic site under steady state conditions. The rate of incorporation was plotted as a function of dGTP concentration (Fig. 5), the Vmax and Km steady state kinetic parameters were obtained from the best fit of the data to the Michaelis-Menten equation, and these parameters are provided in Table 2. The catalytic efficiency (Vmax/Km) of incorporation opposite the abasic site by pol η was reproducibly 2 to 4-fold greater in the presence of wild-type PCNA than in the absence of PCNA, and this was due mainly to a decrease in the Km for nucleotide incorporation. By contrast, the catalytic efficiency was not greater in the presence of the G178S PCNA mutant protein than in its absence. In fact, the catalytic efficiency was reproducibly 2 to 4-fold lower in the presence of the mutant PCNA than in its absence. It should be noted that like wild-type PCNA, the mutant PCNA protein also decreased the Km for nucleotide incorporation in this context. The Vmax for incorporation in the presence of the mutant form of PCNA, however, was ∼10-fold lower than incorporation in its absence. Thus the mutant form of PCNA actually inhibits the ability of pol η to incorporate nucleotides opposite abasic sites.
Fig. 5.
Steady state kinetics of pol η on an abasic site. The rate of nucleotide incorporation was graphed as a function of dGTP concentration for (A) pol η alone, (B) pol η with wild type PCNA protein, (C) and pol η with the G178S PCNA mutant protein. Autoradiographs of the gels showing the incorporation of a single dGTP across from an abasic site at the indicated concentrations of nucleotide are shown above each graph. The solid lines represent the best fits of the data to the Michaelis–Menten equation, and the Vmax and Km steady state parameters are given in Table 2.
Table 2.
Steady state kinetic parameters of pol η–catalyzed nucleotide incorporation.
| Proteins | DNA |
Vmax (nM/min) |
Km (μM) |
Vmax/Km |
Relative efficiency |
|---|---|---|---|---|---|
| Pol η alone | Abasic site | 1.0 ± 0.1 | 34 ± 8 | 0.029 | 1.0 |
| Pol η + wild-type PCNA | Abasic site | 0.70 ± 0.06 | 9.9 ± 3.0 | 0.071 | 2.4 |
| Pol η + mutant PCNA | Abasic site | 0.12 ± 0.01 | 12 ± 1 | 0.010 | 0.34 |
| Pol η alone | Non-damaged | 3.2 ± 0.2 | 0.67 ± 0.12 | 4.8 | 1.0 |
| Pol η + wild-type PCNA | Non-damaged | 4.8 ± 0.3 | 0.32 ± 0.07 | 15 | 3.1 |
| Pol η + mutant PCNA | Non-damaged | 3.4 ± 0.3 | 2.1 ± 0.4 | 1.6 | 0.33 |
The inhibitory effect observed with the mutant PCNA protein requires the mutant protein to be loaded onto the primer-template DNA. We showed this by omitting either replication factor C (RFC, the ATP-dependent PCNA-loading protein) or ATP from the reaction so that the mutant PCNA protein would not be loaded onto the DNA. In these experiments, the efficiencies of nucleotide incorporation by pol η were exactly the same as those measured when no PCNA was present. For example, when we omitted ATP, the catalytic efficiency of incorporation opposite the abasic site in the presence of the unloaded mutant PCNA protein was 0.028, which is identical to the catalytic efficiency of 0.029 determined in the absence of PCNA (Table 2). Thus we observed no inhibition of pol η-catalyzed nucleotide incorporation opposite abasic sites when the mutant form of PCNA was not loaded on the DNA substrate.
To determine whether the inhibition by this mutant PCNA protein is specific to abasic sites, we used steady state kinetics to examine if the mutant form of PCNA could inhibit the incorporation of nucleotides opposite non-damaged DNA. Fig. 6 shows the rate of dCTP incorporation opposite a non-damaged G by pol η as a function of nucleotide concentration. Here again the catalytic efficiency of incorporation opposite the non-damaged template was reproducibly 2 to 4-fold greater in the presence of wild-type PCNA than that in its absence (Table 2). In this case, the increased catalytic efficiency in the presence of wild-type PCNA was due to both an increase in the Vmax and a decrease in the Km for nucleotide incorporation in the presence of wild-type PCNA. The mutant PCNA protein inhibited the ability of pol η to incorporate opposite the non-damaged DNA to a 2 to 4-fold lower level of activity than pol η alone, and this was mainly due to an increase in the Km for nucleotide incorporation. Thus this mutant protein inhibits nucleotide incorporation by pol η opposite both damaged and non-damaged templates.
Fig. 6.
Steady state kinetics of pol η on non-damaged DNA. The rate of nucleotide incorporation was graphed as a function of dCTP concentration for (A) pol η alone, (B) pol η with wild type PCNA protein, (C) and pol η with the G178S PCNA mutant protein. Autoradiographs of the gel showing the single incorporation of dCTP at the indicated concentrations of nucleotide are shown above each graph. The solid lines represent the best fits of the data to the Michaelis–Menten equation, and the Vmax and Km steady-state parameters are given in Table 2.
DISCUSSION
Interactions between PCNA and non-classical polymerases have been shown to be essential for translesion synthesis. Pol η possesses a PCNA-interacting peptide (PIP) motif at its extreme C-terminus (residues 621-628). While a pol η mutant protein truncated at position 624 shows activity in vitro, yeast expressing this truncated version of pol η have the same defect in translesion synthesis as yeast completely lacking pol η (22). Thus the interaction between PCNA and pol η is essential to pol η's function in vivo. This may be because the interaction between PCNA and pol η leads to an increase in the catalytic efficiency (Vmax/Km) of nucleotide incorporation by pol η opposite damaged and non-damaged templates in vitro (22). Similarly, PCNA increases the DNA synthesis activity of pol ζ, which lacks a PIP motif, on both non-damaged DNA and UV-treated DNA (24). Incidentally, this also shows that there are functionally important interactions between non-classical polymerases and PCNA that are not mediated by PIP motifs.
Translesion synthesis by both pol η and pol ζ is severely defective in yeast expressing the G178S mutant form of PCNA. This was clearly demonstrated by experiments in which plasmids containing specific DNA lesions were transformed into various yeast strains such that the transformation efficiency indicated the efficiency of DNA damage bypass (25). The rev6-1 strain (expressing the G178S mutant form of PCNA) bypasses both abasic sites and cis-syn thymine dimers at ∼1 % efficiency compared to wild-type (25). The rev3Δ strain (lacking pol ζ) bypasses thymine dimers at ∼94 % efficiency compared to wild-type; abasic sites are bypassed at ∼5 % efficiency (25). The rad30Δ strain (lacking pol η) bypasses abasic sites at ∼80 % efficiency compared to wild-type; cis-syn thymine dimers are bypassed at ∼15 % efficiency (25). Only in the rad30Δrev3Δ strain are the efficiencies of bypassing either of these lesions as low as they are in the rev6-1 strain (25).
To better understand why the G178S substitution in PCNA leads to a loss of function of both pol η and pol ζ, but no other discernable effects on DNA replication and cell growth, we determined the X-ray crystal structure of the mutant form of PCNA to a resolution of 2.9 Å. We found that while the global structures of the wild-type and mutant protein are the same, there is a significant difference in the structure of a small region of PCNA near the subunit interface. The substituted Ser-178 side chain forms a new hydrogen bond with the backbone carbonyl of Glu-113 on the neighboring subunit. This new hydrogen bonds alters the trajectory the β sheet comprised of strands H1 and I1 (containing Glu-113). This has the effect of moving loop J as much as 6.5 Å in the structure of the mutant protein from its location in the structure of the wild-type protein (Fig. 2).
Since the alteration in loop J is the only significant difference between the wild-type and mutant protein structures, we conclude that the proper positioning of loop J is critical for supporting translesion synthesis by both pol η and pol ζ. Further support for this notion comes from our structural studies of the E113G mutant form of PCNA, which leads to a similar defect in translesion synthesis as does the G178S mutant PCNA protein (26). Glu-113 is at the subunit interface directly across from Gly-178, and the side chain hydroxyl group on the G178S substitution forms a hydrogen bond with the backbone carbonyl of this residue. We have obtained crystals of the E113G mutant form of PCNA and collected X-ray diffraction data to a resolution of 3.6 Å. Comparisons of the electron densities at this resolution of the wild-type, G178S mutant, and E113G mutant proteins show that loop J is also in an aberrant configuration in the structure of the E113G mutant protein as it is in the structure of the G178S protein (see Supplemental Fig. S2). Taken together, these data favor the hypothesis that loop J of PCNA makes an important direct contact with non-classical polymerases and that when loop J adopts an aberrant conformation in the case of these two mutant proteins, this contact is disrupted.
Additional evidence for the importance of loop J comes from previous structure-function studies of PCNA. These studies have shown that several substitutions in and around loop J (residues 105-110) have been found to lead to increased sensitivity to UV radiation and the DNA damaging agent methyl methanesulfonate (MMS) (27). Cells expressing the E104A,D105A double mutant have a severe defect in the ability to grow following UV or MMS treatment (27). Cells expressing the K108A,D109A double mutant have a minor defect in the ability to survive MMS treatment (27). Cells expressing the D109A,R110A double mutant have a minor defect in the ability to survive UV and MMS treatment (27). These studies provide additional compelling evidence that loop J of PCNA is critical for translesion DNA synthesis.
As described above, previous studies have shown that the interaction between PCNA and pol η that is mediated by the PIP motif of the polymerase is essential for translesion synthesis (22). It is important to note that the binding site for the PIP motif is located on the opposite face of the PCNA ring from loop J. The PIP motif binds to a hydrophobic pocket in domain B near the inter-domain connector loop (35). Incidentally, we observe no significant structural differences between the wild-type and G178S mutant PCNA proteins in the regions of domain B or the inter-domain connector loop that bind the PIP motif. Loop J is located at the subunit interface and extends out from the opposite face of the PCNA ring. These observations imply that pol η can simultaneously interact with both faces of the PCNA ring. The feasibility of this model is supported by a structure of a complex containing the “little fingers” domain of E. coli pol IV, which is a Y-family polymerase related to pol η, and the β-clamp processivity factor, which is structurally similar to PCNA. The C-terminal peptide of pol IV (analogous to the pol η PIP motif) binds in a pocket on one face of the β-clamp ring, and the “little fingers” domain of pol IV binds at the subunit interface (36).
To gain further understanding of how the altered conformation of loop J of PCNA affects the function of pol η, we compared the impact of the wild-type and mutant PCNA proteins on pol η's activity. We found that this mutant protein fails to stimulate the activity of pol η on both abasic sites and non-damaged templates. Not only does it fail to stimulate pol η, this mutant form of PCNA actually inhibits the activity of pol η to levels 2 to 4-fold lower than that of pol η in the absence of PCNA. As a result, the activity of pol η in the presence of the G178S PCNA mutant protein is approximately 10-fold lower than its activity in the presence of wild-type PCNA protein. To our knowledge, this is the first amino acid substitution in PCNA that inhibits the activity of a non-classical DNA polymerase. It should be pointed out that the E113G mutant proteins also failed to stimulate the ability of pol η to incorporate nucleotides opposite an abasic site, although no inhibition was observed with this mutant protein (see Supplemental Fig. S3). It is unclear why the G178S mutant protein inhibits pol η and the E113G protein does not. One possibility is that subtle differences in the positions of loop J between these two mutant proteins accounts for this difference in function. While the position of loop J is aberrant in the structures of both mutant proteins, the position of the loop is closer to that of the wild-type protein in the E113G protein structure (see Supplemental Fig. S3).
It should be noted that if RFC or ATP is omitted from the reaction, no inhibition of pol η is observed with the G178S PCNA mutant protein. This shows that the inhibition requires that the mutant PCNA protein be loaded onto the DNA substrate. The precise mechanism by which the G178S mutant form of PCNA inhibits the catalytic activity of pol η is unclear, and this awaits a more detailed understanding of precisely how wild-type PCNA impacts the kinetic mechanism of nucleotide incorporation by pol η. However, one straightforward possibility is that without the requisite interaction between pol η and loop J of PCNA, the presence of PCNA on the DNA sterically interferes with the proper binding and positioning of pol η. This would prevent the formation of a productive polymerase-PCNA complex on the DNA, which may be responsible for the defect in translesion synthesis observed in cells expressing this mutant form of PCNA.
Finally, it had been suggested that the G178S substitution might block translesion synthesis by interfering with PCNA mono-ubiquitination (25), which is required for translesion synthesis (37). This is unlikely to be the case for several reasons. First, the site of mono-ubiquitination, Lys-164, is located in domain B of PCNA, which we have shown here to have the same structure in both the mutant and wild-type proteins. Second, it has previously been shown that the E113G mutant form of PCNA, which appears to block translesion synthesis by the same mechanism as does the G178S mutant protein, is capable of being mono-ubiquitinated in vitro by the Rad6-Rad18 complex (26). It should be pointed out, however, that any ability or inability of the G178S mutant protein to be mono-ubiquitinated is likely not relevant to understanding the mechanism by which this mutant protein blocks translesion synthesis. This is because the data presented here show that this mutant PCNA protein, even in the absence of monoubiquitination, inhibits that catalytic activity of pol η. This implies that the defect in translesion synthesis caused by this mutant protein is independent of its mono-ubiquitination state.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lokesh Gakher for providing technical assistance. We thank Lynne Dieckman, Christine Kondratick, and John Pryor for valuable discussions.
ABBREVIATIONS
The abbreviations used are:
- dNTP
deoxynucleoside triphosphate
- DTT
dithiothreitol
- EDTA
ethylenediamine tetraacetic acid
- MMS
methyl methanesulfonate
- PCNA
proliferating cell nuclear antigen
- PIP
PCNA-interacting peptide
- PMSF
phenylmethanesulphonylfluoride
- Pol
polymerase
- RFC
replication factor C
- RMSD
root mean square deviation
- UV
ultraviolet
- XPV
xeroderma pigmentosum variant form
Footnotes
The project described was supported by Award Number R01GM081433 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Atomic coordinates were deposited in the RCSB Protein Data Bank: ID code 3F1W
SUPPORTING INFORMATION AVAILABLE
Supplemental Figures S1 to S3 providing additional structural and kinetic data are included as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 2.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 3.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 4.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol eta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 5.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase eta. Proc. Natl. Acad. Sci U.S.A. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Washington MT, Prakash L, Prakash S. Mechanism of nucleotide incorporation opposite a thymine-thymine dimer by yeast DNA polymerase eta. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12093–12098. doi: 10.1073/pnas.2134223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 8.Carlson KD, Washington MT. Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase eta. Mol. Cel. Biol. 2005;25:2169–2176. doi: 10.1128/MCB.25.6.2169-2176.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roush AA, Suarez M, Friedberg EC, Radman M, Siede W. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol Gen Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- 11.Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem. 1999;274:15975–15977. doi: 10.1074/jbc.274.23.15975. [DOI] [PubMed] [Google Scholar]
- 12.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 13.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 14.Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 16.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerase iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair. 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 18.Gibbs PEM, McGregor WG, Maher VM, Nisson P, Lawrence CW. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encoded the catalytic subunit of DNA polymerase zeta. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 20.Langston LD, O'Donnell M. DNA polymerase delta is highly processive with PCNA and undergoes collision release upon completing DNA. J. Biol. Chem. 2008;283:29522–29531. doi: 10.1074/jbc.M804488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hingorani MM, O'Donnell M. Sliding clamps: a (tail)ored fit. Curr. Biol. 2000;10:R25–R29. doi: 10.1016/s0960-9822(99)00252-3. [DOI] [PubMed] [Google Scholar]
- 22.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast polymerase eta function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 23.Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol Cel. Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garg P, Stith CM, Majka J, Burgers PMJ. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase zeta. J. Biol. Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Gibbs PEM, Lawrence CW. The Saccharomyces cerevisiae rev6-1 mutation, which inhbits both the lesion bypass and the recombination mode of DNA damage tolerance, is an allele of POL30, encoding proliferating cell nuclear antigen. Genetics. 2006;173:1983–1989. doi: 10.1534/genetics.106.058545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Northam MR, Garg P, Baitin DM, Burgers PMJ, Shcherbakova PV. A novel function of DNA polymerase zeta regulated by PCNA. EMBO J. 2006;25:4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PMJ. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and repair. Mol. Cel. Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Washington MT, Prakash L, Prakash S. Yeast DNA polymerase eta utilizes an induced-fit mechanism of nucleotide incorporation. Cell. 2001;107:917–927. doi: 10.1016/s0092-8674(01)00613-4. [DOI] [PubMed] [Google Scholar]
- 29.Finkelstein J, Antony E, Hingorani MM, O'Donnell M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal. Biochem. 2003;319:78–87. doi: 10.1016/s0003-2697(03)00273-2. [DOI] [PubMed] [Google Scholar]
- 30.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Cryst. D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 31.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Cryst. D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 33.COLLABORATIVE COMPUTATIONAL PROJECT, NUMBER 4 The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Brunning JB, Shampoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure. 2004;12:2209–2219. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 36.Bunting KA, Roe SM, Pearl LH. Structural basis for recruiting of translesion DNA polymerase Pol IV/DinB to the β-clamp. EMBO J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoege C, Pfander B, Moldovan GL, Pyrowolaksi G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 38.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.