Summary
A strong interplay between the voltage-sensor domain (VSD) and the pore domain (PD) underlies voltage-gated channel functions. In a few voltage-sensitive proteins, the VSD has been shown to function without a canonical PD, although its structure and oligomeric state remain unknown. Here using EPR spectroscopy we show that the isolated-VSD of KvAP can remain monomeric in reconstituted bilayer and retain a transmembrane conformation. We find that water-filled crevices extend deep into the membrane around S3, a scaffold conducive to transport of proton/cations is intrinsic to the VSD. Differences in solvent accessibility in comparison to the full-length KvAP, allowed us to define an interacting footprint of the PD on the VSD. This interaction is centered around S1 and S2 and shows a rotation of 70–100° relative to Kv1.2-Kv2.1 chimera. Sequence-conservation patterns in Kv channels, Hv channels and voltage-sensitive phosphatases reveal several near-universal features suggesting a common molecular architecture for all VSDs.
Keywords: EPR spectroscopy, K+ channel
Introduction
Voltage-gated ion channels play a critical role in maintaining the electrical excitability of cells. Understanding the molecular architecture and correlating it to their function lies at the core of ion-channel studies (Bezanilla, 2002; Bezanilla, 2005a; Bezanilla, 2006; Sigworth, 1994; Tombola et al., 2006; Yellen, 1998). Opening and closing of these channels in response to changes in membrane potential is mediated by the interaction of the VSD (formed by segments S1 through S4), and the ion-conducting PD (formed by segments S5 and S6). This functional coupling however does not necessarily require a tight packing between the two domains, as witnessed from the crystal structures of two voltage-gated channels, KvAP and Kv1.2, in which the VSD is placed at the lipid-protein interface and packs rather loosely against the PD (Jiang et al., 2003; Lee et al., 2005; Long et al., 2005). The crystal structure of the isolated-VSD of KvAP revealed that the domain could fold even in the absence of the PD, highlighting the structural independence of the VSD. In addition, high specific binding of the isolated-VSD to the peptide toxin from spider venom that acts on the full-length channel (FL-channel) and to the Fab raised against the FL-channel revealed that the isolated-domain also retains a conformation close to that in the FL-channel, further suggesting a potential functional independence (Jiang et al., 2003; Ruta and MacKinnon, 2004).
The modular nature of the VSD and the PD have been suggested by a number of earlier findings (Kubo et al., 1993); (Schrempf et al., 1995) (Lu et al., 2001; Lu et al., 2002). The structural independence of the two domains is now very well exemplified by recent reports of two classes of VSDs found in the absence of the PD: a VSD homologue attached to a phosphatase domain (Murata et al., 2005); and a voltage-gated-proton channel which appears to be comprised exclusively of the VSD (Ramsey et al., 2006; Sasaki et al., 2006). These proteins also emphasize the fact that the VSDs are not exclusively associated with ion-channels and have a more ubiquitous presence as voltage transducers in a variety of systems. Whether these VSDs can remain and function as monomers or require association to higher oligomeric forms and/or the presence of other auxiliary proteins is yet to be determined.
The crystal structure of the isolated-VSD of KvAP, attached to Fab fragments, is depicted in its monomeric form (Jiang et al., 2003). Though the structure is broadly consistent with several lines of experimental results, to what extent the crystallographic structures obtained in the absence of lipids represent native conformations on the membrane still remain a central issue of controversy. Recent studies also point out the unexpected role of lipid membranes in maintaining the correct relative orientations of the two domains and in their proper functioning (Lee et al., 2005; Long et al., 2007) (Ramu et al., 2006; Schmidt et al., 2006) giving rise to the hypothesis that lipids interact in a rather specific manner with certain regions of the Kv channel, thereby modulating their function. Since membrane lipids appear to form an integral part of the architecture of these domains, understanding the conformation and orientation of the VSD in the context of a lipid environment represents a crucial issue.
Electron paramagnetic resonance (EPR) spectroscopy is a powerful tool to study conformational dynamics of membrane proteins incorporated in a near-native lipid membrane (Hubbell et al., 2000; Hubbell et al., 1998; Hubbell et al., 1996; McHaourab et al., 1996; Perozo et al., 1998; Perozo et al., 1999; Perozo et al., 2002). Our previous work on the VSD of the full-KvAP channel provided the first unequivocal evidence for the peripheral location of S4 in lipid bilayer (Cuello et al., 2004). Guided by these findings our present study aims to address two fundamental questions: First, what is the precise structure of the isolated-VSD in its native environment and how similar is this conformation to that of the sensor in the FL-channel? Second, what is the orientation of the VSD with respect to the PD in the membrane-embedded full-length KvAP and how it compares to the Kv1.2 structures? We use a combination of site-directed spin labeling and EPR spectroscopy to study the structural dynamics of the isolated-VSD of KvAP and extend our findings to further explore the molecular architecture of other isolated-VSDs comprised within the Hv channel and the voltage-sensitive phosphatase families.
Results
Oligomeric state of the VSD in detergent and lipid membrane
The 17kD isolated-VSD (S1-S4 segments) of KvAP (Fig 1A), was expressed and purified as described in Methods. The oligomeric state of the VSD, as determined by gel filtration analysis, was found to be a monomer in the presence of octyl-glucoside as previously reported (Jiang et al., 2003). We further find that in decylmaltoside, the VSD elutes at a position that corresponds to a dimer (Fig 1B). A dimeric configuration was also observed for few cysteine mutants in the S4-S5 linker which formed di-sulfide cross-bridges on the bacterial membrane (Supplemental Fig 1S). This result suggests that the oligomeric state of the VSD is critically governed by the nature of the detergent and raises the possibility that it is likely to be modulated by lipid composition on the membrane.
Figure 1.
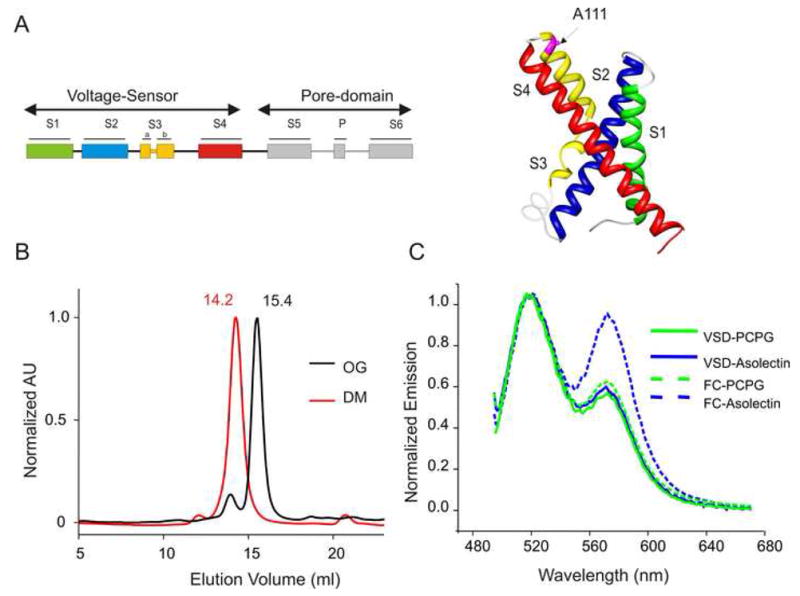
3D topology and the oligomeric state of the isolated-VSD of KvAP. (A) A schematic representation of the full-length KvAP channel showing the location of the VSD and the PD (Left). Ribbon representation of the TM helices as depicted by the crystal structure of the isolated-VSD (Right). (B) Gel filtration chromatography of the isolated-VSD shows a dimeric configuration in DM (red) and a monomeric state OG (black). (C) FRET based assay of the aggregation state of the isolated-VSD and the FL-channel (FC) in asolectin and in a mixture POPC:POPG. Mutant A111C was used to co-label the protein with rhodamine and fluorescein. A prominent rhodamine peak at 565 nm is an indication of protein aggregation on the membrane.
Fluorescence resonance energy transfer (FRET) offers a convenient tool to assess if independent VSD molecules come together within a distance <50 Å on the membrane (Cuello et al., 2004; Vasquez et al., 2007). A single mutant in the S3-S4 linker (A111C) was labeled with either Fluorescein-maleimide (donor) or Tetramethylrhodamine-maleimide (acceptor), and reconstituted into preformed liposomes. The choice of the lipid membranes stems from previous findings showing that asolectin promotes extensive channel-to-channel lateral aggregation in the full-length KvAP channel via S4, while a POPC:POPG mixture eliminates this interaction (represented by dotted lines in Fig 1C) (Cuello et al., 2004). Aggregation of proteins in liposomes is reflected by strong FRET, while non-aggregated proteins produce minimal or no energy transfer. Using this approach, we found that the isolated-VSD remains mono-disperse in both the reconstituted membrane systems (represented by solid lines in Fig 1C). In fact, even overnight incubation at room temperature or freeze/thawing the sample did not produce any significant aggregation, suggesting that the isolated-VSD is very stable in its monomeric form in the tested membrane systems.
Molecular architecture of the isolated-VSD
EPR measurements were carried out on purified and reconstituted spin-labeled cysteine mutants at 130 positions spanning the entire isolated-VSD sequence. Since the experiments were done at 0 mV, the conformation of the protein is likely to represent the activated state of the VSD. Structural analyses were based on the properties of spectral line-shapes of the spin-probe (the mobility parameter, ΔHo−1), reflects the local steric restriction of the spin-label from its neighboring environment and the measured collisional frequencies of relaxing agents (O2 -lipid exposed and NiEdda- water exposed) by power saturation experiments. (Farahbakhsh et al., 1992; Gross and Hubbell, 2002; Perozo et al., 1998). Figure 2A shows the processed EPR data for the isolated-VSD. Residue enviromental parameter profiles are shown for the probe mobility ΔHo−1(black), O2−accessibility ΠO2 (red) and NiEdda-accessibility ΠNiEdda (blue) and together they provide a dynamic structural organization of the isolated-VSD on the membrane in its “activated” conformation. An inspection of ΔHo−1 along the length of the VSD reveals a highly periodic behavior with low mobility every third or fourth residue, as expected from its α-helical structure. The periodic behavior of ΠO2 mirrors the changes in mobility such that positions of high ΠO2 also show large motional dynamics (spin-probe faces an unrestricted membrane environment) and they alternate with positions reflecting restricted mobility and accessibility (faces a proteinacious environment). The location of the putative loop regions are characterized by very high mobility. As seen in the FL-channel (Cuello et al., 2004), ΠNiEdda clearly defines the boundaries for each of the membrane segments. The C- and the N-termini, the S2-S3 and the S3-S4 loops of the VSD show clear accessibility to NiEdda while the S1-S2 loop appears to show more exposure to the lipid environment. The average mobility and accessibility parameters for each of the TM segments appear to be roughly the same as expected of an isolated-four helix bundle that traverses the entire length of the membrane (Fig 2B). This finding that each of the TM segment is exposed to a similar asymmetric environment (with one face of the helix exposed to the membrane while the other to the core of the helix bundle) further strengthens the argument that the VSD in our membrane systems is in its monomeric state. The overall architecture of the isolated-VSD determined in this work agrees remarkably well with the KvAP isolated-VSD crystal structure (Jiang et al., 2003) and the domain can indeed adopt a transmembrane orientation similar to that reported for the FL-channel (Cuello et al., 2004).
Figure 2.
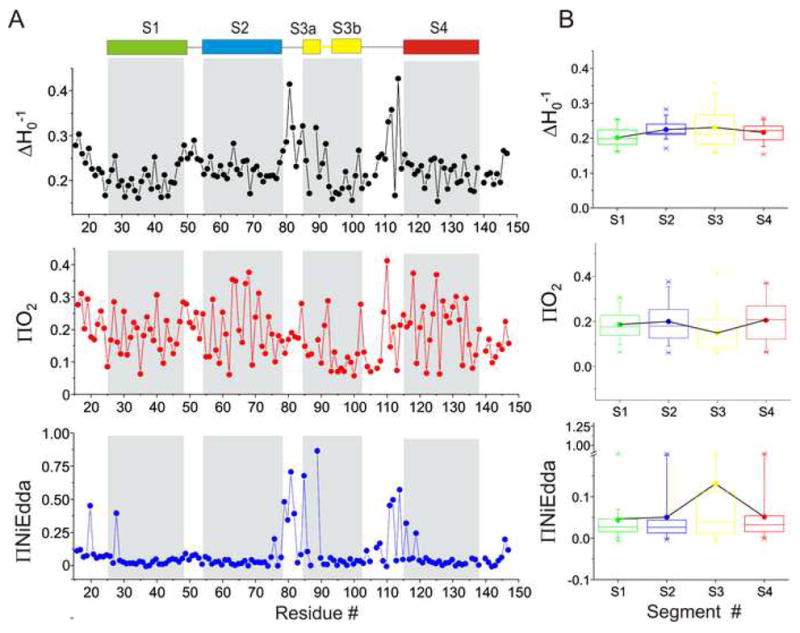
EPR based structural analysis of the isolated-VSD. (A) Mobility ΔHo−1(black), O2 accessibility ΠO2 (red) and NiEdda accessibility ΠNiEdda (blue). The gray regions represent the putative TM segments from the crystal structure. (B) The average mobility and accessibility parameters calculated for each of the TM segments. Each box denotes an individual TM segment with SDs marked as error bars.
Comparison of the residue parameters with the FL-channel
Figure 3A shows an overlap of the spectral line shapes of S1 and S4 in the FL-channel and the isolated-VSD. Significant changes can be seen in many residues in S1, where a decrease in the spectral broadening compared to the FL-channel indicate that these residues now face a less restricted environment. On the other hand the spectral line shapes for S4 in the FL-channel and the isolated-VSD are essentially super imposable which clearly demonstrates that the environment surrounding S4 in the two configurations are identical. These findings are more evident in Figure 3B which shows profound changes in the measured ΠO2 in S1 and S2 with little or no change in S3 and S4, in support to the idea that S3 and most notably S4 in KvAP are fully exposed to the lipid environment in the FL-channel, while S1 and S2 are interfaced with S5 and S6 in the PD. (Cuello et al., 2004).
Figure 3.
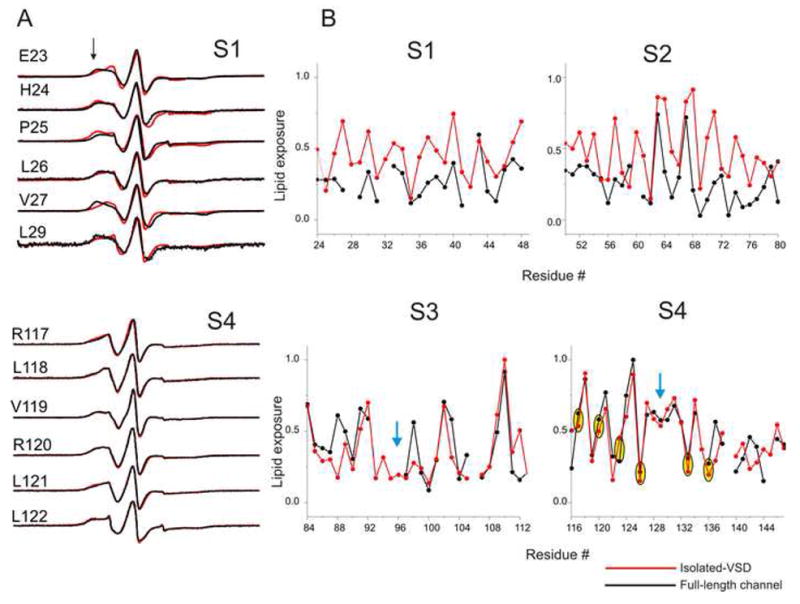
Comparison of the residue-specific enviromental parameters in the FL-channel and the isolated-VSD. (A) An overlap of representative X-band CW EPR spectra of spin-labeled mutants from the S1 and the S4 segments in the FL-channel (black) and the isolated-VSD (red). Black arrow-head shows the location of immobile component of the spectra. (B) Profile of changes in the ΠO2 parameters. The values were normalized to the maximum value measured within each data set. The blue arrow-head marks the break in the α-helical periodicity of the accessibility. Positions of the gating charges in S4 are highlighted by yellow ovals.
A close examination of the α-helical periodicity of ΠO2 reveals a break around residue Pro95 that divides S3 into two short helices S3a and S3b (Fig 3B), in agreement with the crystal structure of the isolated-VSD (Jiang et al., 2003). A break in the periodicity of ΠO2 was also observed in S4 around residues 128–130 with the two short halves twisted with respect to their accessible surface, a pattern although closely matches our findings in the FL-channel (Cuello et al., 2004), is not quite evident in the crystal structure of the isolated-VSD (Jiang et al., 2003).
The next important question that arises is the location of the charges. As in the FL-channel, most of the positions associated to the charges in S4 appear to reveal a protected environment as indicated by their restricted probe dynamics and low accessibility (particularly for spin-labels at positions R123, R126, R133 and R136). The first two arginines (R117 and R120) show intermediate mobility and lipid exposure. Given that the neighboring residues F116 and V119 show considerable water exposure, the first two arginines likely reside in a water/lipid interface. Additionally, as a consequence of the twist in the S4 helix, the last two charges are also placed on the same face of the helix as the rest of the arginines. Besides S4 charges, several acidic residues in S2 and S3 are known to be involved in salt-bridge interactions (Papazian et al., 1995; Planells-Cases et al., 1995; Seoh et al., 1996; Tiwari-Woodruff et al., 1997). The spin-label at these positions (D62, D72 and E93) also reveals environments suggesting that they lie on the face of the helix that allow interaction with the S4 arginines.
The orientation of the individual TM segments can be obtained from the frequency and vector analysis of the residue enviromental parameters. Figure 4A and B shows mapped ΠO2 for S1 and S4 on the isolated-VSD structure along with a plot of mobility and ΠO2 for full-channel and the isolated-VSD in polar coordinates, where the accessibility values are superimposed on a helical wheel such that the calculated moment points to the side of the helix with highest mobility (or ΠO2). In the FL-channel, the sum vectors for S1 mobility and ΠO2 are negligible in comparison to that of the isolated-VSD in which the resultant moment clearly points to the membrane facing side of the helix. On the other hand the two halves of the S4 helix show no change in the magnitude or orientation of the sum vector (Fig 4B). Similarly, the direction of sum vector for the ΠNiEdda in the two halves of S3 point towards water exposed regions (Fig 4C). This orientation is in phase with the mobility vectors, a fact which suggests that these water exposed regions are large enough to allow considerable motional freedom of the spin-label probes. The difference in ΠNiEdda reveals a small tilt with respect to the long axis of the VSD, besides which the position of the isolated-VSD relative to the membrane does not change much in comparison to that in the FL-channel.
Figure 4.
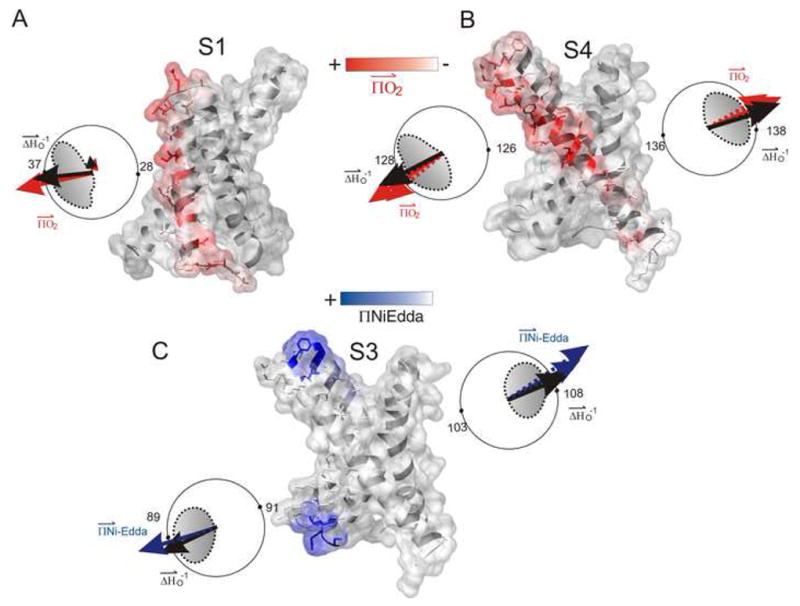
Frequency and vectorial analysis of the structural parameters. The experimentally determined accessibility parameters were mapped on to the crystal structure of the isolated-VSD (1ORS.pdb). ΠO2 is shown in red (A and B) and ΠNiEdda in blue (C). The accessibility parameters are plotted in polar coordinates such that the sum vectors point towards the face of the helix with highest mobility (black) or accessibility (ΠO2- red, ΠNiEdda -blue) values. The solid arrow denotes the isolated-VSD while the broken arrow represents FL-channel. The direction of the calculated moments in the upper and lower halves of the S3 and S4 segment shows a break in the helices accompanied by a twisting of the solvent-exposed surface.
Occurrence of crevices in the isolated-VSD
An inspection of the ΠNiEdda in S3 show that water can penetrate well into the membrane buried regions of the segment, although the segment by itself is transmembrane in orientation (Fig 5). In fact, only a subset of 14 residues in the S3 segment is truly buried within the putative membrane-spanning core, showing no accessibility to NiEdda. In this case, the hydrophobic region of the membrane approaches an apparent “thickness” of < 20 Å in the vicinity of S3. This, along with the high motional freedom of these residues strongly suggests the presence of water filled crevices. However, since the aqueous-accessibility is measured by the collisional quenching of NiEdda, which has a cross-sectional area larger than water, with the spin-probe which in itself is of ~5–7 Å, the extent of reported water penetration into these crevices is most likely an under estimation of the actual depths of penetration. Interestingly, comparison of the FL-channel and the isolated VSD shows that there are no major differences in the depth of NiEdda-accessibility into S3 in the two systems, proving quite convincingly that the occurrence of the crevices is intrinsic to the VSD architecture and does not necessitate the presence of the PD (in the activated state).
Figure 5.
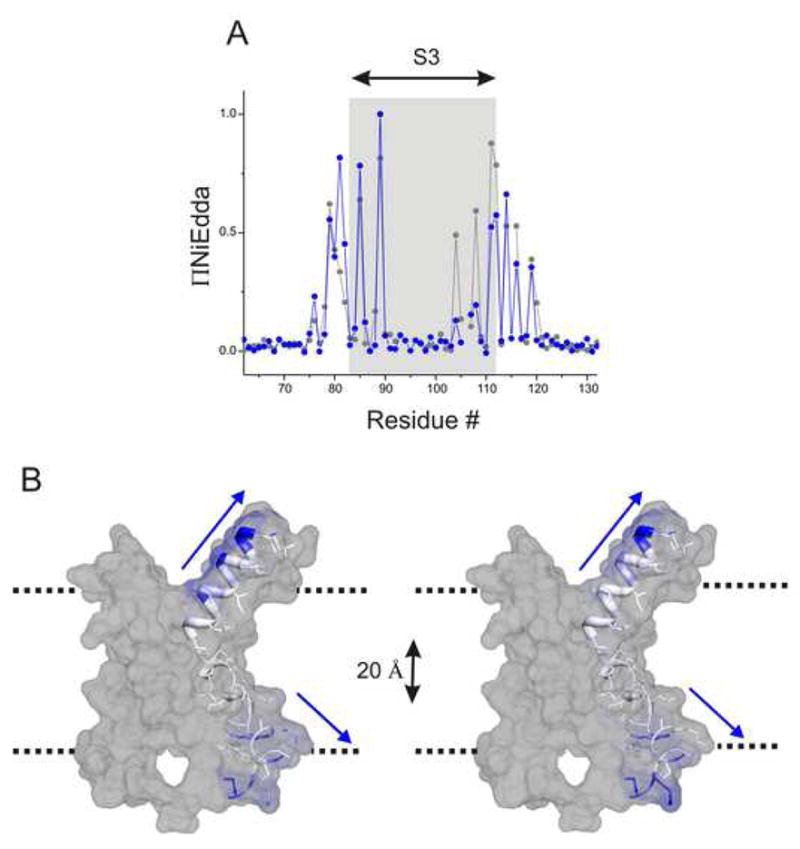
Aqueous crevices within the VSD. (A) Normalized ΠNiEdda of the residues comprising the S3 segment in the isolated-VSD (blue) and the full-channel (black). (B) ΠNiEdda values mapped on the S3 segment to show the depth of water accessible areas within the transmembrane region of the protein. The dotted line denotes the putative limits of the membrane.
A structural footprint of the PD on the VSD
To understand the basis of the electro-mechanical coupling between the VSD the PD, it is important to first learn how the two domains are juxtaposed against each other. The difference in residue enviromental parameters measured for the isolated-VSD and the FL-channel were mapped on the crystal structures of the isolated-VSD. Analysis of the delta values of mobility and ΠO2 shows that the most dramatic effect upon removal of the PD occurs in the vicinity of S1 and S2, indicating that this region interacts with the PD in the FL-channel (Fig. 6). This is in agreement with the EPR measurements showing restricted dynamics and low accessibility for S1 and S2 (Cuello et al., 2004).
Figure 6.
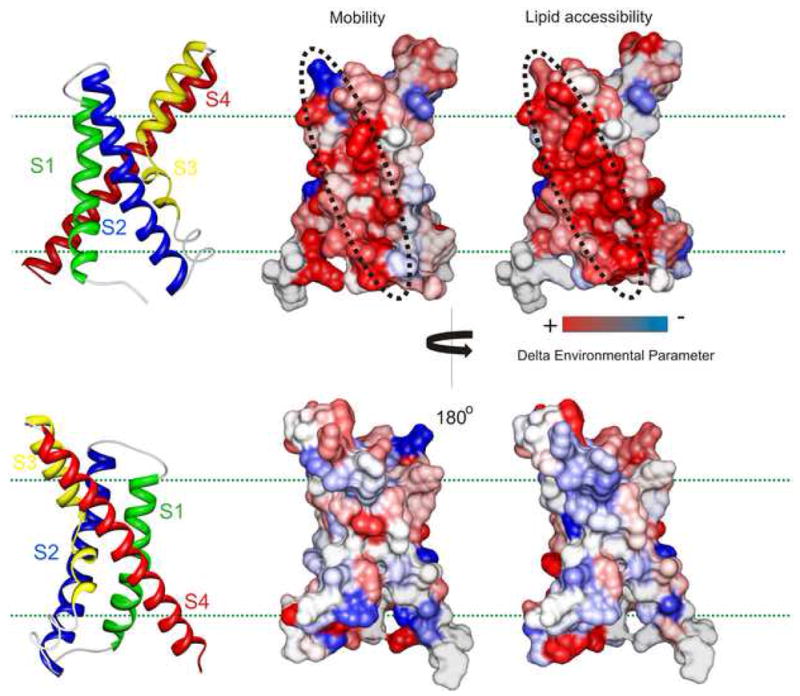
Mapping the molecular interacting surface of the VSD with the PD. Fractional differences in the mobility and ΠO2 measured in the corresponding residues in the isolated-VSD and in the full-channel mapped on the sensor structure and color coded with a gradient from red to blue. Red denotes increase and blue represents decrease in the environmental parameter. Dotted black line marks the surface of highest impact.
The present data set provides a more accurate understanding of the orientation of the VSD with respect to the PD and thus a means to evaluate the current open-state structure of Kv1.2-Kv2.1 chimera (Long et al., 2007) and an open-state model based on the structure of Kv1.2 (Jogini and Roux, 2007). Mapping the delta values of ΠO2 shows a remarkable similarity in the arrangement of individual side-chains, positions with small delta values pointing into the helical core of the VSD while those with large delta values chains pointing away (Fig 7). However, there appears to be a discrepancy in the positioning of the VSD with respect to the PD. Both structures predict that VSD interacts with the PD through S1 and S4, while the EPR data indicates that the interaction with the PD is via S1 and S2 with S3 and S4 clearly placed in the periphery. This arrangement of the two domains requires a rotation ~70–100° (from the intracellular side) about an axis perpendicular to the membrane plane relative to the PD such that S1 and S2 are more closely placed to the PD thereby matching the observed accessibilities (Fig 7).
Figure 7.
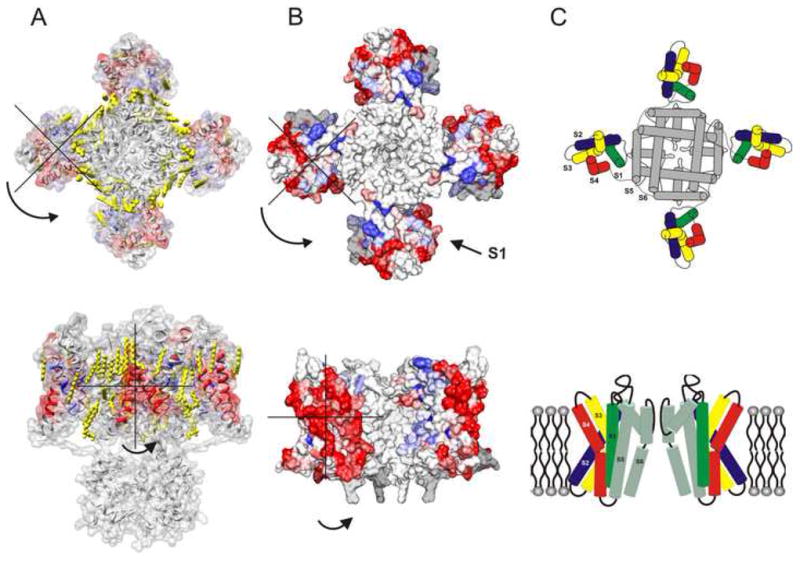
Evaluation of the open state structures for the voltage-gated K channels. The delta values were mapped on to the coordinates of (A) Kv1.2-Kv2.1 chimera structure (2R9R.pdb) and (B) model based on Kv1.2 structure. (C) Model for the open-inactivated state of KvAP based on EPR measurements. The top panel shows the view from the intracellular part of the channel.
Discussion
A thorough understanding of the underlying mechanism of ion-channel functions calls for high-resolution structural information in a native, physiologically relevant environment. But given the complexity of the lipid bilayer environment, emulating the native habitat of membrane proteins under crystallographic conditions still remains a formidable challenge. Therefore, interpreting the conformations of the protein captured by crystallography and equating them to physiologically relevant conformational states is by no means trivial (Ahern and Horn, 2004; Bezanilla, 2005b; Jiang et al., 2003; Minor, 2007; Swartz, 2004; Tombola et al., 2006).
The crystal structure depicts the KvAP isolated-VSD in its monomeric state (Jiang et al., 2003). Our data suggests that this is indeed the case in short chain detergents, such as octyl glucoside, that form compact micelles and thereby favoring the monomeric form of the VSD. On the other hand, in detergents that have longer acyl chain there is a possible mismatch of the hydrophobic micellar diameter and the apolar regions of the TM segments of the protein, which could force the monomers to oligomerize.
Here we show that the activated state of isolated-VSD on the membrane has a configuration very similar to that of the full-channel. Regions in contact with the PD showed large changes in the dynamics and accessibilities, while segments located peripherally show the least amount of change. An almost identical profile for the mobility and lipid exposure, suggest that the location of the S4 within the membrane limits is remarkably similar to that in the FL-channel. Our EPR constraints are consistent with the idea that the first two arginines positions are partly exposed to the lipid (-water interface) but the rest face the same side of the helix as the S2 and S3 charges, in a position conducive to interaction with the S4 arginines (Papazian et al., 1995; Planells-Cases et al., 1995; Seoh et al., 1996; Tiwari-Woodruff et al., 1997). In Shaker the first two arginine positions are accessible to water and forms the basis for proton transport and proton pore (Starace and Bezanilla, 2001; Starace and Bezanilla, 2004; Starace et al., 1997). In KvAP, despite the inaccessibility of R117 and R120 to NiEdda, water exposure of these gating charges cannot be entirely ruled out given that the hydration free-energy for arginine is ~ −60 kcal/mol while that for a spin-label is ~ −1.9 kcal/mol which is close to that of a hydrophobic side chain such as leucine (~2.28 kcal/mol) (Deng, 2004). Therefore unlike arginine, a spin-label at the interface would likely prefer to point into the membrane than “snorkel” and stay exposed to water.
Functional studies in voltage-gated channels have shown the presence of hydrophilic crevices in the vicinity of S4 and several computational studies have also reported similar water penetration into the VSD of Kv1.2 and KvAP. A conceptual advantage of these water-penetrations within the membrane can be in reducing the energetic cost of placing the gating charges in a hydrophobic environment while at the same time focusing the membrane field which will allow an efficient transduction of the electric field into S4 movements. (Cha and Bezanilla, 1997; Freites et al., 2006; Islas and Sigworth, 2001; Larsson et al., 1996; Mannuzzu et al., 1996; Sands and Sansom, 2007; Sorensen et al., 2000; Treptow and Tarek, 2006; Yang et al., 1996). Interestingly, we find the presence of crevices also in the isolated-VSD and they are not considerably different from that in the FL-channel. An upshot of this finding might have mechanistic implications in that the water filled cavities could be a common feature underlying the conductance of protons and other cations through the VSD, and that this pathway may occur in the absence of auxiliary proteins and even in the monomeric state of the VSD.
The availability of structural information of an isolated-VSD in a membrane environment and sequences of several classes of voltage-sensors in the presence and absence of a PD allows for a comprehensive sequence-structure analysis to obtain an architectural blueprint common to these domains. A sequence alignment of families of eukaryotic Kv channel, Hv channel and voltage-sensitive phosphatases (VSP) was used to calculate sequence-variability profiles within the TM segments comprising the VSD (Methods). As shown in Figure 8, the peaks of individual side-chain variability closely match the peaks of ΠO2, This observation is consistent with the side-chains facing the lipid membrane being by far more variable (and therefore tolerant to mutations) and the residues facing within the helical bundle showing strong conservation (highly-impacted upon mutations). It is interesting to note that the acidic residues in S2 and the basic residues in S4 are all predicted to face into the interior of the VSD, in a configuration conducive to interaction with each other as shown to be the case in Kv channels (Papazian et al., 1995; Planells-Cases et al., 1995; Seoh et al., 1996; Tiwari-Woodruff et al., 1997). Most residues in eukaryotic Kv channels that showed high-impact upon perturbations report low variability and ΠO2, and point into the packed helical core (Hong and Miller, 2000; Li-Smerin et al., 2000a; Li-Smerin et al., 2000b; Li-Smerin and Swartz, 2001; Monks et al., 1999). However, a somewhat unexpected pattern was that face of highest changes in the ΠO2 in S1 and S2, reflecting the surface interacting with the PD, also corresponded to side of the helix with largest sequence variability. Though unpredicted, it goes along the notion that the interaction between the two domains is rather loose. Another common feature among these families was found at the position of the conserved proline in the middle of S3, reported to divide the segment into two halves (Hong and Miller, 2000; Li-Smerin et al., 2000a; Li-Smerin and Swartz, 2001) by inducing a bend or a kink in the helix (Blaber et al., 1993; MacArthur and Thornton, 1991; Monne et al., 1999). This position is occupied by glycine/serine or a threonine in the Hv channel and VSP families. These residues have been suggested to destabilize alpha helices, glycine by adopting a wide range of main-chain dihedral angles and serine and threonine by forming hydrogen bonding between the γ O atom and the i-3 or i-4 peptide carbonyl oxygen (Ballesteros et al., 2000; Blaber et al., 1993; Gray and Matthews, 1984; Li-Smerin and Swartz, 2001; MacArthur and Thornton, 1991; Monne et al., 1999). A lack of periodicity in the C-terminal end of S3 following the bend is well predicted by the variability analyses and has also been seen in the ΠO2 and in scanning studies. Strong sequence conservation among the three completely different classes of VSD argues that the sensors are all built along the same basic outlines as observed in isolated-VSD of KvAP. This idea is well supported by a recent study demonstrating the portability of the voltage-sensor motif (S3b-S4) among voltage-sensors of different origin (Alabi et al., 2007).
Figure 8.
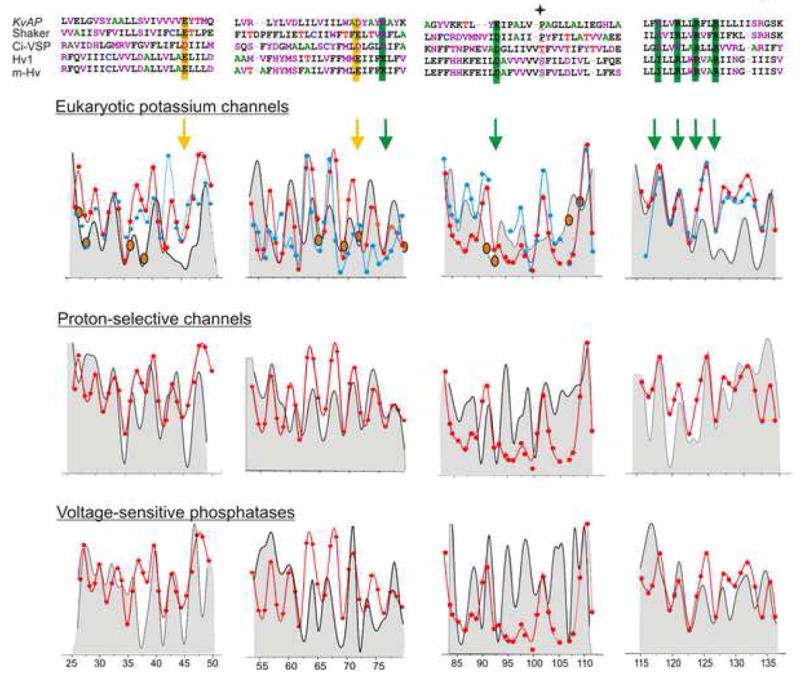
Comparison of the sequence variability within the VSD in eukaryotic Kv channels, Hv channels and the VSP family. Sequence alignment of the TM segments comprising the VSD (Top). Filled star marks the position of the break in S3. An overlap of the calculated sequence variability at each position (grey background) and the corresponding normalized ΠO2 measured within the FL-channel (blue) and the isolated-VSD (red) of KvAP. Positions corresponding to charge residues are marked by arrows. Highly impacted residues in perturbation analyses in eukaryotic Kv channels are shown in orange circles.
Fundamental to elucidating the mechanism of voltage-sensor movement and its coupling to the channel opening is the understanding of how the two domains interact with each other during gating process. EPR measurements predict that the interacting surface in KvAP VSD constitutes S1 and S2. This orientation is further confirmed by LRET measurements in KvAP under similar conditions (Richardson et al., 2006). The Kv1.2 and the Kv1.2-Kv2.1 chimera structures on the other hand suggest that the interaction involves S1 and S4. Results from functional studies, mostly in Shaker, were originally interpreted as involving S3 and S4 at the PD interface. There are three possible explanations for the discrepancies in the predicted interface between the PD and the VSD. First, prediction of the VSD orientation in eukaryotic channel is heavily biased by the results from tryptophan and alanine scanning approaches which predicted a simple amphipathic environment for S1 and S2 therefore placed in the periphery (Hong and Miller, 2000; Li-Smerin et al., 2000a; Li-Smerin and Swartz, 2001; Monks et al., 1999). The S3 and S4 segments on the other hand were predicted to be in a rather protected environment, and therefore placed close to the PD. Analyses of mutation-induced gating perturbations draw on the asymmetric environment surrounding the helix with the premise that mutations are better tolerated on the side of α-helix facing the solvent (lipid/water) than on the side that involved in extensive protein-protein contacts. These results correctly predicted the location of most of the highly impacted residues, which are also the most conserved in variability analysis, and were found to be facing into the packed core of the VDS bundle. However, the interpretation falls short within the region well-tolerated, a condition that could arise either if the side chain faces a lipid membrane or if it is oriented towards a loosely surrounded proteinacious environment. Such a loose packing between the sensor and the pore on the membrane could allow perturbations in this region to be well tolerated. This idea is consistent with the finding that S5 residues interfaced with the VSD are mostly well tolerated by tryptophan mutations, a condition also likely to be true for the interacting surface within the VSD as well. A possible explanation of why S4 (and S3) are so highly impacted could be that this region of the protein undergoes significant structural rearrangement during gating which includes a 180° rotation of the helix, leading to significant changes in the relative exposure to hydrophobic and aqueous environments of the residues in positioned in all the faces of the helix. Though tryptophan substitutions tend to destabilize protein-protein interactions, perturbation analysis in both S4 and S5 reveal blocks of regions that show relative stabilization of the closed or open states suggesting the dynamic nature of the interaction between the two domains (Li-Smerin et al., 2000a; Soler-Llavina et al., 2006).
An alternate explanation for that this arrangement of the VSD relative to the PD might be reflective of a deep open-inactivated state of KvAP, while in the closed (and open) state of the channel the VSD might be rotated such that S4 occurs closer to S5. Such an orientation suggests large conformational changes within the entire VSD during inactivation, as previously proposed (Yarov-Yarovoy et al., 2006). Several studies including a recent fluorescence scanning in the VSD propose that the slow inactivation involves a global rearrangement within all of the TM segments (Cha and Bezanilla, 1997; Loots and Isacoff, 1998; Pathak et al., 2007). Further, prolonged depolarization has been shown to modify the conformation of the voltage sensor, a change likely associated with the process of slow inactivation, such that the channels encounter a higher energy barrier in their transition back to the closed state (Olcese et al., 1997; Olcese et al., 2001). Mackinnon and colleagues claim Kv1.2 to be open (lower gate) with the sensor in its activated state and the selectivity filter similar to that of the conductive filter of KcsA (Long et al., 2005). However, there is no conclusive evidence that indicates that the Kv1.2 crystal structure represents the same deep-inactivated state of KvAP on the membrane sampled by EPR. Recovery experiments of KvAP in lipid bilayer show that the channel’s inactivated state is much more stable (τrecovery 52s) compared to the slow-inactivated state measured in the Shaker family (2–8 s) (Levy and Deutsch, 1996; Rasmusson et al., 1997). Since our EPR measurements were made in lipid membranes at 0 mV, the conformation being sampled would indeed correspond to that of the deep open-inactivated state. Interestingly, tryptophan scanning studies in HERG K+ channels (known to inactivate rapidly) also conclude that the S4 segment is located in a lipid exposed environment (Subbiah et al., 2005)
This idea of the VSD being rotated against the PD in the closed state of KvAP, so as to bring S4 closer to S5, is supported by cross-bridges formed between a cysteine residue introduced at the position of the first arginine residue on S4 with a cysteine introduced near the extracellular end of S5 of a neighboring subunit (Lee et al., 2005) although with less efficiency than reported for in Shaker (Laine et al., 2003). An interesting possibility is that some of these discrepancies might originate by the absence of T1-domain in KvAP which could promote the stabilization of the sensor in the given conformation with the S4 placed in the periphery. Studies show that the truncation of this region alters voltage-gated activation and inactivation rates in several channels (Choe et al., 2002; Cushman et al., 2000; Kurata et al., 2001; Minor et al., 2000) suggesting that the T1 domain indeed affects the way the VSD senses and moves within the membrane field.
Finally, it is also possible that this orientation of the VSD relative to the PD may be the result of an intrinsically different molecular architecture of KvAP. But given the remarkable similarity between the overall organization of the TM segments within the sensor among distantly related systems, and recent studies suggesting that the movement of the sensor in voltage-gated K channels is not that different after all (Campos et al., 2007; Pathak et al., 2007), we tend to favor the idea that the existing arrangement in KvAP pertains to the open-inactivated state of the channel, a conformation that could be more rarely visited by the eukaryotic channels under the voltage-clamp protocols.
Experimental Procedures
Mutagenesis, Protein Expression and Spin labeling
The DNA encoding the isolated voltage-sensor domain of KvAP was cloned into pQE70 expression vector (Qiagen). Cysteine mutants were generated in the entire length of the isolated sensor (16–147). Mutant sensors were expressed and purified as described in the previously published protocols (Jiang et al., 2003). Purified protein was labeled with a methanethiosulfonate spin probe (3-(methane sulfonylthiomethyl)- 2,2,5,5-tetramethylpyrrolidin-1-yloxy (Toronto Research)) at a 10:1 (label/channel) molar ratio and reconstituted at a 250:1 lipid/protein (molar ratio) in mixture of POPC:POPG (3:1) (Cuello et al., 2004).
Fluorescence studies
The A111C mutant was purified and labeled at a 10:1 (label/channel) molar ratio with either Fluorescein-maleimide or Tetramethylrhodamine-maleimide (Molecular probes). The labeled proteins were mixed in equal proportion and reconstituted into preformed liposomes. Fluorescence spectra were measured in a Shimadzu 1501 spectrofluorimeter using protocols reported earlier (Cuello et al., 2004).
EPR spectroscopy and Analysis
Continuous-wave (CW) EPR spectroscopic measurements were performed at room temperature on a Bruker EMX X-band spectrometer equipped with a dielectric resonator and a gas permeable TPX plastic capillary. Spectra were recorded at 2.0 mW incident power, 100 kHz modulation frequency with modulation amplitude of 1.0 G. The motional freedom of the spin label was quantified as a peak-to-peak first derivative width of the central (mI = 0) resonance (ΔHo−1) (McHaourab et al., 1996). As the frequency of nitroxide rotational motion is reduced, as witnessed during the formation of tertiary or quaternary contacts, the line width and hence ΔHo−1 increases for any particular motional geometry. On the contrary, an increase in the probe’s freedom of movement is reflected as a decrease in ΔHo−1. Further, accessibility of a spin-label to lipid or water is estimated from power saturation experiments in which the vertical peak-to-peak amplitude of the central line of the first derivative EPR spectra is measured as a function of increasing incident microwave power (Altenbach et al., 1989; Farahbakhsh et al., 1992; Perozo et al., 1998). These measurements are made for each spin-labeled mutant after equilibration with air (21% O2) or 25 mM NiEdda (Ni(II) ethylenediaminediacetic acid) in the presence of N2. Data were analyz1ed and converted to the accessibility parameter Π according to Farahbakhsh et al. (Farahbakhsh et al., 1992). High accessibility of the spin-probe to O2 (ΠO2) is suggestive of a membrane exposed environment, while high NiEdda accessibilities (ΠNiEdda) is a reflection of the probe being exposed to aqueous solvent. To estimate the periodic behavior of a given residue-specific enviromental parameter, a discrete Fourier transform power spectrum, P(ω), of the amino acid segment is calculated as a function of the angle between two adjacent side chains (ω) (Cornette et al., 1987; Donnelly et al., 1994). To determine the location of the solvent-accessible side of a helix, the resultant vector from the sum of all Π values was calculated (Eisenberg et al., 1984). The angle θ was obtained as the value of the resultant M (ω) evaluated at ω= 100°, taking an arbitrary residue as a reference point (θ= 0). A sliding window (7–9 residues) approach was used in which the periodicity index is estimated for the data set within the window. This method helps in defining the probable limits of a given secondary structures (Cornette et al., 1987).
Sequence Alignment
We constructed an alignment of 88 non-redundant sequences corresponding to the VSD (S1-S4, disregarding the loop sequences) within the eukaryotic Kv channel family spanning a wide variety of species using CLUSTALW. For the Hv channel and the VSP family, we blasted the Hv1 and Ci-VSP sequence respectively against the NCBI non-redundant database. Within the hits for the Hv1 sequence, we selected only those sequences that contained the conserved His residue in S2 and S3. For the Ci-VSP family we choose only those hits that contained both the VSD and the phosphatase motifs. To examine the level of sequence conservation within these classes of protein we used Shannon entropy H(l) at each position l of the TM helical sequence as described by (Larson and Davidson, 2000) using BioEdit software (Hall, 1999)
where H(l) is the uncertainty, also called entropy at position l, and f(b,l) is the frequency at which residue b is found at position l. Less variable positions (especially conserved regions), yield low entropy values in comparison to highly variable positions.
Supplementary Material
Figure S1. Oligomeric state of the mutant A145C. In gel filtration chromatography, mutant A145C in the isolated-VSD of KvAP in octyl-glucoside (red) elutes as two peaks corresponding to a monomer and a dimer band on SDS-PAGE (Inset). In the presence of DTT a single monodisperse peak at the monomer elution volume is seen (black) which also runs as a monomer on SDS-PAGE. In the presence of decylmaltoside without DTT, mutant A145C elutes as a single peak at the elution volume of a dimer.
Figure S2. A comparison of spin-label mobility in S1 in the isolated-VSD and the FL-channel. An overlap of EPR spectral line shapes at the equivalent positions in the isolated-VSD (red) and the FL-channel (Black). Side-chains facing the core of the VSD bundle are shown in yellow and those facing outside (and therefore interacting with the PD in the FL-channel) are shown in red. Note that the residues facing into the VSD do not show any change in the line-shape in the two systems, while those facing the interface show pronounced increase in mobility.
Figure S3. A comparison of spin-label mobility in S2 in the isolated-VSD and the FL-channel. As seen in S1, the side chains facing the interface (red surface) show significant changes in line shapes in the isolated-VSD (red) in comparison to the FL-channel (black). On the other hand positions facing into the VSD core (yellow surface) do not show any significant change.
Figure S4. A plot of the delta values for the measured ΠO2 in the FL-channel and the isolated-VSD. The ΠO2 values were normalized against the maximum value within each data set before calculating the delta differences between the FL-channel and isolated-VSD. The dotted line denotes the average delta parameter for the S1/S2 and the S3/S4 segments.
Figure S5. Sequence alignment of eukaryotic Kv channels, Hv channels and voltage-sensitive phosphatases. An alignment of the residues corresponding to S1-S4 segments. Linker regions between the TM segments were not included.
Acknowledgments
We thank B. Roux, V. Jogini, F. Bezanilla for insightful discussions; V. Vásquez, Julio Cordero for critical reading and comments on the manuscript; R. Mackinnon for the coordinates of the KvAP model and V. Jogini for the coordinates of the Kv1.2 model. This work was supported by US National Institutes of health grants E.P and American Heart Association Postdoctoral Fellowship to S.C.
Footnotes
Supplemental data include four figures and a table showing sequence alignment of eukaryotic Kv channels, Hv channels and voltage-sensitive phosphatases.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahern CA, Horn R. Stirring up controversy with a voltage sensor paddle. Trends Neurosci. 2004;27:303–307. doi: 10.1016/j.tins.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–375. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach C, Froncisz W, Hyde JS, Hubbell WL. Conformation of spin-labeled melittin at membrane surfaces investigated by pulse saturation recovery and continuous wave power saturation electron paramagnetic resonance. Biophys J. 1989;56:1183–1191. doi: 10.1016/S0006-3495(89)82765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros JA, Deupi X, Olivella M, Haaksma EE, Pardo L. Serine and threonine residues bend alpha-helices in the chi(1) = g(−) conformation. Biophys J. 2000;79:2754–2760. doi: 10.1016/S0006-3495(00)76514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F. Voltage sensor movements. J Gen Physiol. 2002;120:465–473. doi: 10.1085/jgp.20028660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F. Voltage-gated ion channels. IEEE Trans Nanobioscience. 2005a;4:34–48. doi: 10.1109/tnb.2004.842463. [DOI] [PubMed] [Google Scholar]
- Bezanilla F. The voltage-sensor structure in a voltage-gated channel. Trends Biochem Sci. 2005b;30:166–168. doi: 10.1016/j.tibs.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Bezanilla F. The action potential: from voltage-gated conductances to molecular structures. Biol Res. 2006;39:425–435. doi: 10.4067/s0716-97602006000300005. [DOI] [PubMed] [Google Scholar]
- Blaber M, Zhang XJ, Matthews BW. Structural basis of amino acid alpha helix propensity. Science. 1993;260:1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- Campos FV, Chanda B, Roux B, Bezanilla F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc Natl Acad Sci U S A. 2007;104:7904–7909. doi: 10.1073/pnas.0702638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha A, Bezanilla F. Characterizing voltage-dependent conformational changes in the Shaker K+ channel with fluorescence. Neuron. 1997;19:1127–1140. doi: 10.1016/s0896-6273(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Choe S, Cushman S, Baker KA, Pfaffinger P. Excitability is mediated by the T1 domain of the voltage-gated potassium channel. Novartis Found Symp. 2002;245:169–175. discussion 175-167, 261-164. [PubMed] [Google Scholar]
- Cornette JL, Cease KB, Margalit H, Spouge JL, Berzofsky JA, DeLisi C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J Mol Biol. 1987;195:659–685. doi: 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- Cuello LG, Cortes DM, Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- Cushman SJ, Nanao MH, Jahng AW, DeRubeis D, Choe S, Pfaffinger PJ. Voltage dependent activation of potassium channels is coupled to T1 domain structure. Nat Struct Biol. 2000;7:403–407. doi: 10.1038/75185. [DOI] [PubMed] [Google Scholar]
- Deng D, Roux B. Hydration of Amino Acid Side Chains: Nonpolar and Electrostatic Contributions Calculated from Staged Molecular Dynamics Free Energy Simulations with Explicit Water Molecules. J Phys Chem B. 2004;108:16567–16576. [Google Scholar]
- Donnelly D, Overington JP, Blundell TL. The prediction and orientation of alpha-helices from sequence alignments: the combined use of environment-dependent substitution tables, Fourier transform methods and helix capping rules. Protein Eng. 1994;7:645–653. doi: 10.1093/protein/7.5.645. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Weiss RM, Terwilliger TC. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc Natl Acad Sci U S A. 1984;81:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farahbakhsh ZT, Altenbach C, Hubbell WL. Spin labeled cysteines as sensors for protein-lipid interaction and conformation in rhodopsin. Photochem Photobiol. 1992;56:1019–1033. doi: 10.1111/j.1751-1097.1992.tb09725.x. [DOI] [PubMed] [Google Scholar]
- Freites JA, Tobias DJ, White SH. A voltage-sensor water pore. Biophys J. 2006;91:L90–92. doi: 10.1529/biophysj.106.096065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TM, Matthews BW. Intrahelical hydrogen bonding of serine, threonine and cysteine residues within alpha-helices and its relevance to membrane-bound proteins. J Mol Biol. 1984;175:75–81. doi: 10.1016/0022-2836(84)90446-7. [DOI] [PubMed] [Google Scholar]
- Gross A, Hubbell WL. Identification of protein side chains near the membrane-aqueous interface: a site-directed spin labeling study of KcsA. Biochemistry. 2002;41:1123–1128. doi: 10.1021/bi015828s. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hong KH, Miller C. The lipid-protein interface of a Shaker K(+) channel. J Gen Physiol. 2000;115:51–58. doi: 10.1085/jgp.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, McHaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4:779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- Islas LD, Sigworth FJ. Electrostatics and the gating pore of Shaker potassium channels. J Gen Physiol. 2001;117:69–89. doi: 10.1085/jgp.117.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- Jogini V, Roux B. Dynamics of the Kv1.2 voltage-gated K+ channel in a Membrane Environment. Biophys J. 2007 doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y, Baldwin TJ, Jan YN, Jan LY. Primary structure and functional expression of a mouse inward rectifier potassium channel. Nature. 1993;362:127–133. doi: 10.1038/362127a0. [DOI] [PubMed] [Google Scholar]
- Kurata HT, Soon GS, Fedida D. Altered state dependence of c-type inactivation in the long and short forms of human Kv1.5. J Gen Physiol. 2001;118:315–332. doi: 10.1085/jgp.118.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine M, Lin MC, Bannister JP, Silverman WR, Mock AF, Roux B, Papazian DM. Atomic proximity between S4 segment and pore domain in Shaker potassium channels. Neuron. 2003;39:467–481. doi: 10.1016/s0896-6273(03)00468-9. [DOI] [PubMed] [Google Scholar]
- Larson SM, Davidson AR. The identification of conserved interactions within the SH3 domain by alignment of sequences and structures. Protein Sci. 2000;9:2170–2180. doi: 10.1110/ps.9.11.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson HP, Baker OS, Dhillon DS, Isacoff EY. Transmembrane movement of the shaker K+ channel S4. Neuron. 1996;16:387–397. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- Lee SY, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc Natl Acad Sci U S A. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DI, Deutsch C. Recovery from C-type inactivation is modulated by extracellular potassium. Biophys J. 1996;70:798–805. doi: 10.1016/S0006-3495(96)79619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Smerin Y, Hackos DH, Swartz KJ. alpha-helical structural elements within the voltage-sensing domains of a K(+) channel. J Gen Physiol. 2000a;115:33–50. doi: 10.1085/jgp.115.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Smerin Y, Hackos DH, Swartz KJ. A localized interaction surface for voltage-sensing domains on the pore domain of a K+ channel. Neuron. 2000b;25:411–423. doi: 10.1016/s0896-6273(00)80904-6. [DOI] [PubMed] [Google Scholar]
- Li-Smerin Y, Swartz KJ. Helical structure of the COOH terminus of S3 and its contribution to the gating modifier toxin receptor in voltage-gated ion channels. J Gen Physiol. 2001;117:205–218. doi: 10.1085/jgp.117.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Loots E, Isacoff EY. Protein rearrangements underlying slow inactivation of the Shaker K+ channel. J Gen Physiol. 1998;112:377–389. doi: 10.1085/jgp.112.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Klem AM, Ramu Y. Ion conduction pore is conserved among potassium channels. Nature. 2001;413:809–813. doi: 10.1038/35101535. [DOI] [PubMed] [Google Scholar]
- Lu Z, Klem AM, Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J Gen Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur MW, Thornton JM. Influence of proline residues on protein conformation. J Mol Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- Minor DL., Jr The neurobiologist’s guide to structural biology: a primer on why macromolecular structure matters and how to evaluate structural data. Neuron. 2007;54:511–533. doi: 10.1016/j.neuron.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor DL, Lin YF, Mobley BC, Avelar A, Jan YN, Jan LY, Berger JM. The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel. Cell. 2000;102:657–670. doi: 10.1016/s0092-8674(00)00088-x. [DOI] [PubMed] [Google Scholar]
- Monks SA, Needleman DJ, Miller C. Helical structure and packing orientation of the S2 segment in the Shaker K+ channel. J Gen Physiol. 1999;113:415–423. doi: 10.1085/jgp.113.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monne M, Nilsson I, Elofsson A, von Heijne G. Turns in transmembrane helices: determination of the minimal length of a “helical hairpin” and derivation of a fine-grained turn propensity scale. J Mol Biol. 1999;293:807–814. doi: 10.1006/jmbi.1999.3183. [DOI] [PubMed] [Google Scholar]
- Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- Olcese R, Latorre R, Toro L, Bezanilla F, Stefani E. Correlation between charge movement and ionic current during slow inactivation in Shaker K+ channels. J Gen Physiol. 1997;110:579–589. doi: 10.1085/jgp.110.5.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olcese R, Sigg D, Latorre R, Bezanilla F, Stefani E. A conducting state with properties of a slow inactivated state in a shaker K(+) channel mutant. J Gen Physiol. 2001;117:149–163. doi: 10.1085/jgp.117.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian DM, Shao XM, Seoh SA, Mock AF, Huang Y, Wainstock DH. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 1995;14:1293–1301. doi: 10.1016/0896-6273(95)90276-7. [DOI] [PubMed] [Google Scholar]
- Pathak MM, Yarov-Yarovoy V, Agarwal G, Roux B, Barth P, Kohout S, Tombola F, Isacoff EY. Closing in on the resting state of the shaker k(+) channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Perozo E, Cortes DM, Cuello LG. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat Struct Biol. 1998;5:459–469. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- Perozo E, Cuello LG, Cortes DM, Liu YS, Sompornpisut P. EPR approaches to ion channel structure and function. Novartis Found Symp. 2002;245:146–158. doi: 10.1002/0470868759.ch10. discussion 158–164, 165-148. [DOI] [PubMed] [Google Scholar]
- Planells-Cases R, Ferrer-Montiel AV, Patten CD, Montal M. Mutation of conserved negatively charged residues in the S2 and S3 transmembrane segments of a mammalian K+ channel selectively modulates channel gating. Proc Natl Acad Sci U S A. 1995;92:9422–9426. doi: 10.1073/pnas.92.20.9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu Y, Xu Y, Lu Z. Enzymatic activation of voltage-gated potassium channels. Nature. 2006;442:696–699. doi: 10.1038/nature04880. [DOI] [PubMed] [Google Scholar]
- Rasmusson RL, Wang S, Castellino RC, Morales MJ, Strauss HC. The beta subunit, Kv beta 1.2, acts as a rapid open channel blocker of NH2-terminal deleted Kv1.4 alpha-subunits. Adv Exp Med Biol. 1997;430:29–37. doi: 10.1007/978-1-4615-5959-7_3. [DOI] [PubMed] [Google Scholar]
- Richardson J, Blunck R, Ge P, Selvin PR, Bezanilla F, Papazian DM, Correa AM. Distance measurements reveal a common topology of prokaryotic voltage-gated ion channels in the lipid bilayer. Proc Natl Acad Sci U S A. 2006;103:15865–15870. doi: 10.1073/pnas.0607532103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruta V, MacKinnon R. Localization of the voltage-sensor toxin receptor on KvAP. Biochemistry. 2004;43:10071–10079. doi: 10.1021/bi049463y. [DOI] [PubMed] [Google Scholar]
- Sands ZA, Sansom MS. How does a voltage sensor interact with a lipid bilayer? Simulations of a potassium channel domain. Structure. 2007;15:235–244. doi: 10.1016/j.str.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- Schrempf H, Schmidt O, Kummerlen R, Hinnah S, Muller D, Betzler M, Steinkamp T, Wagner R. A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. Embo J. 1995;14:5170–5178. doi: 10.1002/j.1460-2075.1995.tb00201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- Sigworth FJ. Voltage gating of ion channels. Q Rev Biophys. 1994;27:1–40. doi: 10.1017/s0033583500002894. [DOI] [PubMed] [Google Scholar]
- Soler-Llavina GJ, Chang TH, Swartz KJ. Functional interactions at the interface between voltage-sensing and pore domains in the Shaker K(v) channel. Neuron. 2006;52:623–634. doi: 10.1016/j.neuron.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Sorensen JB, Cha A, Latorre R, Rosenman E, Bezanilla F. Deletion of the S3-S4 linker in the Shaker potassium channel reveals two quenching groups near the outside of S4. J Gen Physiol. 2000;115:209–222. doi: 10.1085/jgp.115.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starace DM, Bezanilla F. Histidine scanning mutagenesis of basic residues of the S4 segment of the shaker k+ channel. J Gen Physiol. 2001;117:469–490. doi: 10.1085/jgp.117.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- Starace DM, Stefani E, Bezanilla F. Voltage-dependent proton transport by the voltage sensor of the Shaker K+ channel. Neuron. 1997;19:1319–1327. doi: 10.1016/s0896-6273(00)80422-5. [DOI] [PubMed] [Google Scholar]
- Subbiah RN, Kondo M, Campbell TJ, Vandenberg JI. Tryptophan scanning mutagenesis of the HERG K+ channel: the S4 domain is loosely packed and likely to be lipid exposed. J Physiol. 2005;569:367–379. doi: 10.1113/jphysiol.2005.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz KJ. Towards a structural view of gating in potassium channels. Nat Rev Neurosci. 2004;5:905–916. doi: 10.1038/nrn1559. [DOI] [PubMed] [Google Scholar]
- Tiwari-Woodruff SK, Schulteis CT, Mock AF, Papazian DM. Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits. Biophys J. 1997;72:1489–1500. doi: 10.1016/S0006-3495(97)78797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annu Rev Cell Dev Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- Treptow W, Tarek M. Environment of the gating charges in the Kv1.2 Shaker potassium channel. Biophys J. 2006;90:L64–66. doi: 10.1529/biophysj.106.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez V, Cortes DM, Furukawa H, Perozo E. An optimized purification and reconstitution method for the MscS channel: strategies for spectroscopical analysis. Biochemistry. 2007;46:6766–6773. doi: 10.1021/bi700322k. [DOI] [PubMed] [Google Scholar]
- Yang N, George AL, Jr, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K(+) channels. Proc Natl Acad Sci U S A. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellen G. The moving parts of voltage-gated ion channels. Q Rev Biophys. 1998;31:239–295. doi: 10.1017/s0033583598003448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Oligomeric state of the mutant A145C. In gel filtration chromatography, mutant A145C in the isolated-VSD of KvAP in octyl-glucoside (red) elutes as two peaks corresponding to a monomer and a dimer band on SDS-PAGE (Inset). In the presence of DTT a single monodisperse peak at the monomer elution volume is seen (black) which also runs as a monomer on SDS-PAGE. In the presence of decylmaltoside without DTT, mutant A145C elutes as a single peak at the elution volume of a dimer.
Figure S2. A comparison of spin-label mobility in S1 in the isolated-VSD and the FL-channel. An overlap of EPR spectral line shapes at the equivalent positions in the isolated-VSD (red) and the FL-channel (Black). Side-chains facing the core of the VSD bundle are shown in yellow and those facing outside (and therefore interacting with the PD in the FL-channel) are shown in red. Note that the residues facing into the VSD do not show any change in the line-shape in the two systems, while those facing the interface show pronounced increase in mobility.
Figure S3. A comparison of spin-label mobility in S2 in the isolated-VSD and the FL-channel. As seen in S1, the side chains facing the interface (red surface) show significant changes in line shapes in the isolated-VSD (red) in comparison to the FL-channel (black). On the other hand positions facing into the VSD core (yellow surface) do not show any significant change.
Figure S4. A plot of the delta values for the measured ΠO2 in the FL-channel and the isolated-VSD. The ΠO2 values were normalized against the maximum value within each data set before calculating the delta differences between the FL-channel and isolated-VSD. The dotted line denotes the average delta parameter for the S1/S2 and the S3/S4 segments.
Figure S5. Sequence alignment of eukaryotic Kv channels, Hv channels and voltage-sensitive phosphatases. An alignment of the residues corresponding to S1-S4 segments. Linker regions between the TM segments were not included.


