Abstract
Smad ubiquitination regulatory factor-2 (Smurf2) is an E3 ubiqutin ligase that plays a pivotal role in regulating the TGF-β signaling via selectively targeting key components of Smad pathway for degradation. In this study, we have investigated the regulation of Smurf2 expression, its target specificity and the functional implication of its induction in the fibrotic kidney. Immunohistochemical staining revealed that Smurf2 was upregulated specifically in renal tubules of kidney biopsies from patients with various nephropathies. In vitro, Smurf2 mRNA and protein were induced in human proximal tubular epithelial cells (HKC-8) upon TGF-β1 stimulation. Ectopic expression of Smurf2 was sufficient to reduce the steady-state levels of Smad2, but not Smad1, Smad3, Smad4 and Smad7, in HKC-8 cells. Interestingly, Smurf2 was also able to down-regulate the Smad transcriptional corepressors SnoN, Ski and TGIF. Inhibition of the proteasomal pathway prevented Smurf2-mediated down-regulation of Smad2 and Smad corepressors. Functionally, over-expression of Smurf2 enhanced the transcription of TGF-β-responsive promoter and augmented the TGF-β1-mediated E-cadherin suppression, as well as fibronectin and type I collagen induction in HKC-8 cells. These results indicate that Smurf2 specifically targets both positive and negative Smad regulators for destruction in tubular epithelial cells, thereby providing a complex fine-tuning of the TGF-β signaling. It appears that dysregulation of Smurf2 could contribute to an aberrant TGF-β/Smad signaling in the pathogenesis of kidney fibrosis.
Keywords: Smurf2, SnoN, renal fibrosis, TGF-β1, Smad, ubiquitination
Introduction
Transforming growth factor-β (TGF-β) plays a fundamental role in regulating many biological processes such as cell proliferation, apoptosis, differentiation and extracellular matrix production (2, 5). Smad proteins are central components of the TGF-β signaling. After TGF-β binding to its type II serine/threonine kinase receptor (TβRII), type I receptor (TβRI) is activated, resulting in the phosphorylation and activation of the downstream receptor-regulated Smads (R-Smads). Activated R-Smads (Smad2 and Smad3) heteroligomerize with the common partner Smad4 (co-Smad), and then translocate into the nucleus to control the transcription of TGF-β-responsive genes (17). This cascade of signal transduction events from the cell surface to the nucleus is tightly constrained by several negative regulatory mechanisms (15, 18, 19). Smad7, an inhibitory Smad (I-Smad), inhibits R-Smads phosphorylation by blocking their access to TβRI, and/or by promoting the degradation of the receptor complexes. In addition, several Smad transcriptional corepressors, including Ski (Sloan-Kettering Institute proto-oncogene), SnoN (Ski-related novel gene, non Alu-containing) and TGIF (TG-interacting factor), are capable of blocking R-Smads-mediated gene transcription (5, 18). In normal physiologic conditions, both positive and negative components of Smad signaling are delicately balanced and closely regulated, ensuring a homeostasis of TGF-β signal outputs.
Many studies have demonstrated an aberrant regulation of TGF-β signaling in the fibrotic kidney after chronic injury (1, 13, 24). It is well documented that renal fibrogenesis is associated with an increased TGF-β1 expression and post-translational activation, as well as an up-regulation of its receptors in various experimental nephropathies and in kidney biopsies from patients with renal failure (30, 37). Recently, evidence is emerging that several key components of Smad signaling are also dysregulated in the fibrotic kidney. For instance, SnoN and Ski are progressively down-regulated in mouse and rat models of obstructive nephropathy (7, 38). A reduced TGIF expression is also documented in the glomeruli of diabetic animals.(3) Similarly, several studies have revealed an anomalous expression of Smad2, Smad3 and Smad7 in diseased kidney after sustained insults (6, 22, 31). Interestingly, in addition to transcriptional regulation (28), many proteins in Smad signaling are subjected to modulation by the ubiquitin-mediated proteasomal degradation (7, 12, 27), a unique proteolytic pathway that selects, tags, and executes the orderly destruction of a wide variety of physiologically vital proteins such as signal transducers.
Ubiquitin-mediated proteolytic system consists of a highly organized cascade of enzymatic reactions that necessitates a ubiquitin-activating enzyme (E1), ubiquitin-conjugation enzyme (E2) and ubiquitin ligase (E3) (4, 16). The specificity and selectivity of target proteins for degradation are defined by particular E3 ubiquitin ligases. Smad ubiquitination regulatory factor-2 (Smurf2) is an E3 ligase that belongs to the HECT domain ubiquitin ligase family (11, 40), characterized by unique structural motifs that are homologous to the E6-AP COOH terminus. Structurally, Smurf2 features an N-terminal C2 domain, three WW domains and a C-terminal HECT ligase domain (9, 11). Smurf2 can specifically target certain members of Smad proteins and promotes their ubiquitin-dependent degradation, thereby controlling the cellular levels of these signaling mediators. By so doing, Smurf2 could play an important role in regulating numerous biologic processes ranging from embryogenesis, tumorogenesis to fibrotic disease progression.
We and others have recently shown a close association between Smurf2 expression and SnoN degradation in obstructive nephropathy (7, 27), suggesting a potential role of Smurf2 in regulating TGF-β signaling in vivo. In this study, we have continued this line of investigation by delineating Smurf2 regulation, its target specificity and its functional implication in the setting of chronic kidney fibrosis.
Materials and Methods
Human kidney samples and immunohistochemical staining
Human kidney specimens were obtained from diagnostic renal biopsies performed at the University of Pittsburgh Medical Center. Non-tumor kidney tissue from the patients who had renal cell carcinoma and underwent nephrectomy was used as normal controls. All studies involving human tissues were approved by the Institutional Review Board at the University of Pittsburgh. Immunohistochemical staining was performed using established procedures as described previously (29). Briefly, paraffin-embedded human kidney sections (3–4 µm thickness) were prepared by a routine procedure and stained with the specific primary antibody against Smurf2 (sc-25511) (Santa Cruz Biotechnology, Santa Cruz, CA). As a negative control, the primary antibody was replaced with nonimmune normal goat IgG, and no staining occurred.
Cell culture and treatment
Human proximal tubular epithelial cells (HKC, clone 8) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s F-12 medium (1:1) supplemented with 5% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA), as described previously (23, 36). Cells were typically seeded at approximately 70% confluence in complete medium containing 5% FBS for 24 h, and then serum-starved for 16 h, followed by incubation with 2 ng/ml TGF-β1 for various periods of time as indicated. In some experiments, cells were pretreated with either various inhibitors at given concentrations or vehicle (0.1% DMSO) 0.5 h before incubation with TGF-β1. Human recombinant TGF-β1 was purchased from the R & D Systems (Minneapolis, MN). PD98059 (Mek1 inhibitor), wortmannin (phosphatidylinositol 3-kinase inhibitor), SC-68376 (p38 mitogen-activated protein kinase inhibitor) and SP600125 (c-Jun amino-terminal kinase inhibitor) were purchased from Calbiochem (La Jolla, CA).
RNA Isolation and reverse transcriptase (RT)-PCR
Total RNA isolation, reverse transcription of the RNA, and PCR amplification were performed as described (27). Briefly, the first strand cDNA synthesis was carried out by using a Reverse Transcription System kit according to the instructions of the manufacturer (Promega, Madison, WI). PCR amplification was performed using HotStar Taq® Master Mix Kit (Qiagen, Valencia, CA). The primer sequences were as follow: Smurf, 5’-CGCTTGATCCAAAGTGGAAT-3’ (sense), and 5’-GGTTGATGGCATTGGAAAGA-3’ (antisense); collagen I, 5’-CCAAATCTGTCTCCCCAGAA-3’ (sense), and 5’-TCAAAAACGAAGGGGAGATG-3’ (antisense); β-actin, 5’-TCAAGATCATTGCTCCTCCTGAGC-3’ (sense), and 5’-TGCTGTCACCTTCACCGTTCCAGT-3’ (antisense). Relative levels of Smurf2 and collagen I mRNA were calculated after normalizing with house-keeping gene β-actin.
Western blot analysis
Detection of protein expression by Western blot was carried out according to the established protocols described previously (36). The primary antibodies used were as follows: anti-Smurf2 (sc-25511), anti-SnoN (sc-9141), anti-Ski (sc-9140), anti-TGIF (sc-17800), and anti-actin (sc-1616) (Santa Cruz Biotechnology), anti-HA (6E2) (Cell Signaling Technology, Beverly, MA), anti-Smad2 (51–1300) and anti-Smad3 (51–1500) (Zymed Laboratories, South San Francisco,CA), anti-α-tubulin (T-9026) (Sigma), anti-GAPDH (Ambion, Austin, TX), anti-E-cadherin (clone 36) and anti-fibronectin (clone 10) (BD Biosciences, San Diego, CA). Quantitative analysis of Western blot data was performed by measuring the intensity of the band signals with the use of NIH Image analysis software.
Plasmids transient transfection
For transient transfection, HKC-8 cells were seeded in six-well plates at 5 × 105 cells per well. The cells were then co-transfected for 24 h with 1.0 µg human HA-tagged Smad1, HA-tagged Smad2, HA-tagged Smad3, HA-tagged Smad4, HA-tagged Smad7, HA-tagged SnoN, HA-tagged Ski, or HA-tagged TGIF expression vector plus 1.0 µg empty pcDNA3 vector or Flag-tagged Smurf2 expression vector, as described previously (11), using Lipofectamine 2000 reagent according to the instructions specified by the manufacturer (Invitrogen). HA-tagged SnoN and HA-tagged Ski plasmids were provided by Dr. R. Weinberg (Massachusetts Institute of Technology, Cambridge, MA) (26), whereas HA-tagged TGIF expression vector was obtained from Dr. J. Massague (Memorial Sloan-Kettering Cancer Center, New York, NY) (14). In some experiments, transfected cells were also treated with MG132 (2.5 µM) for 24 h. In addition, HKC-8 cells were also transiently transfected with HA-tagged Smad2, HA-tagged Smad3 or Flag-tagged Smurf2 expression vector for 24 h, and then treated with various concentrations of TGF-β1 for 48 or 72 h. The empty pcDNA3 vector was used as a mock transfection control.
Reporter constructs and luciferase Assay
The reporter construct p3TP-Lux was provided by Dr. J. Massague. HKC-8 cells were co-transfected by using Lipofectamine 2000 reagent with p3TP-Lux luciferase reporter construct (1.0 µg) plus Flag-Smurf2 (1.0 µg) expression vector or empty pcDNA3 vector. A fixed amount (0.1 µg) of internal control reporter Renilla reniformis luciferase driven under thymidine kinase (TK) promoter (pRL-TK; Promega) was also co-transfected for normalizing the transfection efficiency. After transfection for 24 h, cells were treated without or with TGF-β1 (2 ng/ml) for an additional 48 h. Luciferase assay was performed using the Dual Luciferase Assay System kit according to the manufacturer’s protocols (Promega). Relative luciferase activity (arbitrary unit) was reported as fold induction over controls after normalizing for transfection efficiency.
Small interfering RNA inhibition experiment
For small interfering RNA (siRNA) inhibition studies, HKC-8 cells were transiently transfected with negative control siRNA (Am4635), Smad2 siRNA (ID#115715) or Smad3 siRNA(ID# 107877) (Ambion, Austin, TX) at the final concentration of 120 nM by using oligofectamine reagent according to the instructions specified by the manufacturer (Invitrogen). After transfection for 72 h, whole-cell lysates were collected for assessing the expression of Smad2 and Smad3 by Western blot analyses. In some experiments, after transfection for 24 h, cells were then serum-starved for 16 h, followed by treated with various concentrations of TGF-β1 for an additional 48 h.
Statistical analysis
Statistical analysis was performed using SigmaStat software (Jandel Scientific Software, San Rafael, CA). Comparisons between groups were made using one-way analysis of variance, followed by the Student-Newman-Keuls test. A P value of less than 0.05 was considered significant.
Results
Smurf2 induction in renal tubules of human fibrotic kidneys
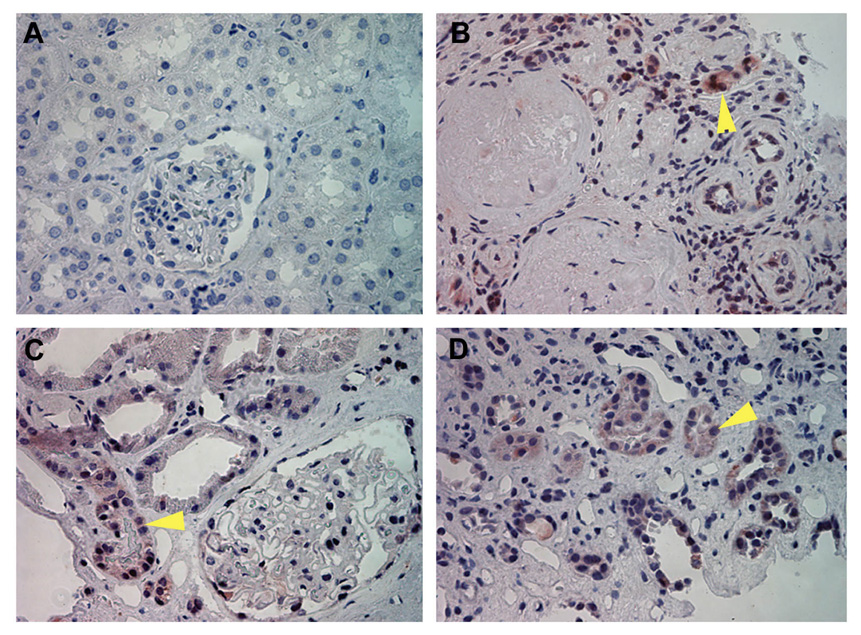
Earlier studies demonstrate that SnoN degradation is closely associated with an up-regulation of Smurf2 in the fibrotic kidney after unilateral ureteral obstruction (27). To explore the clinical relevance of this observation, we investigated Smurf2 protein expression in human kidney biopsies from patients with different nephropathies. As shown in Figure 1A, little Smurf2 expression was detected by immunohistochemical staining in normal control, non-tumor kidney tissues (n=3). However, increased Smurf2 protein was found in fibrotic kidneys with diverse etiologies, such as diabetic nephropathy with focal and segmental glomerulosclerosis (n=1) (Figure 1B), hypertension with moderate interstitial fibrosis (n=4) (Figure 1C) and lupus nephritis with moderate interstitial fibrosis (n=1) (Figure 1D). Smurf2 was localized primarily in renal tubules, but not in glomeruli.
Figure 1.
Smurf2 is induced in renal tubules of human kidney biopsies of patients with different nephropathies. Immunohistochemical staining demonstrated Smurf2 expression and localization in normal control and diseased kidneys. Representative micrographs are shown. (A) Normal kidney (n=3), (B) Diabetic nephropathy with focal and segmental glomerulosclerosis (n=1), (C) Hypertension with moderate interstitial fibrosis (n=4), (D) Lupus nephritis with severe tubular atrophy, moderate interstitial fibrosis (n=1) (×400). Arrowheads indicate the positive staining of Smurf2.
TGF-β1 induces tubular Smurf2 expression in vitro
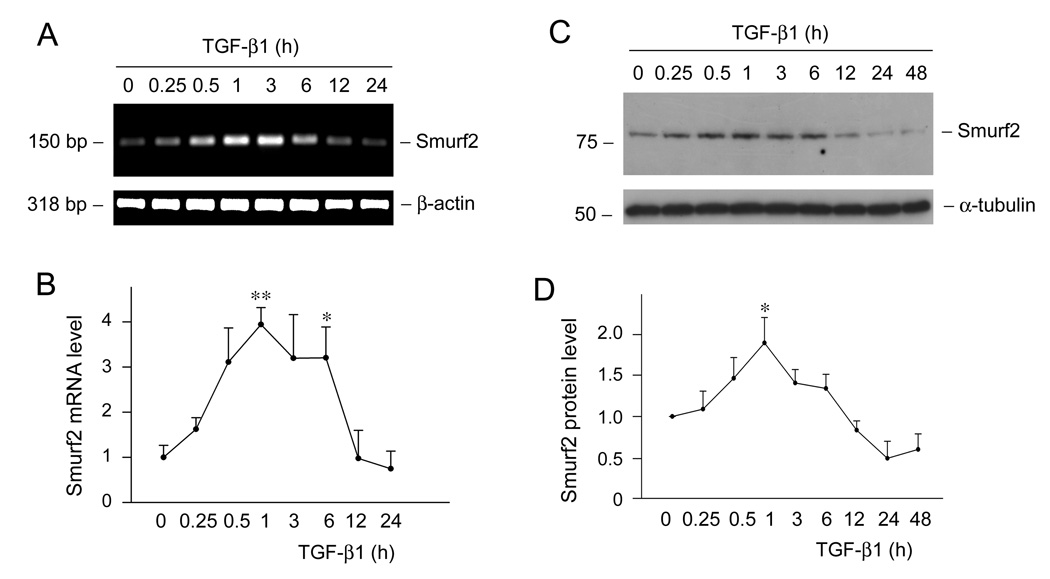
We next investigated the potential inducer that triggers Smurf2 upregulation by using cultured human proximal tubular epithelial cells. As TGF-β1 is induced in virtually all chronic kidney diseases, we sought to examine whether it is responsible for Smurf2 induction under pathologic conditions. Therefore, HKC-8 cells were treated with 2 ng/ml of TGF-β1 for various periods of time as indicated. As presented in Figure 2A, TGF-β1 markedly induces Smurf2 mRNA expression. Quantitative determination revealed a 4-fold induction of Smurf2 mRNA at 1 h after TGF-β1 treatment in HKC-8 cells. Western blot analyses also demonstrated a Smurf2 protein induction in HKC-8 cells by TGF-β1 (Figure 2, C and D). Induction of Smurf2 was transient, and both Smurf2 mRNA and protein returned towards the baseline levels after 6 h incubation with TGF-β1.
Figure 2.
TGF-β1 induces Smurf2 mRNA and protein expression in human proximal tubular epithelial cells. (A, B) Semi-quantitative RT-PCR analyses displayed the induction of Smurf2 mRNA by TGF-β1. HKC-8 cells were treated with 2 ng/ml of TGF-β1 for various periods of time as indicated. Shown in (A) is the representative RT-PCR result. (B) Quantitative determination of Smurf2 mRNA abundance after normalization with β-actin. Data are presented as mean ± SEM of three experiments. *P < 0.05 versus controls. **P < 0.01 versus controls. (C, D) Western blot analyses demonstrated the induction of Smurf2 protein by TGF-β1. Whole-cell lysates were immunoblotted with antibodies against Smurf2 and α-tubulin, respectively. (D) Quantitative determination of the relative abundance of Smurf2 after normalization with α-tubulin. Data are presented as mean ± SEM of three experiments. *P < 0.05 versus controls.
Smurf2 induction by TGF-β1 requires Smad signaling
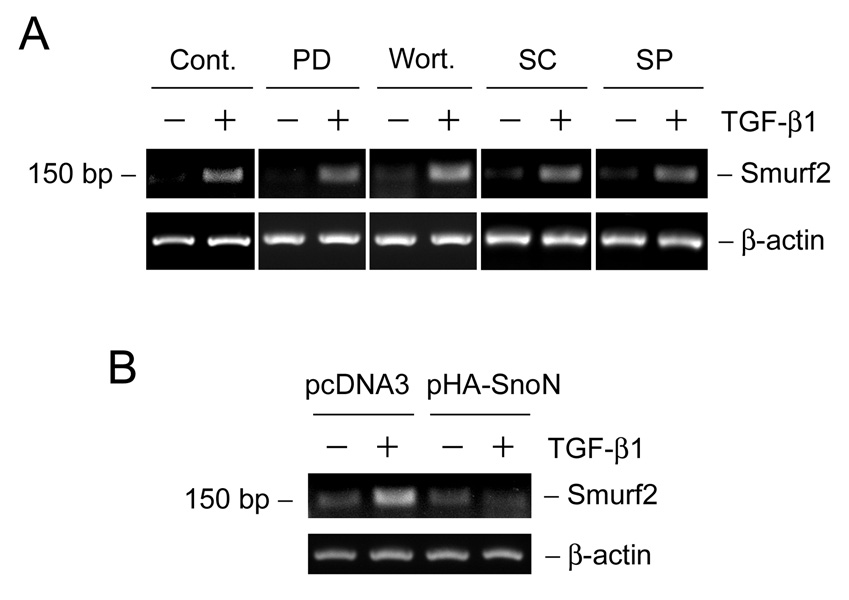
We further explored the potential mechanism of Smurf2 induction by delineating the critical signal pathway involved. As shown in Figure 3A, pharmacologic inhibitor of Mek1 kinase (PD98059), PI-3-kinase (wortmannin), p38 mitogen-activated protein kinase (MAPK) (SC68376) or c-Jun amino-terminal kinase (JNK) (SP600125) did not block Smurf2 induction after TGF-β1 treatment in HKC-8 cells. To examine the potential involvement of Smad signaling, HKC-8 cells were transiently transfected with SnoN expression vector or empty pcDNA3 vector, and then treated without or with TGF-β1. We found that over-expression of Smad corepressor SnoN completely abolished the TGF-β1-mediated Smurf2 mRNA induction in HKC-8 cells, indicating its dependency on Smad signaling.
Figure 3.
Smad signaling is required for the TGF-β1-mediated induction of smurf2 in tubular epithelial cells. (A) HKC-8 cells were pretreated with either various chemical inhibitors or vehicle (DMSO) as indicated for 30 min, followed by incubation in the absence or presence of TGF-β1 (2 ng/ml) for 1 h. PD, PD98059 (10 µM); wort., wortmannin (10 nM); SC, SC68376 (20 µM); SP, SP600125 (20 µM). (B) HKC-8 cells were transiently transfected with SnoN expression vector or empty pcDNA3 vector for 24 h, and then treated without or with TGF-β1 (2 ng/ml) for 1 h. The steady-state levels of Smurf2 mRNA were assessed by RT-PCR analyses.
Smurf2 specifically targets Smad2 and Smad corepressors for degradation
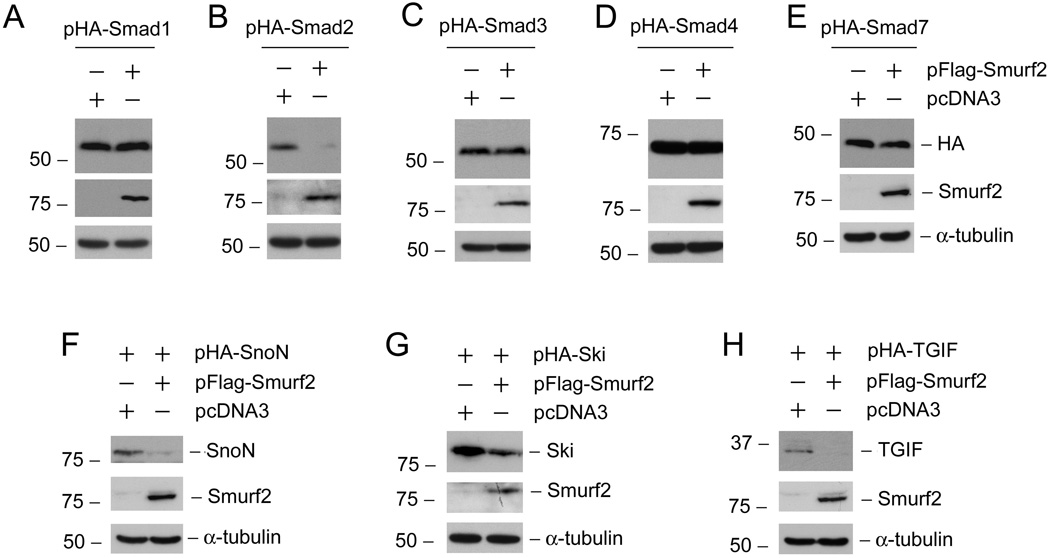
Smurf2 is an E3 ligase that regulates the lifespan of intracellular mediators in TGF-β signal circuit. To identify and test the specificity of Smurf2 targets, we co-transfected HKC-8 cells with HA-tagged Smad proteins and Flag-tagged Smurf2 expression vector or empty pcDNA3 vector, respectively, followed by analyzing HA-tagged Smads abundances with Western blotting. As shown in Figure 4 (A–E), co-transfection with Smad2 and Smurf2 expression vectors completely abolished HA-tagged Smad2 expression in HKC-8 cells, whereas Smurf2 did not significantly affect the expression and protein abundance of Smad1, Smad3, Smad4 and Smad7. We next examined the effects of Smurf2 on the abundance of Smad transcriptional corepressors by using the same approach. As presented in Figure 4 (F–H) , Smurf2 expression clearly down-regulated SnoN, Ski and TGIF protein expression. Altogether, these results indicate that Smurf2 specifically down-regulate the protein expression of Smad2, SnoN, Ski and TGIF in tubular epithelial cells.
Figure 4.
Differential effects of Smurf2 on the abundance of various Smads and their transcriptional corepressors in tubular epithelia cells. HKC-8 cells were transiently co-transfected with 1.0 µg HA-tagged Smads, or their corepressors plus either 1.0 µg Flag-tagged Smurf2 expression vector or empty pcDNA3 vector as indicated for 24 h. Whole cell lysates were immunoblotted with antibodies against HA, SnoN, Ski, TGIF, Smurf2 and α-tubulin, respectively.
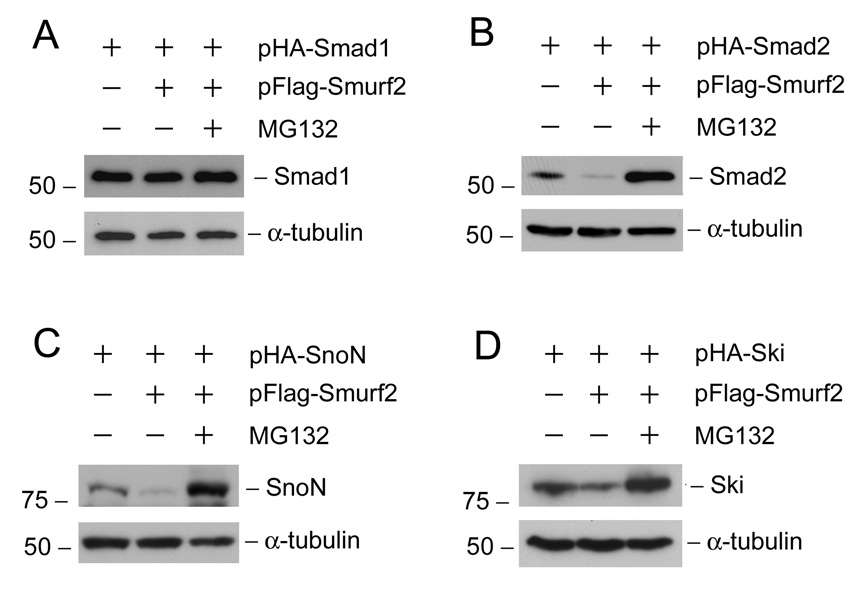
To investigate the mechanism by which Smurf2 inhibits Smad2 and Smad corepressors expression, we sought to examine whether blockade of the proteasomal-dependant degradation prevents the Smurf2-mediated down-regulation of these proteins in tubular epithelial cells. As shown in Figure 5, proteasomal degradation inhibitor MG132 completely restored Smad2, SnoN and Ski expression after ectopic expression of Smurf2. Notably, the protein abundance of Smad2, SnoN and Ski in the groups with Smurf2 over-expression and MG132 treatment was even greater than that in control groups without exogenous Smurf2 and MG132 (lane 3 vs. lane 1, Figure 5, B–D). This probably reflects that endogenous Smurf2 is also involved in the HA-tagged Smad2, SnoN and Ski degradation. As a negative control, Smad1 was not subjected to Smurf2-mediated degradation, and thus MG132 treatment caused little alteration in its abundance in HKC-8 cells (Figure 5A).
Figure 5.
Down-regulation of Smad2 and Smad corepressors by Smurf2 is mediated by proteasomal-dependant degradation in tubular epithelial cells. HKC-8 cells were transiently co-transfected with 1.0 µg HA-tagged Smad2, SnoN or Ski expression vector plus either 1.0 µg Flag-tagged Smurf2 expression vector or empty pcDNA3 vector as indicated. Immediately after transfection, cells were treated without or with 2.5 µM MG132 for 24 h. The HA-tagged Smad1expression vector was also transfected as a negative control. Whole cell lysates were immunoblotted with antibodies against HA, SnoN, Ski and α-tubulin, respectively.
Ectopic expression of Smurf2 enhances the TGF-β1-mediated gene transcription and fibrogenic action
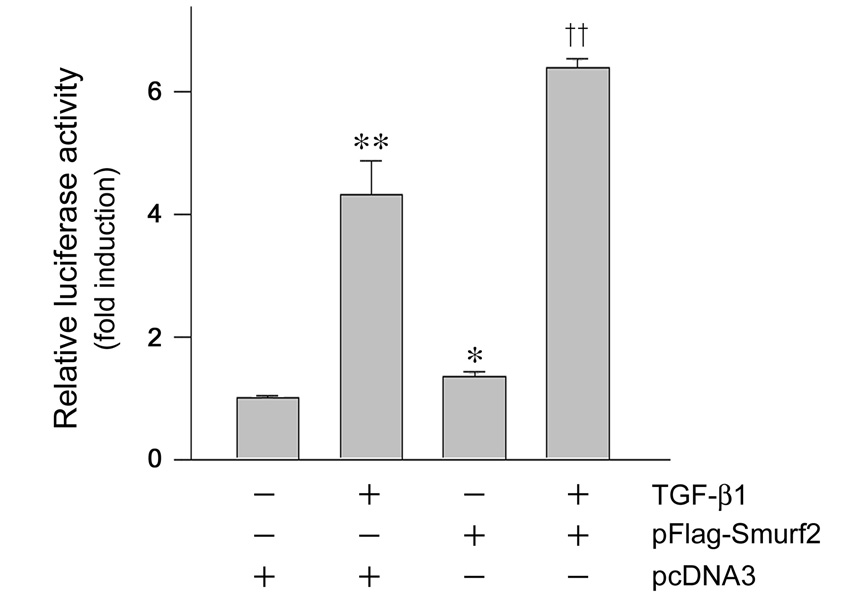
The observation that Smurf2 concurrently targets both Smad2 and Smad corepressors makes its influence on TGF-β signaling uncertain. To clarify this, we first examined the effects of Smurf2 on TGF-β1-mediated gene transcription in a promoter reporter assay. HKC-8 cells were transiently co-transfected with p3TP-Lux luciferase reporter construct and Flag-tagged Smurf2 expression vector or empty pcDNA3 vector, followed by incubation without or with TGF-β1. As shown in Figure 6, forced expression of Smurf2 promoted p3TP-Lux luciferase reporter activity in the absence or presence of TGF-β1. These data suggest that despite its impact on both positive (Smad2) and negative (SnoN, Ski and TGIF) regulators of TGF-β signaling, the net effect of Smurf2 appears to enhance the TGF-β-mediated gene transcription.
Figure 6.
Overexpression of Smurf2 enhances the TGF-β1-mediated gene transcription. HKC-8 cells were transiently co-transfected with p3TP-Lux luciferase reporter construct and Flag-tagged Smurf2 expression vector or empty pcDNA3 vector for 24 h, followed by incubation without or with 2 ng/ml of TGF-β1 for 48 h. Relative luciferase activities (arbitrary unit) are calculated after normalization of transfection efficiency, and presented as mean ± SEM of three experiments. *P < 0.01, **P < 0.001 versus pcDNA3 group in the absence of TGF-β1; ††P< 0.001 versus pcDNA3 group in the presence of TGF-β1.
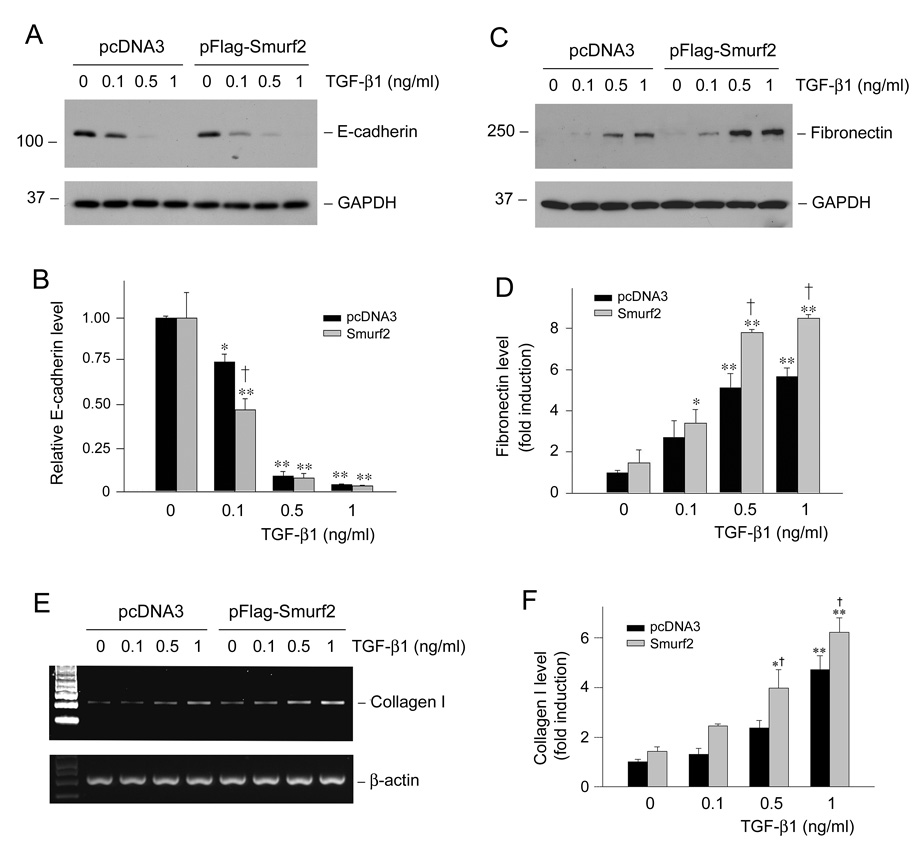
To ascertain the effect of Smurf2 on expression of the fibrosis-related genes, we examined its influence on E-cadherin suppression, as well as on fibronectin and type I collagen induction triggered by TGF-β1. As demonstrated in Figure 7, ectopic expression of exogenous Smurf2 moderately sensitized TGF-β1’s effect and promoted E-cadherin suppression. Likewise, Smurf2 also enhanced the TGF-β1-mediated fibronectin (Figure 7, C and D) and type I collagen (Figure 7, E and F) induction. These data suggest that increased expression of Smurf2 promotes the fibrogenic action of TGF-β1 in tubular epithelial cells.
Figure 7.
Overexpression of Smurf2 amplifies the TGF-β1-mediated fibrogenic action in tubular epithelial cells. HKC-8 cells were transiently transfected with Flag-tagged Smurf2 expression vector or empty pcDNA3 vector for 24 h, followed by incubation without or with various amounts of TGF-β1 as indicated for 72 h. Representative Western blot analyses show the expression of E-cadherin (A) and fibronectin (C), respectively. Graphical presentations show the relative abundance of E-cadherin (B) and fibronectin (D) after normalization with GAPDH. Representative RT-PCR analyses (E) and graphical presentation (F) show the expression of type I collagen mRNA after various treatments. Relative levels (fold induction over pcDNA3 group in the absence of TGF-β1) are presented as mean ± SEM of three experiments. *P < 0.05, **P < 0.001 versus pcDNA3 group in the absence of TGF-β1. †P < 0.05 versus pcDNA3 group in the presence of TGF-β1.
Both Smad2 and Smad3 are indispensable for TGF-β1-mediatd fibronection induction
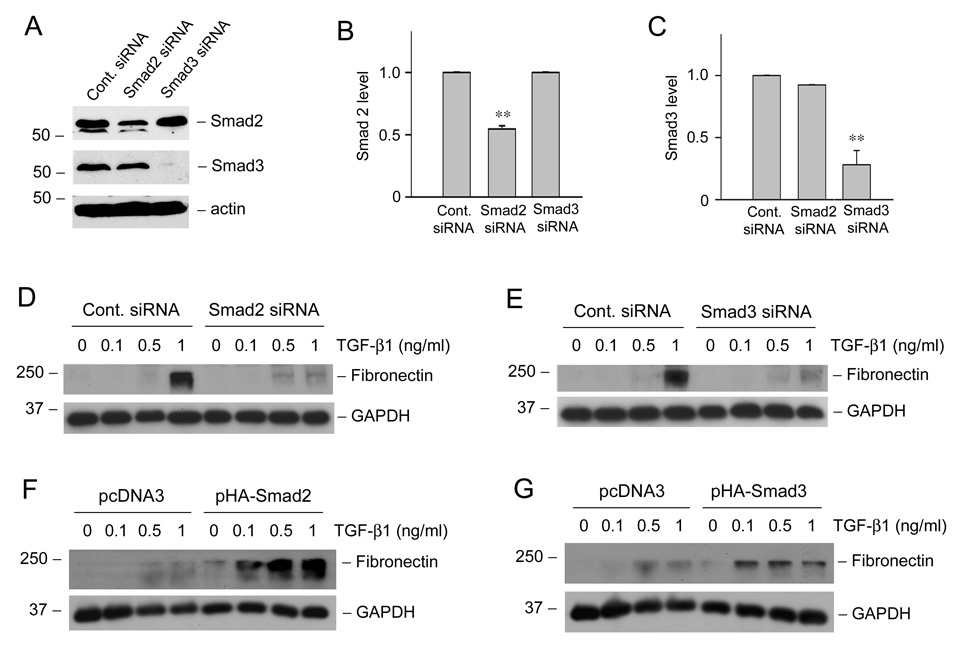
Smad2 and Smad3 display both common and distinctive functions in transmitting TGF-β signaling. We next sought to test whether Smad2 or Smad3 is indispensable for TGF-β1-mediated matrix production by using siRNA approach. HKC-8 cells were transiently transfected with control, Smad2 or Smad3 siRNA, respectively. Specific suppression of Smad2 and Smad3 expression was confirmed by Western blot analysis (Figure 8, A–C). As shown in Figure 8D, knockdown of Smad2 largely abrogated fibronectin induction by TGF-β1 in HKC-8 cells. Similarly, down-regulation of Smad3 by siRNA inhibition also essentially abolished the TGF-β1-mediated fibronectin expression (Figure 8E). Consistently, we found that overexpression of Smad2 or Smad3 via transfection markedly enhanced the fibronectin expression induced by TGF-β1 in HKC-8 cells. Therefore, both Smad2 and Smad3 are indispensable for TGF-β1-mediated matrix production by tubular epithelial cells.
Figure 8.
Smad2 and Smad3 mediate fibronection induction by TGF-β1 in tubular epithelial cells. (A–C) Knockdown of Smad2 and Smad3 expression by siRNA. HKC-8 cells were transiently transfected with control siRNA, Smad2 siRNA or Smad3 siRNA (120 nM). Representative Western blots show the expression levels of Smad2 and Smad3 (A) at 72 h after transfection. Graphic presentations of the relative abundance of Smad2 (B) and Smad3 (C) protein levels after normalization with actin are also given. Data (relative to the controls = 1.0) are presented as mean ± SEM of four experiments. ** P <0.01 versus control siRNA group. (D, E) Knockdown of Smad2 and Smad3 expression inhibited the TGF-β1-mediated fibronectin induction. HKC-8 cells were transiently transfected with control siRNA, Smad2 siRNA or Smad3 siRNA (120 nM) for 24 h, followed by incubation without or with different concentrations of TGF-β1 for 48 h. (F, G) Overexpression of Smad2 and Smad3 enhance TGF-β1-mediated fibronectin expression in tubular epithelial cells. HKC-8 cells were transiently transfected with empty pcDNA3 vector, HA-tagged Smad2 or HA-tagged Smad3 expression vector for 24 h, respectively, and then treated with TGF-β1 for an additional 48 h. Whole-cell lysates were immunoblotted with anti-fibronectin and anti-GAPDH antibodies, respectively.
Discussion
The ubiquitin-mediated proteasomal degradation pathway is an evolutionary conserved enzymatic cascade that controls the lifespan of many physiologically important proteins such as cell cycle regulators, signal transducers and transcription factors (4, 16). Smurf2, an E3 ubiquitin ligase, is involved in the regulation of the stability of numerous components of TGF-β signaling (11, 40). The results presented in this study demonstrate that Smurf2 protein is specifically up-regulated in renal tubules of the kidney biopsies from patients with various nephropathies. Smurf2 selectively targets Smad2 and Smad transcriptional corepressors Ski, SnoN and TGIF for degradation in human tubular epithelial cells. Furthermore, increased Smurf2 renders tubular epithelial cells susceptible to TGF-β1 stimulation, leading to an amplified, more efficient TGF-β signal transmission. These findings suggest that dysregulation of Smurf2 expression affects the profibrotic TGF-β/Smad signaling, and thus could contribute to the development and progression of human kidney fibrotic diseases.
Smurf2 exhibits an overall 83% sequence identity with Smurf1 (11), another E3 ubiquitin ligase of the C2-WW-HECT domain family that was originally identified through its interacting with Smad1 in a yeast two-hybrid screening (41). While Smurf1 contains two WW domains, Smurf2 comprises three WW domains, resulting from a 30 amino-acid insertion downstream of the phospholipid/calcium-binding C2 domain (11). Accumulating evidence indicates that Smurf1 and Smurf2 may be functionally distinct in terms of target specificity. Smurf1 is indispensable for the regulation of intracellular mediators of the bone morphogenetic proteins (BMP) signaling by specifically targeting Smad1 and Smad5 for degradation (41). Smurf2, as shown in this study as well as in a previous report (11), preferentially targets Smad2, a major R-Smad of TGF-β signaling, for ubiquitination and proteasomal degradation. In this regard, it appears clear that Smurf1 and Smurf2 differentially control the BMP and TGF-β signaling, respectively, by selectively targeting different Smads for destruction. It is of interest to point out that while Smurf2 is upregulated in the fibrotic kidney, Smurf1 expression is barely detectable and not significantly altered (27). This is in agreement with the notion that an altered TGF-β signaling is the driving force that promotes kidney fibrotic lesions (2, 13). Notably, Smurf2 is preferentially up-regulated in the tubular interstitium in human nephropathies (Figure 1), which may reflect that this ubiquitin ligase could play an important role in regulating TGF-β signaling in that region.
One of the intriguing findings in the present study is that Smurf2 can target three Smad transcription corepressors, Ski, SnoN and TGIF, for degradation. Ski and SnoN are two closely related nuclear proteins that bind to Smads and antagonize TGF-β signaling (15). Studies demonstrate that Smad corepressors blocks Smad-mediated gene transcription by a multitude of mechanisms (15, 20, 33, 35). Corepressor proteins can physically bind to and interact with activated R-Smad in the nuclei, thereby sequestering their trans-activation ability. SnoN and Ski may also displace or prevent Smads from binding to the general transcriptional coactivator p300/CBP. Furthermore, members of Smad co-repressor family proteins have the ability to directly interact with other transcriptional corepressors such as the nuclear hormone receptor corepressor N-CoR and mammalian Sin3 ortholog, leading to repression of a TGF-β transcriptional response (33, 34). Regardless of the mechanisms involved, that Smurf2 simultaneously targets three corepressor proteins Ski, SnoN and TGIF for degradation underscores its predominant role in controlling TGF-β/Smad signaling. By down-regulating the cellular levels of corepressors, Smurf2 may have profound effect on Smad signal transmission. Notably, up-regulation of Smurf2 is accompanied by significant down-regulation of Smad corepressor proteins, but not mRNA levels, in the fibrotic kidney in vivo (7, 27).
Both Smad2 and Smad3 are key mediators of TGF-β signaling, and they share a high degree of homology, with a 92% identity in the amino acid sequence (5, 18). Knockdown of one Smad (either Smad2 or Smad3) appears not to significantly affect the other Smad abundance (Figure 8). Interestingly, Smurf2 appears to have little effect on Smad3 degradation. This may suggest that Smurf2 could selectively abolish a subset of TGF-β responses mediated by Smad2 but not Smad3. It appears, however, that both Smad2 and Smad3 are required for mediating fibronectin expression in tubular epithelial cells (Figure 8), although other studies suggest that they exhibit distinct activities in transmitting the fibrogenic action of TGF-β1 in some cell types (21). Earlier studies indicate that in tubular epithelial cells, Smad3 mediates the E-cadherin suppression induced by TGF-β1, while Smad2 is responsible for mediating matrix metalloproteinase-2 induction. Both Smad2 and Smad3 are critical for TGF-β1 to induce de novo expression of α-smooth muscle actin in tubular cells (21). Therefore, Smad2 is also an important, positive effector of TGF-β signaling that plays a role in mediating matrix production and tissue scarring in the pathogenesis of kidney fibrosis.
We show here that Smurf2 upregulation results in an enhanced TGF-β signaling (Figure 6 and Figure 7), which is consistent with a previous report demonstrating that knockdown of Smurf2 increases intracellular SnoN protein, leading to a reduced TGF-β response in tubular epithelial cells (27). These observations suggest that despite that Smurf2 targets both positive and negative regulators, the net effect of Smurf2 is to promote Smad signaling. This outcome is likely attributable to several mechanisms. One explanation could be related to the fact that Smurf2 concomitantly targets all three corepressors, whereas it only aims at Smad2 but leaves Smad3 alone. Alternatively, Smad corepressors may play a predominant role in ultimately controlling TGF-β signaling, so that the functional impact of loss of corepressors may be greater than that of Smad2 obliteration. The reason why Smurf2 concurrently targets both positive and negative Smad regulators in a paradoxical way remains unclear, but it could provide a complex fine-tuning of the TGF-β signaling.
It should be emphasized that the biology of Smurf E3 ligases may go beyond the regulation of Smads and their corepressors. Studies have shown that Smurf2 and Smurf1 can regulate cell polarity and the formation of cellular protrusions via its ability to target the GTPase Rap1B, RhoA for degradation (25, 32). Despite little effect of Smurf2 on Smad7 in tubular epithelial cells, previous studies demonstrate that Smurf2 can interact with Smad7 to form a complex, which then is exported from the nucleus to the cytoplasm and subsequently destined to the TGF-β receptor complex at the cell surface. Once bound to the receptor complex, Smurf2 cause its degradation (10). In such way, Smad7 function as an adaptor for Smurf2-mediated degradation of its substrate. Consistent with this, recent studies indicate that Smad7 binds β-catenin and induces its degradation by recruiting Smurf2 to the Smad7/β-catenin complex in keratinocytes (8). Interestingly, Smurf2 upregulation also activates telomere-dependent senescence by a novel mechanism distinct from its E3 activity (39).
In summary, our present study has demonstrated that Smurf2 is up-regulated specifically in renal tubules of the fibrotic kidney, which is likely mediated by TGF-β1. Increased Smurf2 renders tubular epithelial cells susceptible to TGF-β1 stimulation, creating a vicious cycle between Smurf2 induction and TGF-β action. Smurf2 concomitantly targets both positive and negative regulators of Smad signaling for degradation, thereby providing a complex fine-tuning of TGF-β signal transmission. However, because of these complexities, strategies that manipulate specific Smurf2-targeted genes (such as SnoN), rather than Smurf2 itself, might be a better way to confine hyperactive TGF-β/Smad signaling and to combat the fibrotic conditions.
Acknowledgments
We thank Drs. J. Massague and R. Weinberg for providing experimental reagents. This work was supported by the National Institutes of Health Grants DK061408, DK064005 and DK071040.
References
- 1.Border WA, Noble NA. TGF-beta in kidney fibrosis: a target for gene therapy. Kidney Int. 1997;51:1388–1396. doi: 10.1038/ki.1997.190. [DOI] [PubMed] [Google Scholar]
- 2.Bottinger EP, Bitzer M. TGF-beta signaling in renal disease. J Am Soc Nephrol. 2002;13:2600–2610. doi: 10.1097/01.asn.0000033611.79556.ae. [DOI] [PubMed] [Google Scholar]
- 3.Dai C, Liu Y. Hepatocyte growth factor antagonizes the profibrotic action of TGF-beta1 in mesangial cells by stabilizing Smad transcriptional corepressor TGIF. J Am Soc Nephrol. 2004;15:1402–1412. doi: 10.1097/01.asn.0000130568.53923.fd. [DOI] [PubMed] [Google Scholar]
- 4.Debigare R, Price SR. Proteolysis, the ubiquitin-proteasome system, and renal diseases. Am J Physiol Renal Physiol. 2003;285:F1–F8. doi: 10.1152/ajprenal.00244.2002. [DOI] [PubMed] [Google Scholar]
- 5.Feng XH, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 6.Fukasawa H, Yamamoto T, Togawa A, Ohashi N, Fujigaki Y, Oda T, Uchida C, Kitagawa K, Hattori T, Suzuki S, Kitagawa M, Hishida A. Down-regulation of Smad7 expression by ubiquitin-dependent degradation contributes to renal fibrosis in obstructive nephropathy in mice. Proc Natl Acad Sci U S A. 2004;101:8687–8692. doi: 10.1073/pnas.0400035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukasawa H, Yamamoto T, Togawa A, Ohashi N, Fujigaki Y, Oda T, Uchida C, Kitagawa K, Hattori T, Suzuki S, Kitagawa M, Hishida A. Ubiquitin-dependent degradation of SnoN and Ski is increased in renal fibrosis induced by obstructive injury. Kidney Int. 2006;69:1733–1740. doi: 10.1038/sj.ki.5000261. [DOI] [PubMed] [Google Scholar]
- 8.Han G, Li AG, Liang YY, Owens P, He W, Lu S, Yoshimatsu Y, Wang D, Ten Dijke P, Lin X, Wang XJ. Smad7-induced beta-catenin degradation alters epidermal appendage development. Developmental cell. 2006;11:301–312. doi: 10.1016/j.devcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Izzi L, Attisano L. Regulation of the TGFbeta signalling pathway by ubiquitin-mediated degradation. Oncogene. 2004;23:2071–2078. doi: 10.1038/sj.onc.1207412. [DOI] [PubMed] [Google Scholar]
- 10.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 11.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 12.Liu FY, Li XZ, Peng YM, Liu H, Liu YH. Arkadia-Smad7-mediated positive regulation of TGF-beta signaling in a rat model of tubulointerstitial fibrosis. American journal of nephrology. 2007;27:176–183. doi: 10.1159/000100518. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y. Renal fibrosis: New insights into the pathogenesis and therapeutics. Kidney Int. 2006;69:213–217. doi: 10.1038/sj.ki.5000054. [DOI] [PubMed] [Google Scholar]
- 14.Lo RS, Wotton D, Massague J. Epidermal growth factor signaling via Ras controls the Smad transcriptional co-repressor TGIF. Embo J. 2001;20:128–136. doi: 10.1093/emboj/20.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo K. Ski and SnoN: negative regulators of TGF-beta signaling. Curr Opin Genet Dev. 2004;14:65–70. doi: 10.1016/j.gde.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23:4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 17.Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 18.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 19.Mehra A, Wrana JL. TGF-beta and the Smad signal transduction pathway. Biochem Cell Biol. 2002;80:605–622. doi: 10.1139/o02-161. [DOI] [PubMed] [Google Scholar]
- 20.Melhuish TA, Gallo CM, Wotton D. TGIF2 interacts with histone deacetylase 1 and represses transcription. J Biol Chem. 2001;276:32109–32114. doi: 10.1074/jbc.M103377200. [DOI] [PubMed] [Google Scholar]
- 21.Phanish MK, Wahab NA, Colville-Nash P, Hendry BM, Dockrell ME. The differential role of Smad2 and Smad3 in the regulation of pro-fibrotic TGFbeta1 responses in human proximal-tubule epithelial cells. The Biochemical journal. 2006;393:601–607. doi: 10.1042/BJ20051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poncelet AC, Schnaper HW, Tan R, Liu Y, Runyan CE. Cell phenotype-specific down-regulation of Smad3 involves decreased gene activation as well as protein degradation. J Biol Chem. 2007;282:15534–15540. doi: 10.1074/jbc.M701991200. [DOI] [PubMed] [Google Scholar]
- 23.Racusen LC, Monteil C, Sgrignoli A, Lucskay M, Marouillat S, Rhim JG, Morin JP. Cell lines with extended in vitro growth potential from human renal proximal tubule: characterization, response to inducers, and comparison with established cell lines. J Lab Clin Med. 1997;129:318–329. doi: 10.1016/s0022-2143(97)90180-3. [DOI] [PubMed] [Google Scholar]
- 24.Schnaper HW, Hayashida T, Hubchak SC, Poncelet AC. TGF-beta signal transduction and mesangial cell fibrogenesis. Am J Physiol Renal Physiol. 2003;284:F243–F252. doi: 10.1152/ajprenal.00300.2002. [DOI] [PubMed] [Google Scholar]
- 25.Schwamborn JC, Muller M, Becker AH, Puschel AW. Ubiquitination of the GTPase Rap1B by the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. Embo J. 2007;26:1410–1422. doi: 10.1038/sj.emboj.7601580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Sun Y, Liu X, Ng-Eaton E, Lodish HF, Weinberg RA. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling. Proc Natl Acad Sci U S A. 1999;96:12442–12447. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan R, Zhang J, Tan X, Zhang X, Yang J, Liu Y. Downregulation of SnoN expression in obstructive nephropathy is mediated by an enhanced ubiquitin-dependent degradation. J Am Soc Nephrol. 2006;17:2781–2791. doi: 10.1681/ASN.2005101055. [DOI] [PubMed] [Google Scholar]
- 28.Tan R, Zhang X, Yang J, Li Y, Liu Y. Molecular basis for the cell type-specific induction of SnoN expression by hepatocyte growth factor. J Am Soc Nephrol. 2007;18:2340–2349. doi: 10.1681/ASN.2007010128. [DOI] [PubMed] [Google Scholar]
- 29.Tan X, Li Y, Liu Y. Paricalcitol attenuates renal interstitial fibrosis in obstructive nephropathy. J Am Soc Nephrol. 2006;17:3382–3393. doi: 10.1681/ASN.2006050520. [DOI] [PubMed] [Google Scholar]
- 30.Teteris SA, Menahem SA, Perry G, Maguire JA, Dowling JP, Langham RG, Thomson NM, Stein AN. Dysregulated growth factor gene expression is associated with tubulointerstitial apoptosis and renal dysfunction. Kidney Int. 2007;71:1044–1053. doi: 10.1038/sj.ki.5002176. [DOI] [PubMed] [Google Scholar]
- 31.Togawa A, Yamamoto T, Suzuki H, Fukasawa H, Ohashi N, Fujigaki Y, Kitagawa K, Hattori T, Kitagawa M, Hishida A. Ubiquitin-dependent degradation of Smad2 is increased in the glomeruli of rats with anti-thymocyte serum nephritis. Am J Pathol. 2003;163:1645–1652. doi: 10.1016/S0002-9440(10)63521-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wranax JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- 33.Wotton D, Knoepfler PS, Laherty CD, Eisenman RN, Massague J. The Smad transcriptional corepressor TGIF recruits mSin3. Cell Growth Differ. 2001;12:457–463. [PubMed] [Google Scholar]
- 34.Wotton D, Lo RS, Swaby LA, Massague J. Multiple modes of repression by the Smad transcriptional corepressor TGIF. J Biol Chem. 1999;274:37105–37110. doi: 10.1074/jbc.274.52.37105. [DOI] [PubMed] [Google Scholar]
- 35.Wu JW, Krawitz AR, Chai J, Li W, Zhang F, Luo K, Shi Y. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-beta signaling. Cell. 2002;111:357–367. doi: 10.1016/s0092-8674(02)01006-1. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Liu Y. Blockage of tubular epithelial to myofibroblast transition by hepatocyte growth factor prevents renal interstitial fibrosis. J Am Soc Nephrol. 2002;13:96–107. doi: 10.1681/ASN.V13196. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Liu Y. Dissection of key events in tubular epithelial to myofibroblast transition and its implications in renal interstitial fibrosis. Am J Pathol. 2001;159:1465–1475. doi: 10.1016/S0002-9440(10)62533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Zhang X, Li Y, Liu Y. Downregulation of Smad transcriptional corepressors SnoN and Ski in the fibrotic kidney: an amplification mechanism for TGF-beta1 signaling. J Am Soc Nephrol. 2003;14:3167–3177. doi: 10.1097/01.asn.0000099373.33259.b2. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Cohen SN. Smurf2 up-regulation activates telomere-dependent senescence. Genes Dev. 2004;18:3028–3040. doi: 10.1101/gad.1253004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]