Abstract
Tumor necrosis factor receptors (TNFR) are single transmembrane-spanning glycoproteins that bind cytokines and trigger multiple signal transduction pathways. Many of these TNFRs rely on interactions with TRAF proteins that bind to the intracellular domain of the receptors. CD40 is a member of the TNFR family that binds to several different TRAF proteins. We have determined the crystal structure of a 20-residue fragment from the cytoplasmic domain of CD40 in complex with the TRAF domain of TRAF3. The CD40 fragment binds as a hairpin loop across the surface of the TRAF domain. Residues shown by mutagenesis and deletion analysis to be critical for TRAF3 binding are involved either in direct contact with TRAF3 or in intramolecular interactions that stabilize the hairpin. Comparison of the interactions of CD40 with TRAF3 vs. TRAF2 suggests that CD40 may assume different conformations when bound to different TRAF family members. This molecular adaptation may influence binding affinity and specific cellular triggers.
The question of how tumor necrosis factor receptor (TNFR) signaling controls events such as immune responses, Ig class-switching, activation of NF-κB, or regulation of apoptosis is central to understanding the role of these receptors in disease, including autoimmune disorders, allergic reactions, inflammatory responses, and cancer. The cytoplasmic domains of TNFRs vary in length from 40 to 200 residues with little sequence homology and can be broadly classified into those that contain “death domains” (TNFR1, Fas, DR3, DR4, and DR5) and those that do not (TNFR2, CD27, CD30, CD40, LTβR, 4–1BB, OX-40, ATAR, and RANK).
CD40 is a well characterized member of the TNFR family that is expressed on all B lymphocytes as well as activated endothelial cells, antigen-presenting dendritic and monocytic cells, and synovial fibroblasts. CD40 signals are critical for B cell proliferation, growth, and differentiation (1–3). The receptor is also expressed on epithelial carcinomas and has been shown to promote spontaneous and chemotherapeutic drug-induced apoptosis in epithelial cancer cell lines (4, 5). We have determined the crystal structure of a portion of the CD40 cytoplasmic domain bound to a downstream signaling molecule, TRAF3.
TNFR-associated factors (TRAFs) are recently discovered adaptor proteins that connect TNFRs to downstream signaling pathways including the NF-κB and c-JUN N-terminal kinase (JNK) pathways (6–9). Six TRAF proteins have been identified to date and are numbered sequentially in order of their discovery. From amino acid sequence homology, it was determined that some features are conserved throughout the family. TRAFs 2–6 have ring fingers and zinc finger motifs (10, 11), followed by the TRAF domain that is conserved in all TRAFs. This domain mediates binding to the cytoplasmic domains of TNFRs. Gene knockout studies in mice have demonstrated critical roles for several of the TRAF proteins in signal transduction pathways stimulated by TNFRs (12–15). Moreover, mutational analyses of TRAF-binding sites within the cytoplasmic domains of TNFRs have provided evidence that interactions of TRAFs with these receptors are critical for many TNF-induced signaling and cellular responses (9, 16–21).
Materials and Methods
Crystallization and Data Collection.
The TRAF3 TRAF domain was cloned, expressed, and purified by procedures to be described elsewhere (C.-Z.N., K.W., J. Zheng, M. Havert, J.C.R., and K.R.E., unpublished work). Crystals grew in two morphologically indistinguishable forms in Tris buffer at pH 6.5–8.3. Crystals formed in space group P6322 with cell dimensions a = b = 83.8 Å, c = 212.6 Å, γ = 120° but diffracted only to 3.5 Å. Crystals also formed in space group R32 with a = b = 84.5 Å, c = 319.5 Å, γ = 120° and were used for data collection. There is one monomer in the asymmetric unit. Crystals were transferred quickly to cryoprotectant solutions containing 35% (vol/vol) glycerol and frozen. A complete native data set to 2.6-Å resolution was collected at −165°C at the Stanford Synchrotron Research Laboratory on beamline BL9-1. Diffraction images were recorded on a MAR345 detector (λ = 0.98 Å; MAR Research, Hamburg). Data were reduced, scaled, and processed with the programs denzo and scalepack (22). A summary of data collection statistics is presented in Table 1.
Structure Solution and Refinement.
The structure of the TRAF3 TRAF domain was solved by molecular replacement with amore (23) and the atomic coordinates of the TRAF2 TRAF domain (24) as the probe model, followed by simulated annealing in crystallography and NMR system (25). To improve the phasing and to minimize bias from the molecular replacement solution, phases from a heavy atom single isomorphous replacement were obtained by soaking crystals in thimerosol. A complete data set to 3.5 Å was collected at Stanford Synchrotron Research Laboratory on beamline BL7-1. The heavy atom site was located as a clear Patterson peak, and single isomorphous replacement phases were calculated with the program solve (26). Phases from the single isomorphous replacement and molecular replacement solutions were combined with the program mlphare (27). The final electron density map showed clear solvent boundaries, and electron density was well defined for residues 300–504. The structure was refined with alternating cycles of simulated annealing and positional refinement and manual model building in o (28). Finally, temperature factors (B values) were included in the refinement. The R factor for the final model, including 137 well ordered solvent molecules, was 25% (Rfree = 29%). The final refinement statistics are presented in Table 1. Graphic images for the figures were created with spock (29) and raster-3d (30).
Peptide Cocrystallization.
A synthetic peptide, KTAAPVQETLHGSQPVTQEDG, corresponding to residues 247–266 (plus an N-terminal lysine added to increase solubility) from the cytoplasmic domain of CD40 was soaked into crystals of TRAF3. Diffraction data were collected from crystals of the bound TRAF3 TRAF domain in space group P6322 at Stanford Synchrotron Research Laboratory on beamline BL7-1 at −165°C by using a MAR345 detector. The data set from 8.0 to 3.5 Å was used for a molecular replacement solution implementing the TRAF3 atomic coordinates as probe model in amore (23). The final R factor for the TRAF3–peptide complex was 26% (Rfree = 34%).
Results and Discussion
Structure of TRAF3.
TRAF3, also known as CD40bp (19), CAP-1 (31), LAP-1 (32), and CRAF-1 (17), binds to the CD40 receptor, to other related receptors, including TNFR-2, CD30, and LT-βR, and to the transforming oncoprotein of Epstein–Barr virus latent-infection membrane protein 1 (LMP-1). Binding of TRAF3 to these receptors modulates activation of NF-κB (9). Moreover, TRAF3 gene knockout mice are depleted in T cells and exhibit an impaired T-dependent immune response (14). These studies suggest that TRAF3 is required for hematopoietic and immune system development and T cell activation.
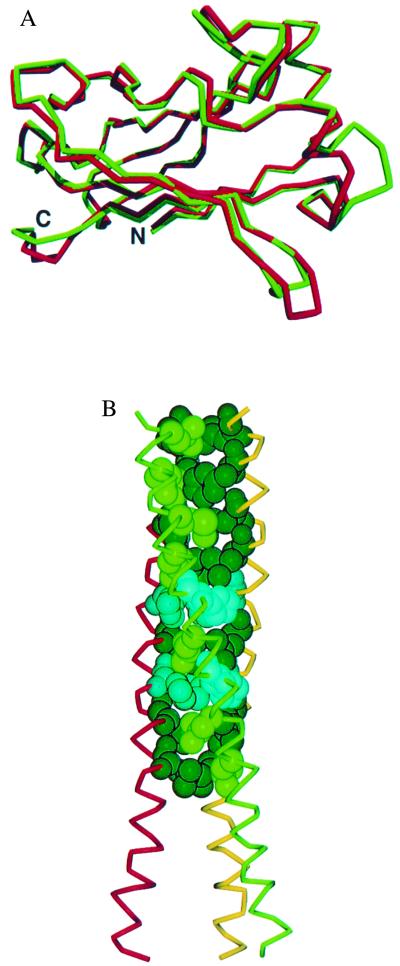
The TRAF domain of TRAF3 is composed of two structural features: an N-terminal α-helical segment (TRAF-N domain) extending from residues 277 to 347 and a C-terminal domain (TRAF-C domain) that is independently folded into an eight-stranded β-sandwich. The folding pattern is similar to that reported recently for TRAF2 (24, 33) as shown in Fig. 1A. TRAF3 is a trimer in the crystal with three identical subunits related by crystallographic 3-fold symmetry. The trimer is stabilized by hydrophilic (Y418–T347, H331–K493, and R334–D332) and hydrophobic interactions (L421–M435–F491) between the β-sandwiches as well as coiled-coil interactions between the intertwined N-terminal helical segments (Fig. 1B). The elongated helical segments extend 77 Å away from the TRAF-C domain cluster, forming the stalk of the mushroom-shaped trimer (see Fig. 3).
Figure 1.
The overall structure of the TRAF3 subunit is composed of an elongated helix followed by an eight-stranded β-sandwich (TRAF domain). (A) Superimposition of the polypeptide backbone of the C-terminal TRAF domain of TRAF3 (red) and TRAF2 (24). The rms deviation between corresponding α-carbons is 1 Å. Strong homology (59% identical) exists between TRAF3 and TRAF2 in this domain. The sandwich is formed by two layers of β-sheet, each with four antiparallel strands. The topology of this β-sandwich is found thus far only in TRAF domains. (B) The TRAF-N domain of TRAF3 is a long amphipathic α-helix that forms a coiled-coil when TRAF3 trimerizes (see Fig. 3). Residues 300–347 were ordered in the electron density map. The coiled-coil interactions are stabilized by nine heptad repeats of hydrophobic residues. This hydrophobic pattern is interrupted at residues 324 and 331 where three histidines are found in the interior of the coiled-coil. The side chains of these histidines extend out of the coiled-coil. The TRAF3 fragment is considerably longer at the N terminus than the TRAF2 fragment, where residues from the N terminus were missing or disordered (24, 33). This model provides structural details for most of the helical TRAF-N region.
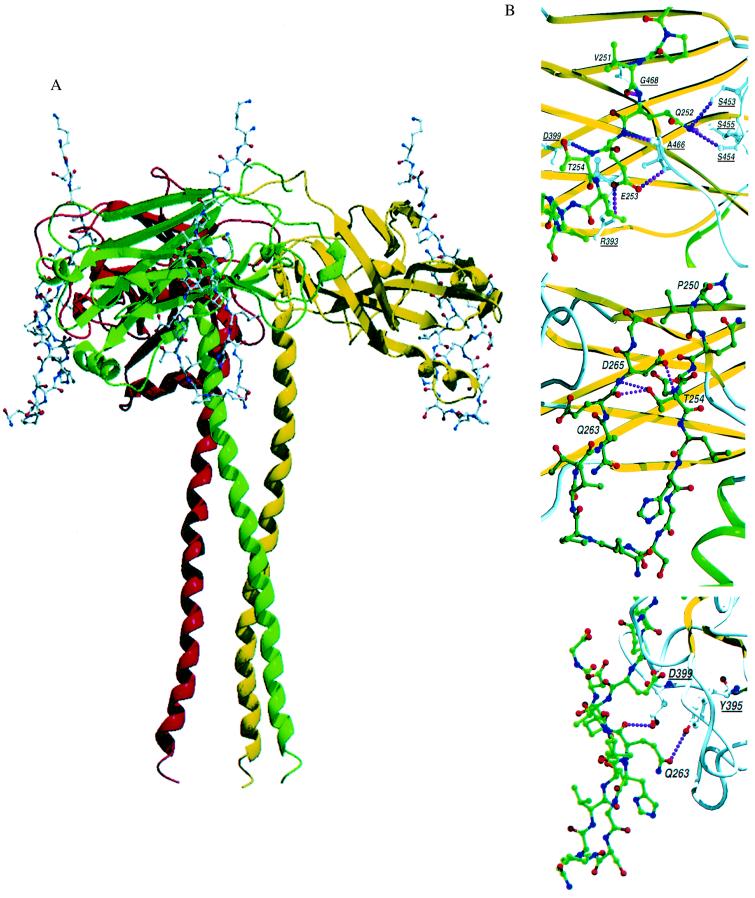
Figure 3.
TRAF3/CD40 interactions. (A) Schematic drawing of the TRAF3 trimer with the polypeptide backbone of TRAF3 presented as a ribbon model and the CD40 peptide shown as a ball and stick model. One CD40 fragment binds to each TRAF3 monomer at the edge of the TRAF3 domain, crossing one β-sheet. No conformational changes were seen when comparing TRAF3 alone or bound to CD40. (B) Close-up view of the CD40 fragment bound to TRAF2 and TRAF3. Residues in CD40 are labeled, and critical contact residues in TRAF3 are also marked and underlined for identification. Interactions within 3.0 Å that are proposed to dictate specific recognition of CD40 and TRAF3 or TRAF2 are shown as dotted lines. The images show intramolecular hydrogen bonds within the CD40 fragment that stabilize the reverse turn (Middle) and direct contacts between CD40 and TRAF3 (Bottom). These images can be contrasted with the contacts in TRAF2 (Top; 1CZZ-; based on figure 3 of Ye et al. in ref. 34).
Structure of CD40 Fragment.
In the complex, one CD40 peptide binds to the edge of each of the three TRAF domains, making no contacts to other TRAF domain monomers. When bound to TRAF3, the CD40 fragment assumes a hairpin configuration with two extended segments and a reverse turn centered at residues 258–260 (see Figs. 2 and 3). A consensus sequence PxQx(T/S), proposed for TRAFs 1–3 binding to CD40, CD30, and LMP-1 receptors, is 250PVQET254 in CD40. Conserved residues within this sequence lie in a crevice across the face of the β-sandwich in position to make hydrophobic (P250) and hydrophilic contacts with TRAF3 (Fig. 3B). Glu-252 does not make direct contact with TRAF3, but within the range of motion, the side chain could form hydrogen bonds with one or more conserved serines (TRAF3 residues 454, 455, and/or 456). This interaction was proposed as a critical anchoring point in TRAF2 for specific recognition of TNFRs (33, 34); however, from the current structure, it does not seem to be essential for TRAF3/CD40 binding.
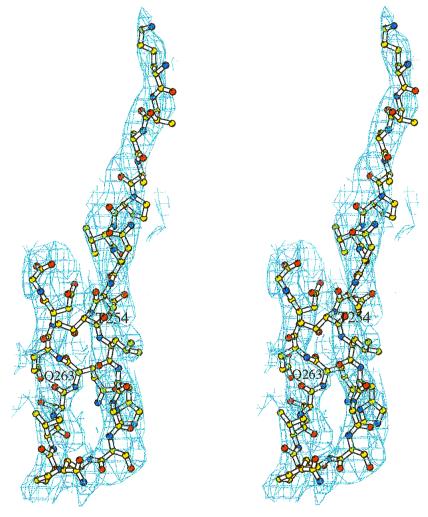
Figure 2.
Model for the CD40 fragment displayed in the electron density map. The 3.5-Å 2Fo − Fc map was contoured at 0.75σ and is displayed in stereo. For modeling, the peptide was fitted to Fo − Fc difference maps and omitmaps (36). Clear electron density was visible for the backbone to position the bound peptide; however, density was weak for some side chains. The reverse turn configuration was clearly defined.
It has been suggested that differences in binding of TRAF3 versus TRAF2 to CD40 are influenced by residues distal to the PxQxT sequence (9, 35). Because of these suggestions, we cocrystallized a CD40 fragment long enough to examine these residues as well. The cytoplasmic portion of CD40 alone contains little regular secondary structure in solution (as determined by NMR; results not shown), consistent with secondary structure predictions of this region. In the complex studied here, the CD40 peptide assumes an ordered configuration when bound to TRAF3. The two strands flanking the reverse turn do not interact with regular hydrogen bonding typical of β-hairpins. Instead, the configuration is apparently stabilized by hydrogen bonds involving T254 in the PxQxT consensus sequence. The threonine side chain makes intrapeptide hydrogen bonds with the main chain of D265 and E264 on the adjacent strand (Fig. 3B). It is known from mutagenesis studies that substitution of any residue other than serine for T254 abolishes binding of TRAF2 and TRAF3 to CD40 (9, 20, 35). From the present structural analysis, it is clear that a side chain with a hydroxyl group is required to maintain a stable CD40 hairpin configuration rather than to make contact with TRAF3. If a similar hairpin configuration exists in interaction with TRAF2, a comparable structural requirement may be necessary but not sufficient to maintain the hairpin structure, because residues beyond 258 were not ordered in the complex of the same CD40 peptide with TRAF2 (34). In the TRAF2 studies, CD40 T254 or its equivalent in the related LMP-1 receptor was located near protein atoms in a position to hydrogen bond with TRAF2 D399 (33, 34).
Another difference in TRAF3 versus TRAF2 interactions with CD40 is revealed in comparing E253 in the respective complexes. When binding to TRAF2, this residue makes a bidentate ion pair with TRAF2 residues R393 and Y395 (33, 34). In contrast, CD40 E253 does not contact R393 and Y395 in TRAF3. However, a different critical contact is made with one of these residues. The side chain of Q263, on the other side of the CD40 hairpin, forms hydrogen bonds with side chain atoms of Y395 in TRAF3 (see Fig. 3B). Another interaction near this point is made when the carboxyl group of D399 in TRAF3 contacts the main chain carbonyl atom of Q263. Consistent with these structural observations, substitution of alanine for Glu-263 significantly reduces binding of CD40 to TRAF3 but not to TRAF2 (9, 20). C-terminal truncations of 15 residues from the cytoplasmic portion of CD40, corresponding to removal of Q263 and the following residues, abolished binding of TRAF3 to CD40 but only reduced binding to TRAF2 (9).
TRAF3/CD40 Recognition.
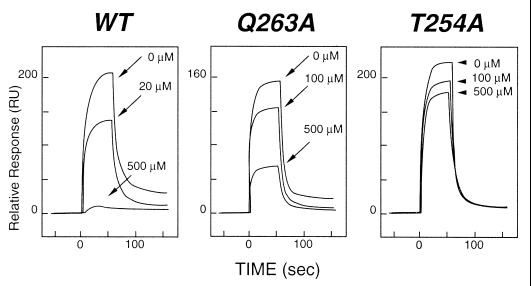
The importance of CD40 T254 and Q263 for TRAF3 binding was evaluated further in binding studies by using surface plasmon resonance for a series of synthetic peptides that contained alanines substituted systematically at each of 17 positions in the CD40 hairpin. The assay tested inhibition of binding of wild-type CD40 cytoplasmic domain to TRAF3 by the mutant peptides (Fig. 4). The results confirmed that T254 and Q263 are critical for TRAF3 binding. The role of these two residues is different. T254 stabilizes the reverse turn by forming hydrogen bonds with two residues that are located on the opposite strand of the CD40 hairpin. These contacts are with residues C-terminal to Q263; thus, the CD40 T254 intramolecular contact plays an important role in establishing the configuration of the reverse turn and thus the orientation of Q263. In contrast, the direct contact between Q263 and TRAF3 is critical for TRAF3/CD40 recognition. Q263 contacts TRAF3 Y395 and D399. These residues are conserved in TRAFs 1, 2, 3, and 5. Therefore, it is theoretically possible for identical contacts to be made by all of these TRAF molecules with CD40. However, as stated previously, differences have been reported in binding of different TRAFs to CD40 (9). In the absence of ordered structure for a comparable fragment of CD40 in complex with TRAF2, it is not possible to evaluate the extent of interactions that occur distal to the PVQET motif. The observation that C-terminal deletion of 11 or 13 residues (equivalent to K267–end or D265–end) reduces binding of CD40 to TRAF2 but not TRAF3 (9) further suggests that CD40 may assume different conformations on binding to these two TRAF molecules. This molecular adaptation would be consistent with the observation that substitution of alanine for glutamine at position 263 affects binding of CD40 to TRAF3 but not TRAF2. Further comparative structural studies are needed to define the specific recognition of CD40 and other TNFRs by individual TRAFs.
Figure 4.
Inhibition of TRAF3 binding to CD40 by synthetic peptides. Binding was measured by using surface plasmon resonance with a BIACORE 3000 instrument (Biacore AB, Uppsala). A recombinant fragment representing the entire cytoplasmic domain of CD40 was cloned as a glutathione S-transferase fusion protein, expressed in Escherichia coli, and purified by affinity chromatography on a glutathione-Sepharose (Amersham Pharmacia) column. The fusion partner was removed by thrombin digestion, and the CD40 fragment was purified by ion-exchange chromatography. The CD40 domain was immobilized on a Biacore CM5 sensor chip, and the TRAF3 TRAF domain was injected at 3.5 μM. The Left sensorgram shows the relative response for binding interactions between immobilized CD40 and TRAF3 and inhibition of this binding by a synthetic peptide from the cytoplasmic domain of CD40 (250PVQETLHGCQPVTQEDG266). A series of peptides with alanine substituted singly for each residue in the fragment was tested for inhibition of binding of TRAF3 to CD40. Mutant peptides were injected at concentrations ranging from 20 to 500 μM. As can be seen from the Center and Right sensorgrams, striking changes in inhibition were observed when alanine was substituted for Q263 or T254, suggesting reduced binding of the mutant fragments. Substitution of alanine for other residues in the peptide did not significantly alter levels of inhibition observed with the wild-type (WT) peptide.
Table I.
Summary of crystallographic data and refinement
| Native | Complex (with CD40 peptide) | |
|---|---|---|
| Space Group | R32 | P6322 |
| Unit cell dimensions | a = b = 84.53 Å, c = 319.5 Å | a = b = 83.76 Å, c = 212.6 Å |
| Molecules/asymmetric unit | 1 monomer | 1 monomer |
| Data completeness | 98.7% (50–2.8 Å) | 86.2% (50–3.5 Å) |
| Observed reflections | 59,634 | 34,451 |
| Unique reflections | 11,198 | 5,283 |
| Rmerge* | 5.6% (26%) | 10.6% (30%) |
| Final refinement statistics | ||
| R factor† | 25.3% (8.0–2.8 Å) | 26.2% (8.0–3.5 Å) |
| Rfree‡ | 29.4% (8.0–2.8 Å) | 34.7% (8.0–3.5 Å) |
| Average B factor | 42.5 | 24.1 (protein + peptide) |
| Number of protein atoms | 1,626 | 1,626 |
| Number of CD40 peptide atoms | None | 154 |
| Number of water molecules | 137 | None |
| rms deviation bond lengths | 0.007 Å | 0.008 Å |
| rms deviation bond angles | 1.31° | 1.48° |
Rmerge = ∑(|I − 〈I〉|)/∑(I), where I is the observed intensity and 〈I〉 is the mean intensity of all symmetry-related reflections.
R factor = ∑||Fo| − |Fc||/∑|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
Rfree was calculated by using a random set containing 5% of observations that were omitted during refinement.
Acknowledgments
We thank the staff at the Stanford Synchrotron Research Laboratory for help at the beamlines. We thank Lars Brive for advice on molecular imaging and Debin Huang for helpful discussions. We appreciate the efforts of Kosi Gramatikoff for graphics and Sara Harmon for preparing the manuscript for publication. This work was supported by National Institutes of Health Grant CA69381, a grant from Zaiya Pharmaceuticals to J.C.R., and Deutsche Forschungsgemeinschaft Grant EL-818 to E.L.
Abbreviation
- TNFR
tumor necrosis factor receptor
Note Added in Proof.
While pursuing further binding analyses with the TRAF3 crystals, we observed twinning of the crystals. It was then recognized that the data used for this study were derived from twinned crystals, space groups R3 and P321. The refinement was calculated again by using simulated annealing for the data with hemihedral twinning (in crystallography and NMR system program). The final R factor was reduced to 22.4% (Rfree = 28.6%). The structures were identical within 0.3-Å rms deviation for all atoms. Coordinates deposited are those from refinement of the twinned data.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1FLK and 1FLL).
References
- 1.Foy T M, Aruffo A, Bajorath J, Buhlmann J E, Noelle R J. Annu Rev Immunol. 1996;14:591–617. doi: 10.1146/annurev.immunol.14.1.591. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Bazan F, Blanchard D, Briere F, Galizzi J P, van Kooten C, Liu Y J, Rousset F, Saeland S. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 3.Clark E A, Ledbetter J A. Nature (London) 1994;367:425–428. doi: 10.1038/367425a0. [DOI] [PubMed] [Google Scholar]
- 4.Eliopoulos A G, Dawson C W, Mosialos G, Floettmann J E, Rowe M, Armitage R J, Dawson J, Zapata J M, Kerr D J, Wakelam M J, et al. Oncogene. 1996;13:2243–2254. [PubMed] [Google Scholar]
- 5.Eliopoulos A G, Stack M, Dawson C W, Kaye K M, Hodgkin L, Sihota S, Rowe M, Young L S. Oncogene. 1997;14:2899–2916. doi: 10.1038/sj.onc.1201258. [DOI] [PubMed] [Google Scholar]
- 6.Natoli G, Costanzo A, Ianni A, Tempelton D J, Woodgett J R, Balsano C, Levrero M. Science. 1997;275:200–203. doi: 10.1126/science.275.5297.200. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z G, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 8.Grammer A C, Swantek J L, McFarland R D, Miura Y, Geppert T, Lipsky P E. J Immunol. 1998;161:1183–1193. [PubMed] [Google Scholar]
- 9.Leo E, Welsh K, Matsuzawa S, Zapata J M, Kitada S, Mitchell R S, Ely K R, Reed J C. J Biol Chem. 1999;274:22414–22422. doi: 10.1074/jbc.274.32.22414. [DOI] [PubMed] [Google Scholar]
- 10.Freemont P S. Ann NY Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 11.Schwabe J W, Klug A. Nat Struct Biol. 1994;1:345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- 12.Lomaga M, Yeh W C, Sarosi I, Duncan G S, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, et al. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakano H, Sakon S, Koseki H, Takemori T, Tada K, Matsumoto M, Munechika E, Sakai T, Shirasawa T, Akiba H, et al. Proc Natl Acad Sci USA. 1999;96:9803–9808. doi: 10.1073/pnas.96.17.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Cheng G, Baltimore D. Immunity. 1996;5:407–415. doi: 10.1016/s1074-7613(00)80497-5. [DOI] [PubMed] [Google Scholar]
- 15.Yeh W C, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa J, Ferrick D, Hum B, Iscove N, et al. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 16.Akiba H, Nakano H, Nishinaka S, Shindo M, Kobata T, Atsuta M, Morimoto C, Ware C F, Malinin N L, Wallach D, et al. J Biol Chem. 1998;273:13353–13358. doi: 10.1074/jbc.273.21.13353. [DOI] [PubMed] [Google Scholar]
- 17.Cheng G, Cleary A M, Ye Z S, Hong D I, Lederman S, Baltimore D. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 18.Hostager B S, Bishop G A. J Immunol. 1999;162:6307–6311. [PubMed] [Google Scholar]
- 19.Hu H M, O'Rourke K, Boguski M S, Dixit V M. J Biol Chem. 1994;269:30069–30072. [PubMed] [Google Scholar]
- 20.Lee H H, Dempsey P W, Parks T P, Zhu X, Baltimore D, Cheng G. Proc Natl Acad Sci USA. 1999;96:1421–1426. doi: 10.1073/pnas.96.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leo E, Zapata J M, Reed J C. Eur J Immunol. 1999;29:3908–3913. doi: 10.1002/(SICI)1521-4141(199912)29:12<3908::AID-IMMU3908>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z. In: CCP4 Study Weekend: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. Warrington, United Kingdom: Daresbury Laboratories; 1993. pp. 55–62. [Google Scholar]
- 23.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 24.Park Y C, Burkitt V, Villa A R, Tong L, Wu H. Nature (London) 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- 25.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger T C, Kim S-H, Eisenberg D. Acta Crystallogr A. 1987;43:1–5. [Google Scholar]
- 27.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend. Wolf W, Evans P R, Lestie A G W, editors. Warrington, U.K.: SERC Daresbury Laboratory; 1991. pp. 80–86. [Google Scholar]
- 28.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Christopher J A. spock. College Station, TX: Texas A&M University; 1998. [Google Scholar]
- 30.Bacon D J, Anderson W F. J Mol Graphics. 1988;6:219–220. [Google Scholar]
- 31.Sato T, Irie S, Reed J C. FEBS Lett. 1995;358:113–118. doi: 10.1016/0014-5793(94)01406-q. [DOI] [PubMed] [Google Scholar]
- 32.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 33.McWhirter S M, Pullen S S, Holton J M, Crute J J, Kehry M R, Alber T. Proc Natl Acad Sci USA. 1999;96:8408–8413. doi: 10.1073/pnas.96.15.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye H, Park Y C, Kreishman M, Kieff E, Wu H. Mol Cell. 1999;4:321–330. doi: 10.1016/s1097-2765(00)80334-2. [DOI] [PubMed] [Google Scholar]
- 35.Pullen S S, Dang T T A, Crute J J, Kehry M R. J Biol Chem. 1999;274:14246–14254. doi: 10.1074/jbc.274.20.14246. [DOI] [PubMed] [Google Scholar]
- 36.Bhat T N, Cohen G H. J Appl Crystallogr. 1984;17:244–248. [Google Scholar]