Abstract
The mechanosensitive channel of small conductance (MscS) is part of a coordinated response to osmotic challenges in E. coli. MscS opens as a result of membrane tension changes, thereby releasing small solutes and effectively acting as an osmotic safety valve. Both, the functional state depicted by its crystal structure and its gating mechanism remain unclear. Here, we combine site-directed spin labeling, electron paramagnetic resonance (EPR) spectroscopy, and molecular dynamics simulations with novel energy restraints based on experimental EPR data to investigate the native transmembrane and periplasmic molecular architecture of closed MscS in a lipid bilayer. In the closed conformation, MscS shows a more compact transmembrane domain than in the crystal structure, characterized by a realignment of the transmembrane segments towards the normal of the membrane. The previously unresolved NH2-terminus forms a short helical hairpin capping the extracellular ends of TM1 and TM2 and in close interaction with the bilayer interface. The present three-dimensional model of membrane-embedded MscS in the closed state represents a key step in determining the molecular mechanism of MscS gating.
Cells rely on a tight regulation of the balance between internal and external osmotic forces to survive. In bacteria and archaea, osmoregulation is achieved by accumulation and release of solutes through active transport systems and mechanosensitive (MS) channels, respectively 1; 2; 3. MS channels act as indirect osmosensors in the emergency response to hypo-osmotic challenges. When water enters the cell, the turgor pressure increases and the membrane expands reducing its lateral pressure which leads to MS channels opening. The subsequent release of solutes through the open MS channels alleviates turgor pressure and prevents cell lysis 4. The E. coli inner membrane has several MS channels involved in this complex response 5; 6. These channels have been classified, according to their single-channel conductance and the tension required to activate them, as MS channel of Large, Small, and Mini conductance, or MscL, MscS/MscK, and MscM, respectively 4.
MscL and MscS x-ray crystal structures 7; 8 have provided an important framework to understand the gating of MS channels and interpret experimental results. However, while both MscL and MscS seem to respond to similar bilayer deformations and the MscL structure has lead to explicit modeling of its open and closed conformations 9; 10; 11; 12; 13; 14, the MscS gating mechanism and the functional state depicted by its crystal structure remain unclear. The MscS crystal is arranged as a homoheptamer with three transmembrane (TM) segments per subunit (TM1, TM2, and TM3A–B) and a large cytoplasmic domain featuring seven side “windows” and a narrow opening at the most distal region from the membrane (Figures 1A, B). The first 26 residues of the NH2-terminus, a region that appears to play a role in gating and stability, are not resolved in the crystal structure. As the narrowest point along the permeation path is ~ 7 Å in diameter, it was originally suggested that this conformation represented the open state of MscS 8, its dimensions putatively supporting its 1 nS single channel conductance and slightly selectivity towards anions. Molecular dynamics (MD) and continuum simulations have systematically shown that the crystal conformation exhibits low conductance, rectification at high voltages, and high anionic selectivity 15; 16; 17; 18; 19; 20. Hence, while the current consensus is that the MscS crystal conformation is unlikely to represent the fully open state, its precise functional assignment remains to be established. It is then clear that to determine the conformations of the closed and open states and thereby understand the mechano-gating process underlying MscS function in vivo, additional structural and dynamical information of MscS in a native-like membrane is required.
Figure 1.
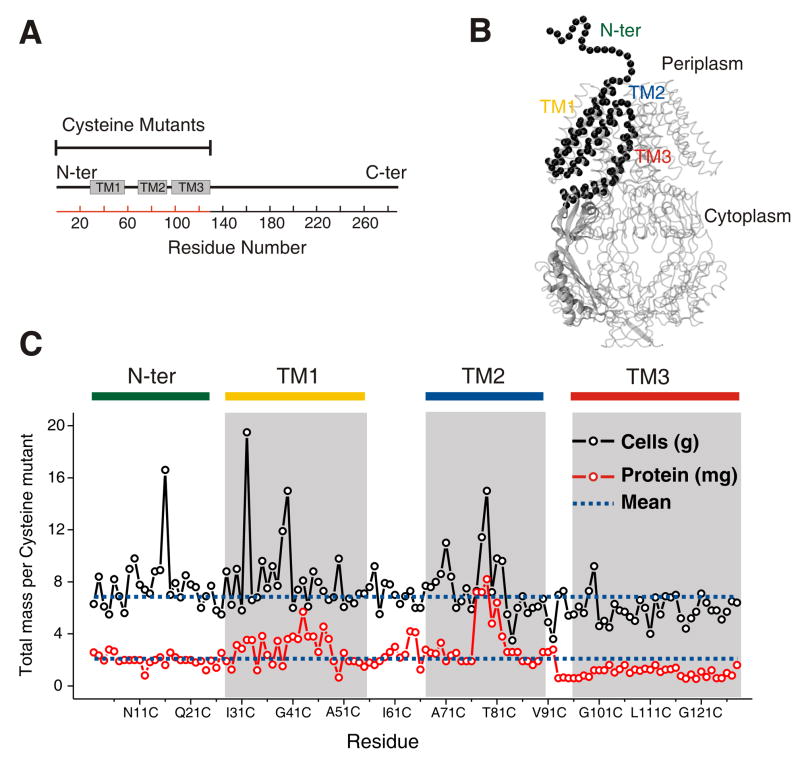
MscS architecture and cysteine mutants expression (A) Linear representation of MscS topology. The TM helices (as suggested from the crystal structure) of the MscS monomer (TM1, TM2, and TM3) are represented by rectangles. The scale corresponds to the amino acid residue numbering. The red line denotes the residues that were mutated to cysteine. (B) Single MscS subunit showing amino acid residues (black spheres) probed through site directed spin labeling of cysteines. A single MscS monomer is represented as part of the heptamer according to the crystal structure of the channel. The NH2-terminus segment (26 residues) is represented by a schematic line. (C) Expression of cysteine mutants. The total amount of cell and protein obtained is shown for each residue after expressing single cysteine mutants in vector pET28 (containing MscS with His6 epitope at the NH2-termini), in E. coli Rosetta™ cells. Grayed areas represent the membrane embedded areas derived from the MscS crystal structure.
Electron paramagnetic resonance (EPR) spectroscopy is a powerful tool for structural analysis of soluble and membrane proteins in native-like conditions. This technique is particularly useful for judging crystal structures obtained in non-native environments 9; 21; 22; 23. Site-directed spin labeling (SDSL) and EPR spectroscopy gives three types of structural information: spin-label motion, solvent accessibility, and distance measurements 21; 24. It is therefore possible to determine whether a spin-labeled position is exposed to the solvent or the membrane, or buried within the protein. Altogether, the EPR data in the context of a lipid bilayer offers an extensive collection of accessibilities constraints that considerably decreases the number of possible folds, in order to derive along with computational methods three-dimensional models of proteins, in their native environments and in different conformational states 25; 26; 27.
Here, we have analyzed the structural dynamic properties of membrane-reconstituted MscS in the closed state conformation, as determined by SDSL, EPR spectroscopy and computational modeling. The experimental data was used to refine the crystal structure under known physiological conditions, and to construct a model of the closed conformation of MscS that includes the previously unresolved NH2-terminus. The new model shows significant differences regarding the position of the TM1-TM2 hairpin, and the arrangement of TM2 with respect to TM3. We find that the NH2-terminus is a mixed helical hairpin, placed at the periplasmic interface of the lipid bilayer, away from the channel central axis of symmetry. Our experimental data suggest that the MscS closed state is in a different and a more compact conformation than the one trapped in the crystal structure.
RESULTS
Expression and purification of MscS cysteine mutants
Individual residues along the complete TM domain of MscS were sequentially mutated to cysteine and subsequently spin labeled (WT MscS is cysteine less). The targeted regions are highlighted in Figure 1A along a schematic representation of the MscS sequence, where individual rectangles represent the approximate length of the α-helical TM segments according to the crystal structure 8. The mutated residues (2 to 128) are also shown in Figure 1B as black spheres on a ribbon diagram of the MscS monomer, and over a schematic line in the structurally-undetermined NH2-terminus.
All MscS cysteine mutants were expressed, purified in reducing conditions, labeled, and reconstituted into pre-formed DOPC:POPG liposomes, as previously described 28. None of the mutations were associated with significant gain of function (GOF) phenotypes that would result in lethal leakage of solutes during cell growth, since the cultures optical density did not decrease upon induction with IPTG. Instead, some cysteine substitutions at conserved sites 8 promoted cell growth (W16C at the NH2-terminus; V32C and V40C at TM1; and A79C at TM2).
Mutant expression yield was in general below WT MscS (~ 4 mg, Figure 1C), and even lower for mutations at the NH2-terminus and the TM3 helix. Despite these differences, heptameric oligomerization was always achieved. This suggests that mutant purification under reducing conditions did not affect the overall architecture of the channel, i.e. not compromising the heptameric structure in the formation of spontaneous disulfide bridges, as suggested by gel filtration analysis and SDS-PAGE gels (Figure S1). Mutations on the narrowest part of the MscS permeation pathway (positions 93–101, 107 and 115) promoted some dissociation of the heptamer into an unfolded channel, which represented a small fraction of the total protein 28. For these particular TM3 mutants, EPR spectroscopy was performed on the fraction that corresponded to the heptamer, as determined from gel filtration analysis (Figure S1).
Environmental parameters of MscS TM segments
Spin labeled mutants were subject to CW-EPR spectroscopic measurements on liposomes, under zero transbilayer pressure and in absence of a TM voltage. It is expected that under these conditions the spectroscopic signal will come overwhelmingly from MscS in its closed state (Figure S2), with negligible contributions from other conformations. Nitroxide mobility and accessibility to O2 and NiEdda were measured for each MscS spin labeled mutant, and accessibility to the lipid Ni(II) chelate (DOGS-NTA) was evaluated in selected mutants at the NH2-terminus, TM1–TM2 loop, and at the end of the TM3B. It must be noted, however, that we have been unable to estimate inter-subunit distances or proximities in the present data set. This is likely due to under-labeling of samples caused by the presence of genomic MscS monomers (Figure S3). The high natural abundance of these cysteine-less monomers in the population of heptameric channels sharply reduce the probability of finding spin-spin coupling between adjacent subunits, and limits our ability to determine inter-subunit distances. However, this fact does not affect the quality and usefulness of the environmental data from under-labeled channels.
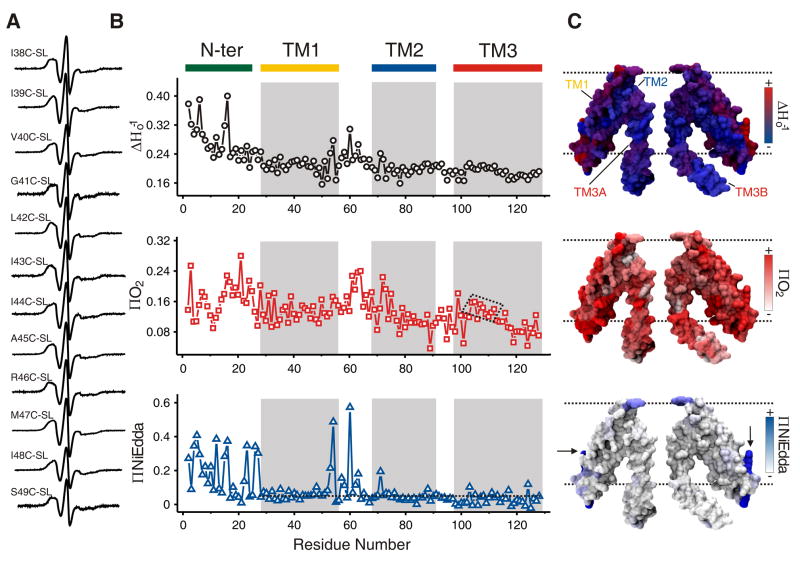
A representative set of EPR spectra obtained for MscS′ TM1 is shown in Figure 2A. The fully processed EPR environmental data set for the NH2-terminus and TM segments is shown in Figure 2B. Mobility and O2 accessibility parameters indicate a clear trend for the dynamics and membrane exposure of the TM domain. Excluding the first 26 residues of the NH2-terminus (see analysis below), residues located towards the NH2-terminal region of TM segments, periplasmic face of TM1 and cytoplasmic face of TM2, are more mobile and more exposed to O2 than those at the COOH-terminal. Average mobility and O2 accessibility (top and middle panels in Figure 2B, respectively) were found to be higher in TM1 and at the NH2-terminal segment of TM2 than those obtained for TM3. On the other hand, the EPR data suggests that the environment of TM3 helices is sterically restricted, as reflected by a lower O2 exposure and lower overall dynamics than seen for the remaining TM helices (see box plots in Figure 5B). This behavior is expected for a segment that lines the conduction pathway of an ion channel in its closed conformation. A patch of the TM3 segment (positions 104 to 112) exhibits higher mobility and O2 accessibility than the TM3 average (dotted box on middle panel of Figure 2B). This can be interpreted to support the hypothesis that the ion flow may be blocked by the hydrophobicity of the surface that contours the pore in the closed state 16.
Figure 2.
EPR spectroscopy of MscS (A) Representative X-band EPR spectra of consecutively spin-labeled mutants from TM1 segment of MscS reconstituted in DOPC:POPG liposomes. All spectra were obtained from samples with the same protein to lipid ratio, and using a loop-gap resonator with the microwave power set to 2 mW. (B) Residue-specific environmental parameter profiles obtained for the NH2 terminal and TM segments: mobility parameter ΔHo−1 (top, black circles), O2 accessibility parameter ΠO2 (middle, red squares, the dotted box pinpoints the patch of higher O2 accessibility in TM3), and NiEdda accessibility parameter ΠNiEdda (bottom, blue triangles, the broken line represents the NiEdda average for the TMs). Grayed areas represent the TM segment assignment derived from the MscS crystal structure. (C) Residue environmental parameter profiles mapped onto a molecular surface of two representative MscS monomers. Mobility parameter ΔHo−1 (top panel), O2 accessibility parameter ΠO2 (middle panel), and NiEdda accessibility parameter ΠNiEdda (bottom panel). Arrows on bottom panel pinpoint residues with high NiEdda accessibility in the middle of the bilayer.
Figure 5.
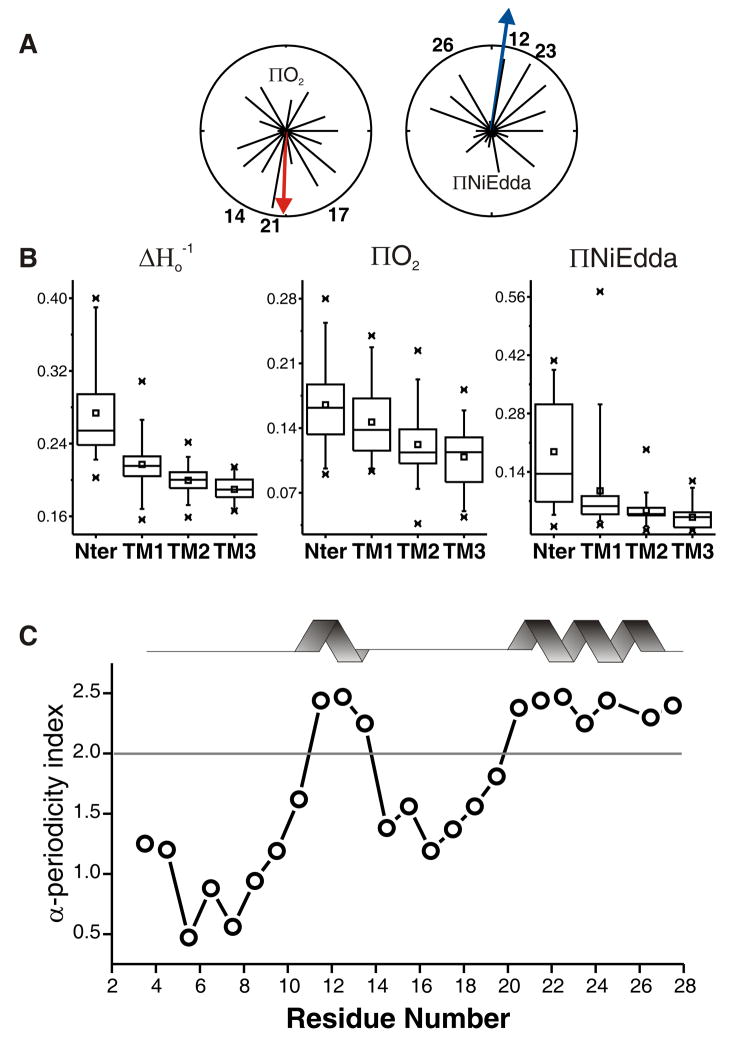
Structural Analysis of the NH2-terminal segment. (A) Helical wheel representation of the NH2-termini. Environmental parameters have been superimposed in a polar coordinate representation. A resultant vector was calculated for the O2 accessibility (left panel, red arrow) and NiEdda accessibility (right panel, blue arrow). (B) Box plot analysis of the NH2-termini and TM segments. Squares represent the mean, boxes the data distribution, and bars the standard deviation. (C) Windowed periodicity analysis for the NiEdda parameter ΠNiEdda. The α-helical periodicity index was calculated as described earlier 36 with an angular range from 80°–120° and a sliding window of seven residues. The horizontal line represents the threshold at which the periodicity of the windowed segment is significantly α-helical (10 to 14, and 20 to 27).
Although the MscS crystal structure was obtained in detergent, the EPR-derived environmental data mapped onto its surface (Figures 2C and 8A) reveals interesting correlations between the two types of structural data. First, probe dynamics and exposure to aqueous environment in the TM1–TM2 connecting loop (upper and bottom panel in Figure 2B–C) appears to be inconsistent with the boundary between TM1 and TM2 segments predicted by the crystal structure 8. Positions exposed to NiEdda at the TM1–TM2 hairpin appear to be in the non-polar region of the membrane (bottom panel in Figure 2C), while highly mobile and O2 exposed residues tend to locate at the cytoplasmic region of TM2 (upper and middle panel in Figure 2C, and left panel in Figure 8A). As NiEdda-accessible residues seem to cluster toward the COOH-terminal end of TM1 (bottom of Figure 2C and middle panel in Figure 8A), we would suggest that to match the membrane boundaries to the experimental data, these should shift about two turns of the helix towards the extracellular face of the channel in relation to putative boundaries based on the crystal structure.
Figure 8.
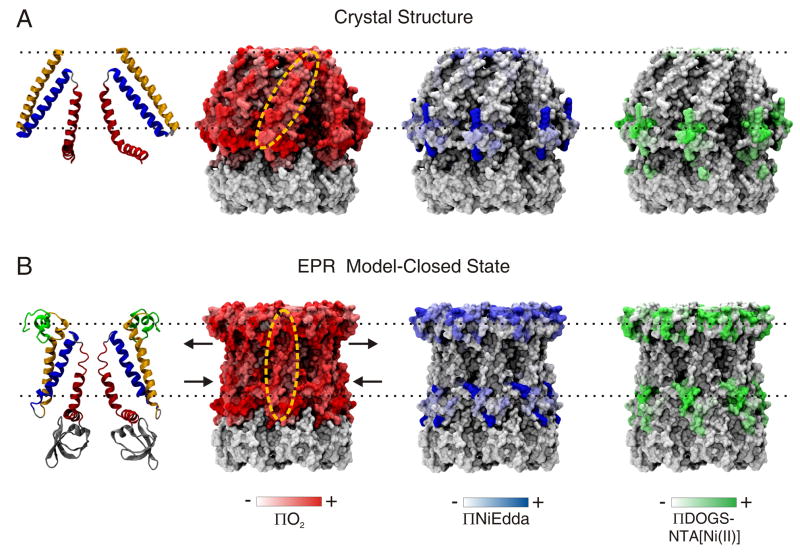
Comparison of MscS crystal structure and outcome of EPR based refinement. (A) Crystal structure model. Left panel, ribbon representation of the TM segments of the MscS (two subunits are shown for clarity). Individual TM segments are color-coded as follows: TM1, yellow; TM2, blue; and TM3, red. Right most panels show residue environmental parameter profiles mapped onto molecular surfaces of the x-ray model (B) Closed state model from EPR based refinement. Left panel, ribbon representation of the MscS closed state model. Individual TM segments are color-coded as above, except for the NH2-terminus in green. Right most panel show residue environmental parameter profiles mapped onto molecular surfaces of the EPR refined MscS model. The arrows show the key rearrangement in the TM segments of MscS in a native closed state.
Residue solvent accessibilities obtained using hard-sphere methods on the crystal structure were used to calculate TM1 and TM2 environmental moments, which were compared with the O2 accessibilities (ΠO2) resultant moment obtained from EPR data (Figure S4). The position of the resultant vectors agrees with the TM1 being exposed to two environments (lipidic and proteinaceous), and the TM2 COOH-terminal region facing a proteinaceous homogenous environment. For TM3, environmental moments could not be calculated, since overall mobility and accessibilities were quite low compared to those of the other TM segments. Within this segment, however, residues G104C-SL to Q112C-SL exhibited higher O2 accessibility than the TM3 average. Although the overall topology suggested by EPR environmental parameters in a membrane environment is consistent with the one depicted by the available MscS crystal structure, the data suggests that the closed conformation of MscS has to feature a TM2 helix that is packed against TM3. This suggests a more compact arrangement of the TM helices relative to each other and shifted downwards in relation to the plane of the bilayer.
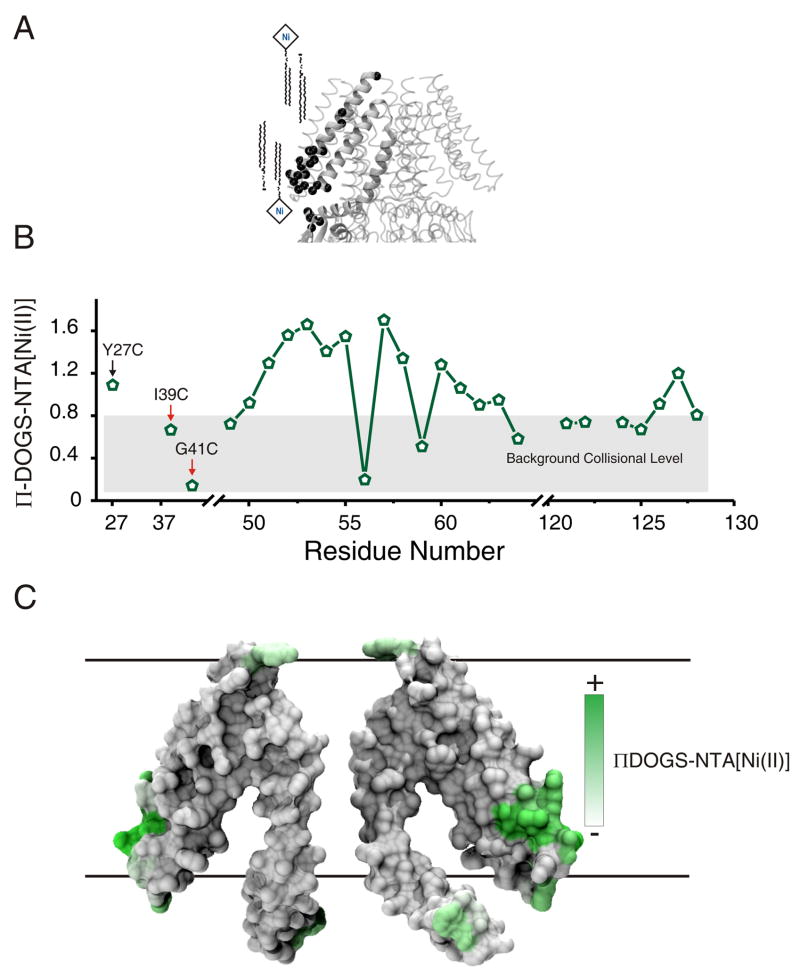
To assist us in predicting the position of MscS TM domain with respect to the membrane, we used DOGS-NTA[Ni(II)]lipids as a probe of residue interfacial positioning. These synthetic lipids confine the Ni(II) relaxing agent within a region of 14 Å at the membrane aqueous interface (14 Å being the maximum distance in the lipid extended configuration, Figure 3A) 29. Figure 3B and C shows the DOGS-NTA collisional frequency data obtained for positions at the end of TM1 and the beginning of TM2. We used two contrasting residues in the middle of the bilayer as experimental controls in order to set the background collisional levels, I39C-SL is one of the most O2 exposed positions in the core of the membrane while G41C-SL is one of the most buried ones (see red arrows on Figure 3B). According to these environmental parameters, the TM1–TM2 loop (positions 50–61) and positions M126C-SL, F127C-SL, and R128C-SL are at the membrane-aqueous interface, with positions N53C-SL, R54C-SL, K60C-SL, and A63C-SL largely exposed to the aqueous compound NiEdda.
Figure 3.
Identification of MscS interfacial residues with DOGS-NTA[Ni(II)]lipids (A) Ribbon diagram of the TM segments of the MscS crystal structure (a single MscS monomer is highlighted) embedded in a schematic membrane consisting of DOPC:POPG and DOGS-NTA[Ni(II)]lipids (few lipid molecules are shown for clarity). Black spheres show the location of cysteine mutants that were used to obtain the data. (B) Residue environmental parameter profile derived from Y27C (black arrow), TM1–TM2 loop and TM3B. Experimental controls (I39C and G41C) are identified with red arrows. Gray box highlights the region that corresponds to background collisional level. (C) Results mapped onto a molecular surface of MscS′ TM segments.
Conformation of the NH2-terminal segment
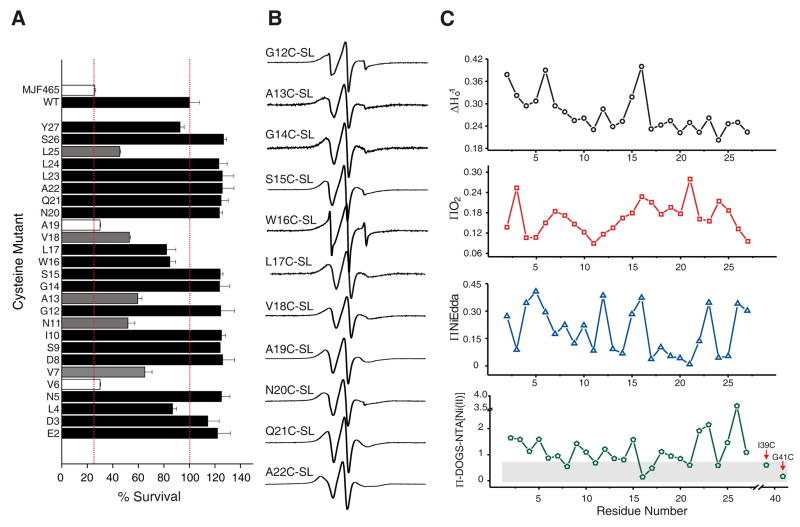
The majority of the studies performed on MscS, so far, have focused on its TM segments 30; 31; 32; 33 or its large cytoplasmic domain 34; 35, but not on the structurally unknown NH2-terminus. The MscS NH2-terminal segment, comprising 26 residues, was not resolved in the MscS crystal structure 8. However, functional experiments show that NH2-terminal truncation affects protein expression but not the ability of the channel to protect the cell during hypo-osmotic challenges 30. To establish any functional effect of the mutated cysteines on the channel function, after protein over expression, cells were subjected to a sudden osmotic downshock and cell survival assay 28. The majority of the cysteine mutants (73%) had no effect on the response of MscS to hypo-osmotic shock, 19% had a partially impaired response (V7C, N11C, A13C, V18C, and L25C) and only 8% experienced severe loss of function (LOF) mutants (V6C and A19C), i.e., channels that did not open upon increase in membrane tension by the osmotic downshock. Regardless of their particular functional phenotype, all of the NH2-terminal mutants were found to be heptamers after purification. These LOF phenotypes (likely caused by channels locked in a closed state) were not disregarded in the present analysis, since this study only covers the closed conformation of MscS.
To accurately evaluate the structure of the NH2-terminal region, we used an expression vector with the His-tag on the COOH-termini 28. Under these conditions, the NH2-terminus appears as a highly mobile fragment (Figure 4B, and 5B) with well defined segments of secondary structure. The spectral line shape and mobility values are of the motional regime found on residues at the surface of soluble proteins 21. The NH2 terminus mobility profile (Figure 4C top panel, black trace) shows a slight decrease in probe dynamics, from residue E2C-SL to the point of initiation of TM1, with the exception of positions V6C-SL and W16C-SL, which have high mobility values, an indication of local unfolding (loop or hinge region). O2 accessibility, shown in Figure 4C, mainly reveals a valley (S9C-SL up to A13C-SL) and a peak (W16C-SL up to L25C-SL) of accessibilities, whereas NiEdda clearly reveals an α-helical pattern. Calculation of the Fourier Transform power spectrum of the NiEdda accessibility profile gives a statistically significant peak of angular frequency of 100° (Figure S6), confirming the presence of α-helical structure. Figure 4C (bottom panel) shows the DOGS-NTA collisional frequency suggesting that the NH2-termini is located at the membrane interface. One segment of the NH2-termini being exposed to the hydrated side of the membrane (positions 4–6, 12, 22–23, and 26–27) while another one is exposed to the membrane fatty acid chains (positions 3, 7–9, 17–22). S15C-SL and W16C-SL have high accessibilities to both contrasting agents, but W16C-SL does not have much accessibility to the Ni(II) lipid chelate, this could account for the S15C-SL facing the membrane interface and the W16C-SL pointing towards the pore.
Figure 4.
Functional and residue-specific environmental parameter profiles of NH2-terminal cysteine mutants (A) Functional assays of cells containing NH2-terminal cysteine mutants over-expressed in pQE70 vector. MJF465 and WT MscS were used as a negative and positive control, respectively. Red dotted lines represent the mean value of WT MscS (right) and the mean value of MJF465 (left). (B) X-band EPR spectra of consecutively spin-labeled mutants from the NH2-terminal segment of MscS reconstituted in DOPC:POPG liposomes. (C) Residue-specific environmental parameter profiles obtained for the NH2-terminal segment: mobility parameter ΔHo−1 (top, black circles), O2 accessibility parameter ΠO2 (middle, red squares), NiEdda accessibility parameter ΠNiEdda (middle, blue triangles), and interfacial accessibility parameter ΠDOGS-NTA[Ni(II)]lipids (bottom, green rhombuses). Gray box highlights the region that corresponds to background collisional level.
We have used this set of environmental data to assign the secondary structure elements within the NH2-terminus. Figure 5A shows individual environmental moments for the residues in this mutant set and the vectors were superimposed on a helical wheel plot. The resultant O2 moment points to the face with the highest accessibility (positions G14C-SL, L17C-SL, and Q21C-SL) therefore to the hydrophobic part of the membrane, while the NiEdda moment is oriented about 180° away, mostly defined by positions G12C-SL, L23C-SL, and S26C-SL. Overall, these results establish the NH2-terminus as an amphipathic helix lying at the membrane interface. A comparison between the NH2-terminus and the three TM segments (Figure 5B) leads us to suggest that this amphipathic segment, as a whole, is likely extending away from the channel pore, since its average mobility and accessibilities are higher than those of the TM domain. Application of sliding α-periodicity index window helped us define statistically significant α-helical regions within the NH2-terminal segment 36; 37 (Figure 5C). This information was used to carry out an unbiased assignment of two α-helical segments, from I10 to G14 and N20 to Y27. These assignments served as the starting point for further refinement of our final three-dimensional MscS model.
Three-dimensional model for the NH2-terminus and TM segments of MscS in the closed state
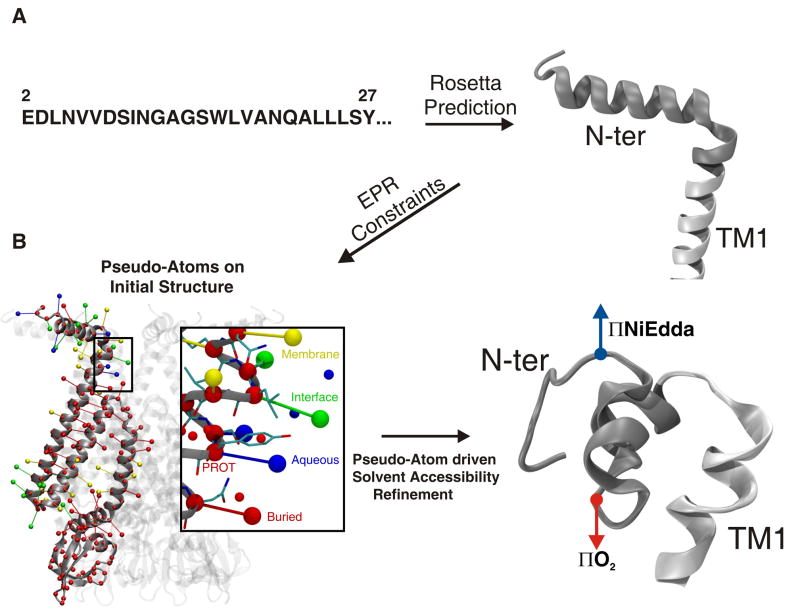
The present data set provides information-rich constraints (128 constraints, each with three degree of freedom) to refine the molecular architecture of closed MscS in its membrane environment. Using the available crystal structure of MscS as a starting point, we generated a structural model of the NH2-terminus of MscS in the closed state driven by the EPR environmental data. The starting model was based on a seven-fold symmetric structure that included an NH2-terminus conformation predicted by the program Rosetta 38; 39, as well as the MscS crystal (pdb core 2OAU 40) covering residues 27 to 178 (Figure 6A). Constraints on the secondary structure of the NH2-terminus (α-helix between residues 10 and 14, and 20 to 27) were incorporated based on frequency analysis of the angular periodicity components of the NiEdda accessibility profile (see Figure 5C). Structural refinement was carried out using a newly developed approach that takes advantage of EPR determined solvent accessibility restraints in MD simulations 41(Sompornpisut et al, 2007; unpublished data). In this approach, pseudo-atoms representing EPR spin-label probes are attached to each MscS residue (from positions 1 to 178, Figure 6B) and interact with pseudo-atoms that represent NiEdda and O2 molecules (Figure 6B inset). Their interactions were chosen to enforce the environments detected in the EPR experiments (aqueous, lipidic, buried in protein, membrane interface, or undetermined) as summarized in a phase diagram shown in Figure S7.
Figure 6.
Topology and structural model of MscS′ NH2-terminal domain, obtained by the pseudo-atom driven solvent accessibility refinement. (A) Secondary structure prediction, from the linear sequence (left panel) of residues 1 to 178, using the software Rosetta 38; 39. The right panel shows a conformation of the NH2-terminal domain obtained after energy minimization of the obtained model. The resulting structure was used in the EPR based refinement of the MscS closed conformation. (B) Left panel, cartoon representation of MscS model used as initial structure for refinement. The model includes residues 1 to 178. Pseudo-atoms representing the spin label attached to each residue are shown for one subunit and color coded as follows: red particles represent buried residues, blue particles represent aqueous residues, yellow particles represent residues facing the membrane, and green particles represent residues at the water-membrane interface. Red particles are shown for each C-α atom representing PROT particles attached to each residue. Inset shows protein residues in thin licorice representation and pseudo-spin probes in CPK representation. Unbounded particles in red and blue represent O2 and NiEdda virtual environment particles, respectively. View of the NH2-terminal model after pseudo-atom driven solvent accessibility refinement is shown on the right panel. Residues 1 to 28 are shown in gray, resultant vectors for NiEdda and O2 accessibilities are shown in blue and red respectively.
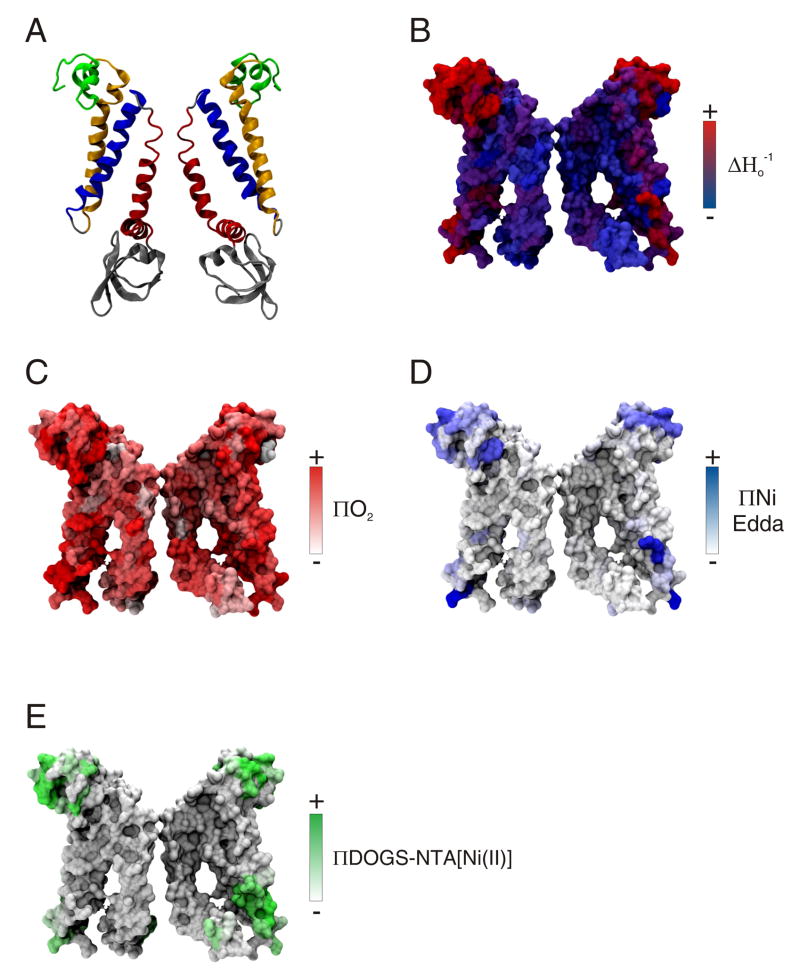
MD simulations including the EPR constraints and probes mentioned above, together with local adjustments to ensure orientation of residues at the membrane interface, permitted a dynamic refinement of MscS. The resulting MscS model, shown in Figures 6B (NH2-terminus) and 7, satisfies the mobility and accessibility data by: compacting the TM1-TM2 helices against the pore, featuring a reorientation of these two helices (of ~ 9°) with respect to the normal of the membrane, and narrowing the pore lined by the TM3 helices. Interestingly, within the channel context, the NH2-terminus conformation exhibits an amphipathic segment with α-helical regions at the membrane interface, which must face away from the center of the channel in order to fit the mobility and accessibility data. As expected, the experimental data mapped onto the surfaces of the resulting model of the MscS closed state are consistent with the obtained conformation (Figures 7A–E).
Figure 7.
MscS closed conformation obtained from EPR based refinement (A) Ribbon representation of the MscS closed state model (two subunits are shown for clarity). Individual TM segments are color-coded as follows: NH2-terminus, green, TM1, yellow; TM2, blue; and TM3, red. (B–E) Mobility parameter ΔHo−1, O2 accessibility parameter ΠO2, NiEdda accessibility parameter ΠNiEdda, and interfacial accessibility parameter ΠDOGS-NTA[Ni(II)]lipids, respectively.
The stability of the resulting symmetric model was then probed through all-atom MD simulations of MscS in an explicit, fully-hydrated membrane environment (Figure S8A). The structure remained reasonably stable during six nanoseconds of free dynamics (without any symmetric constraint, Figure S8B). The root-mean square deviation (RMSD) of the complete model and of the TM domain alone reached values of 3.5 and 2.6 Å, respectively (Figure S8E). The larger value observed for the complete structure likely reflects the high mobility of the NH2-terminus. In fact, four out of seven subunits keep their initial architecture, but partially lose their initial secondary structure.
The RMSD values observed for the EPR-based model were found to be similar to those observed for the MscS crystal structure (pdb code 2OAU), which closes asymmetrically when simulated in a relaxed membrane environment (see Figure S8C-D). Two additional simulations of the EPR-based closed model were performed at +/− 1.2 Volts (3 ns each). The simulations demonstrated that the structure indeed represents a conformation of very low conductance (data not shown). Water penetration into the periplasmic mouth of the channel observed throughout simulations, along with the NiEdda accessibility data suggests that the periplasmic channel region might be even more occluded than what the EPR-based model predicts. Finally, it is interesting to note that the model predicts formation of a salt bridge between residues 62 and 128 of adjacent subunits, likely relevant for MscS function 15.
DISCUSSION
Membrane proteins are a fundamental part of the cell machinery, serving as switches, sensors, and bridges between the extracellular and intracellular space. However, establishing the correlation between structure and mechanism is fraught with difficulty. X-ray crystallography can provide a detailed atomistic view of a membrane protein, but crystallization in absence of a native-like lipid environment has the potential to induce a non-physiological conformation 42. Particularly, for the MscS channel it is very important to know its conformation in the membrane, since its gating relies on the direct interaction with the lipid bilayer 33. Determining what functional state is depicted by the crystal conformation is extremely challenging, particularly when large conformational changes arise from external stimuli. MS channels undergo such large rearrangements in their structure upon application of mechanical stimuli, as has been shown in MscL 11. Unambiguous identification of MscS’s closed, open, and inactive conformations is an essential step in the quest to understand its gating mechanism and function in bacterial cells and mechanosensation, in general.
The present study focuses on determining the three-dimensional fold of MscS NH2-terminus and TM domain, when MscS is in its closed conformation, embedded in a lipid bilayer. EPR spectroscopy analysis of spin labeled mutants provided information on the topology, secondary, and tertiary structure of MscS under native conditions. This information was used, along with MD simulations, to evaluate a recently further refined crystal structure 40 and propose a model of MscS in a closed conformation.
The EPR data shows that there are general similarities between the overall structure and topology of MscS in a biological membrane and in the crystal conformation. However, some differences point to the fact that the MscS crystal structure does not represent the true closed state, unlike MscL where differences between the crystal structure and the one suggested by the EPR data are more subtle 9. The accessibilities to the water-soluble compound NiEdda indicate that positions at the cytoplasmic end of TM1, that were predicted to be in contact with fatty acid chains, were able to collide with this aqueous reagent. Additional evidence for the lipid exposure of MscS came from the results obtained with the DOGS-NTA[Ni(II)]lipids. These results, when combined with information obtained from mobility, O2, and NiEdda accessibilities, clearly localizes the protein within the lipid bilayer. The thickness of a DOPC membrane, ~ 50 Å 43, was used along with the accessibilities obtained with DOGS-NTA[Ni(II)]lipids to determine how MscS sits in the lipid bilayer.
According to the crystal structure, the TM2 helix should be asymmetrically solvated, i.e., one surface of this helix should be facing TM1 (buried), while the opposing surface should be solvated by lipids found in crevices between TM1–TM2 hairpins. However, our model suggests that, in the closed state, the TM2 NH2-terminal faces the lipid bilayer while its COOH-terminal region is less exposed to the membrane and likely faces TM3, placing TM2 spatially closer to TM3 than what is predicted by the crystal. The TM2 helix is also predicted to be longer, beginning at position T64 instead of F68 (crystal). It remains to be elucidated if the rotation in the membrane plane of the TM3B helix (Figure 8B) affects the architecture of the cytoplasmic domain and its openings.
While overall exposure to O2 and NiEdda in TM3 is low when compared to TM1 and TM2, a patch of higher O2 accessibilities than the TM3 average (positions 104 to 112), was observed (Figure 2B–C). This hydrophobic area could account for a partially open pore but functionally closed with a region of the permeation path that contains no water and, thus, blocks ion conduction. Multiple computational studies of water dynamics inside MscS have shown that such scenario is possible 15; 16; 18; 20 (Figure S8B). Other ion channels trapped in the closed conformation such as MscL 7, the nicotinic acetylcholine receptor 44, KcsA 45 and KirBac1.1 46, have been shown to maintain a non-conducting pore even if the dimensions of the permeation path are large enough to allow passage of dehydrated ions.
The MscS NH2-terminal segment model presented here forms a cap at the periplasmic face of the channel. In order to satisfy our mobility and accessibility data the final model displays an NH2-terminal domain that points away from the permeation pathway and includes 42% of α-helical structure. The function of this segment is unknown, but substitutions at W16 affects the response to negative pressure under patch clamp conditions 47, and deletions on the NH2-terminal segment affect the organization, and perhaps incorporation, of MscS in the lipid bilayer 30. We can then suggest that, the NH2-terminal could also play a potential role in membrane-modulated gating due to its amphipathic nature.
Using a novel refinement method with energy restraints exploiting the information from EPR data, we have rearranged the x-ray structure into a model that represents the closed state. While the final model incorporates as much information as possible from the experiments, there are two limitations of the modeling process that should be noted. First, prediction of a three-dimensional model based only on sequence is difficult, particularly for a multimeric membrane protein. Second, the EPR derived data presented here represents an average over a large population of channels and, therefore, asymmetry as observed in the crystal structure and MD simulations 15; 17; 18; 20 cannot be detected. Thus, the presented model is a symmetrized version of MscS. Nevertheless, the power of our approach comes from the pattern of global accessibilities more than the absolute values of a given position, along with the fact that the membrane has well defined environments that reduce the number of models that can properly fit the experimental constraints. Further structural, spectroscopical, and computational studies will be required to overcome these limitations and to also establish the open state of MscS, and whether the conformation depicted by the crystal structure is physiologically relevant.
A summary of the proposed closed conformation of the MscS TM domain in a lipid bilayer and a comparison to the crystal conformation is presented in Figure 8. TM helices are more perpendicular to the plane of the membrane than the helices in the crystal structure. TM1 has two faces, one exposed to the membrane and the other one to TM2. TM2 is surrounded by TM1 and TM3, except for its cytoplasmic region that has some exposure to the membrane. In our refined model, the pore is apparently narrower, but how much closer the TM3 helices are to each other cannot be determined in a quantitative manner. The nine degree inward motion of TM1–TM2 favoring a more compact structure, and the almost horizontal TM3B helix observed during refinement are both in agreement with the motions observed in previous MD studies 15.
Finally, the present study also provides some hints to an additional, physiologically relevant function of MscS. Some cysteine mutants produced twice the amount of cells than the WT and other cysteine mutants (after IPTG induction). The respective residues, most of them hydrophobic (W16, V32, V40 and A79), are conserved throughout the MscS family 8. Residue W16 has already been shown to participate in the positioning of MscS on the membrane 30, and it does not seem to dramatically affect MscS function (see Figure 4A and refs. 30,47). In contrast, when residue V40 was substituted by aspartic acid or lysine, a significant GOF phenotype (in which cells were not able to grow) was observed, whereas cysteine substitution was tolerated by the cell 48. Additionally, cells lacking MS channels exhibit a faster growth rate than other E. coli strains (data not shown). Eukaryotic MS channels have been shown to play an important role during mitosis, as the mechanical properties of their membrane changes during the cell cycle 49; 50; 51. Perhaps MscS also plays a role in bacterial cell division.
EXPERIMENTAL PROCEDURES
Mutagenesis, expression, spin labeling, and reconstitution of MscS
Cysteine mutants were generated for residues 2–128 in MscS, covering the NH2-terminus, TM1, TM2, and TM3. Mutagenesis was performed using the QuickChange™ Site-Directed Mutagenesis Kit (Stratagene). Mutant channels were expressed, purified, labeled and reconstituted as previously described 28 using a pET28 vector containing MscS with His6 epitope at the NH2-termini and a pQE70 vector with His6 epitope at the COOH-termini, for the study of the TM segments and the NH2-terminus, respectively. MscS-pET28 was used to express the channel in E. coli Roseta™ and MscS-pQE70 was used for expression in E. coli M15. The samples were reconstituted at a 750:1 lipid/channel (DOPC:POPG, 6:1) molar ratio by dilution in PBS in the presence of Bio-beads. To determine the interfacial residues a mixture of DOPC:POPG:DOGS-NTA[Ni(II)]lipids, in a molar ratio of 6:2:3, respectively, was used.
EPR spectroscopy and data analysis
All EPR data were obtained at room temperature. X-band CW EPR spectra were performed as previously described 22. Spectra were obtained in a Bruker EMX spectrometer fitted with a loop gap resonator under the following conditions: 2 mW incident power, 100 kHz modulation frequency and 1 G modulation. For DOGS-NTA[Ni(II)]lipids experiments a dielectric resonator was used. The accessibility parameters were quantified as previously reported 22; 29; 52. Briefly, collision frequency is directly proportional to the accessibility of the paramagnetic reagent to the nitroxide, and is defined as
where P1/2(DPPH) is the microwave power that saturated the signal relative to that without saturation for a 2,2-diphenyl-1-picrylhydrazyl crystal (molecule with a single unpaired electron), ΔP1/2(X) is the difference in P1/2 of a given sample exposed to O2 or NiEdda and N2. ΔHo is an estimate of probe mobility derived from the spectral line-shape, determined by the degree of averaging of the anisotropic g value 53.
Functional assays
Downshock assays were performed as described earlier 54, using the E. coli strain MJF465 (kindly donated by Ian Booth 4. Method modifications and details were provided earlier 28. In brief, cultures growing in LB and 500 mM NaCl were induced at an OD600 of 0.6 with 1 mM IPTG; after being induced for 1 h, the cells were adjusted to an OD600 of 1.0, pelleted down, and then resuspended in 50 μL of fresh LB and 500 mM NaCl. The downshock was made by diluting the cells 1:40 in various shock media ranging from 0 to 500 mM NaCl (pH 7). After 5 min, the cells were serially diluted and 5 μL aliquots were plated on LB agar. Plated cells were grown overnight, and the colonies were counted and normalized against the OD600 (measured right before the downshock).
NH2-terminal model and pseudo-atom driven solvent accessibility refinement of MscS
An initial, symmetrized model of MscS was built from monomer C of the refined MscS crystal structure (pdb code 2AOU) 40 including residues 1 to 178. The model included a conformation of the NH2-terminus obtained using the software Rosetta 38; 39 and applying energy minimization in vacuum (Figure 6A). In addition to protein atoms, two EPR virtual particles were harmonically attached to each C-α atom of the protein, following the method described by 41(Sompornpisut et al, 2007; unpublished data). The first EPR particle, labeled PROT, was attached on top of the C-α atom, while the second EPR particle, representing a spin label probe, was attached through a bond with a resting length of 6 Å (Figure 6B).
The water or lipid exposed pseudo-spin probes were assigned attractive or repulsive interactions to additional O2 and NiEdda virtual environment particles, distributed so as to mimic the membrane location, while the buried pseudo-spins were allowed to artificially overlap with the backbone core (PROT particles). Assignment of interactions was performed based on EPR accessibilities as indicated on the text.
Structural refinement simulations were performed using the CHARMM program version c32a2 55 modified to implement the novel methodology. The extended atom PARAM19 force field was modified to reduce the effect of charged residues in vacuum according to the scheme of ref. 56. Harmonic restraints were applied to keep O2 particles away from the MscS pore, to keep residues 5, 23, 53, 57, 60, and 127 at the membrane-water interface, and to force side-chains of residues 23 and 27 to face away from the center of the channel. In addition, RMSD constraints were applied (to maintain the initial secondary structure) to residues 10 to 14, 20 to 26, 29 to 57, 68 to 91, 96 to 113, 114 to 128, and 129 to 178. Multiple minimization and equilibration cycles were performed. After ten million steps (1 fs/time step), the NH2-terminus was rotated and further equilibration was carried out.
Accession Codes
Protein Data Bank: Coordinates have been deposited with accession codes XXXX
Supplementary Material
Acknowledgments
The authors thank J. Cordero-Morales, P. Sompornpisut, S. Chakrapani, C. Ptak, L. Cuello, V. Jogini, and H. Raghuraman for providing comments and experimental advice. This work was supported by the National Institutes of Health to E.P. (NIH grant No. GM063617), to K.S. (NIH grants No. P41-RR05969, No.1 R01 GM067887), and to B.R. (NIH grant No GM-62342). M.S and K.S. also acknowledge supercomputer time provided through NSF grant LRAC MCA93S028. Data mapping and molecular images were done using VMD 57, its data-import plugin, and Tachyon 58. The authors declare that they have no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wood JM. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol Mol Biol Rev. 1999;63:230–62. doi: 10.1128/mmbr.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamill OP, Martinac B. Molecular basis of mechanotransduction in living cells. Physiol Rev. 2001;81:685–740. doi: 10.1152/physrev.2001.81.2.685. [DOI] [PubMed] [Google Scholar]
- 3.Poolman B, Blount P, Folgering JH, Friesen RH, Moe PC, van der Heide T. How do membrane proteins sense water stress? Mol Microbiol. 2002;44:889–902. doi: 10.1046/j.1365-2958.2002.02894.x. [DOI] [PubMed] [Google Scholar]
- 4.Levina N, Totemeyer S, Stokes NR, Louis P, Jones MA, Booth IR. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. Embo J. 1999;18:1730–7. doi: 10.1093/emboj/18.7.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinac B, Buechner M, Delcour AH, Adler J, Kung C. Pressure-sensitive ion channel in Escherichia coli. Proc Natl Acad Sci U S A. 1987;84:2297–301. doi: 10.1073/pnas.84.8.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berrier C, Besnard M, Ajouz B, Coulombe A, Ghazi A. Multiple mechanosensitive ion channels from Escherichia coli, activated at different thresholds of applied pressure. J Membr Biol. 1996;151:175–87. doi: 10.1007/s002329900068. [DOI] [PubMed] [Google Scholar]
- 7.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science. 1998;282:2220–6. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 8.Bass RB, Strop P, Barclay M, Rees DC. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science. 2002;298:1582–7. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 9.Perozo E, Kloda A, Cortes DM, Martinac B. Site-directed spin-labeling analysis of reconstituted Mscl in the closed state. J Gen Physiol. 2001;118:193–206. doi: 10.1085/jgp.118.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukharev S, Betanzos M, Chiang CS, Guy HR. The gating mechanism of the large mechanosensitive channel MscL. Nature. 2001;409:720–4. doi: 10.1038/35055559. [DOI] [PubMed] [Google Scholar]
- 11.Perozo E, Cortes DM, Sompornpisut P, Kloda A, Martinac B. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 2002;418:942–8. doi: 10.1038/nature00992. [DOI] [PubMed] [Google Scholar]
- 12.Perozo E, Kloda A, Cortes DM, Martinac B. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat Struct Biol. 2002;9:696–703. doi: 10.1038/nsb827. [DOI] [PubMed] [Google Scholar]
- 13.Anishkin A, Gendel V, Sharifi NA, Chiang CS, Shirinian L, Guy HR, Sukharev S. On the conformation of the COOH-terminal domain of the large mechanosensitive channel MscL. J Gen Physiol. 2003;121:227–44. doi: 10.1085/jgp.20028768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gullingsrud J, Schulten K. Gating of MscL studied by steered molecular dynamics. Biophys J. 2003;85:2087–99. doi: 10.1016/s0006-3495(03)74637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sotomayor M, Schulten K. Molecular dynamics study of gating in the mechanosensitive channel of small conductance MscS. Biophys J. 2004;87:3050–65. doi: 10.1529/biophysj.104.046045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anishkin A, Sukharev S. Water dynamics and dewetting transitions in the small mechanosensitive channel MscS. Biophys J. 2004;86:2883–95. doi: 10.1016/S0006-3495(04)74340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sotomayor M, van der Straaten TA, Ravaioli U, Schulten K. Electrostatic properties of the mechanosensitive channel of small conductance MscS. Biophys J. 2006;90:3496–510. doi: 10.1529/biophysj.105.080069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spronk SA, Elmore DE, Dougherty DA. Voltage-dependent hydration and conduction properties of the hydrophobic pore of the mechanosensitive channel of small conductance. Biophys J. 2006;90:3555–69. doi: 10.1529/biophysj.105.080432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vora T, Corry B, Chung SH. Brownian dynamics investigation into the conductance state of the MscS channel crystal structure. Biochim Biophys Acta. 2006;1758:730–7. doi: 10.1016/j.bbamem.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Sotomayor M, Vasquez V, Perozo E, Schulten K. Ion Conduction through MscS as Determined by Electrophysiology and Simulation. Biophys J. 2007;92:886–902. doi: 10.1529/biophysj.106.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 22.Perozo E, Cortes DM, Cuello LG. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat Struct Biol. 1998;5:459–69. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 23.Cuello LG, Cortes DM, Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–5. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 24.Columbus L, Kalai T, Jeko J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in alpha-helices: analysis by variation of side chain structure. Biochemistry. 2001;40:3828–46. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 25.Koteiche HA, McHaourab HS. Folding pattern of the alpha-crystallin domain in alphaA-crystallin determined by site-directed spin labeling. J Mol Biol. 1999;294:561–77. doi: 10.1006/jmbi.1999.3242. [DOI] [PubMed] [Google Scholar]
- 26.Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–8. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 27.Cortes DM, Cuello LG, Perozo E. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol. 2001;117:165–80. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasquez V, Cortes DM, Furukawa H, Perozo E. An Optimized Purification and Reconstitution Method for the MscS Channel: Strategies for Spectroscopical Analysis. Biochemistry. 2007;46:6766–6773. doi: 10.1021/bi700322k. [DOI] [PubMed] [Google Scholar]
- 29.Gross A, Hubbell WL. Identification of protein side chains near the membrane-aqueous interface: a site-directed spin labeling study of KcsA. Biochemistry. 2002;41:1123–8. doi: 10.1021/bi015828s. [DOI] [PubMed] [Google Scholar]
- 30.Miller S, Bartlett W, Chandrasekaran S, Simpson S, Edwards M, Booth IR. Domain organization of the MscS mechanosensitive channel of Escherichia coli. Embo J. 2003;22:36–46. doi: 10.1093/emboj/cdg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller S, Edwards MD, Ozdemir C, Booth IR. The closed structure of the MscS mechanosensitive channel. Cross-linking of single cysteine mutants. J Biol Chem. 2003;278:32246–50. doi: 10.1074/jbc.M303188200. [DOI] [PubMed] [Google Scholar]
- 32.Edwards MD, Li Y, Kim S, Miller S, Bartlett W, Black S, Dennison S, Iscla I, Blount P, Bowie JU, Booth IR. Pivotal role of the glycine-rich TM3 helix in gating the MscS mechanosensitive channel. Nat Struct Mol Biol. 2005;12:113–9. doi: 10.1038/nsmb895. [DOI] [PubMed] [Google Scholar]
- 33.Takeshi N, Sokabe M, Yoshimura K. Lipid-protein interaction of the MscS mechanosensitive channel examined by scanning mutagenesis. Biophys J. 2006 doi: 10.1529/biophysj.106.084541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koprowski P, Kubalski A. C termini of the Escherichia coli mechanosensitive ion channel (MscS) move apart upon the channel opening. J Biol Chem. 2003;278:11237–45. doi: 10.1074/jbc.M212073200. [DOI] [PubMed] [Google Scholar]
- 35.Schumann U, Edwards MD, Li C, Booth IR. The conserved carboxy-terminus of the MscS mechanosensitive channel is not essential but increases stability and activity. FEBS Lett. 2004;572:233–7. doi: 10.1016/j.febslet.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 36.Cornette JL, Cease KB, Margalit H, Spouge JL, Berzofsky JA, DeLisi C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J Mol Biol. 1987;195:659–85. doi: 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- 37.Rees DC, DeAntonio L, Eisenberg D. Hydrophobic organization of membrane proteins. Science. 1989;245:510–3. doi: 10.1126/science.2667138. [DOI] [PubMed] [Google Scholar]
- 38.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268:209–25. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 39.Bonneau R, Strauss CE, Rohl CA, Chivian D, Bradley P, Malmstrom L, Robertson T, Baker D. De novo prediction of three-dimensional structures for major protein families. J Mol Biol. 2002;322:65–78. doi: 10.1016/s0022-2836(02)00698-8. [DOI] [PubMed] [Google Scholar]
- 40.Steinbacher S, Bass R, Strop P, Rees DC. Current Topics in Membranes. Mechanosensitive Ion Channels. Part A. Vol. 58. Elsevier Inc; 2007. Structures of the prokaryotic mechanosensitive channels MscL and MscS. [Google Scholar]
- 41.Sompornpisut P, Perozo E, Roux B. Structural Refinement of Membrane Proteins on the Basis of Experimentally-determined Solvent Accessibility from Spin-Label Electron Paramagnetic Resonance Spectroscopy; 51st Annual Meeting; Biophysical Society, Baltimore, Maryland. 2007. [Google Scholar]
- 42.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 43.Wiener MC, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. III. Complete structure. Biophys J. 1992;61:434–47. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–55. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 45.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 46.Kuo A, Gulbis JM, Antcliff JF, Rahman T, Lowe ED, Zimmer J, Cuthbertson J, Ashcroft FM, Ezaki T, Doyle DA. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 2003;300:1922–6. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 47.Rasmussen A, Rasmussen T, Edwards MD, Schauer D, Schumann U, Miller S, Booth IR. The Role of Tryptophan Residues in the Function and Stability of the Mechanosensitive Channel MscS from Escherichia coli. Biochemistry. 2007;46:10899–908. doi: 10.1021/bi701056k. [DOI] [PubMed] [Google Scholar]
- 48.Okada K, Moe PC, Blount P. Functional design of bacterial mechanosensitive channels. Comparisons and contrasts illuminated by random mutagenesis. J Biol Chem. 2002;277:27682–8. doi: 10.1074/jbc.M202497200. [DOI] [PubMed] [Google Scholar]
- 49.Zhou XL, Kung C. A mechanosensitive ion channel in Schizosaccharomyces pombe. Embo J. 1992;11:2869–75. doi: 10.1002/j.1460-2075.1992.tb05355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rundle DR, Gorbsky G, Tsiokas L. PKD2 interacts and co-localizes with mDia1 to mitotic spindles of dividing cells: role of mDia1 IN PKD2 localization to mitotic spindles. J Biol Chem. 2004;279:29728–39. doi: 10.1074/jbc.M400544200. [DOI] [PubMed] [Google Scholar]
- 51.Haswell ES, Meyerowitz EM. MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr Biol. 2006;16:1–11. doi: 10.1016/j.cub.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 52.Farahbakhsh ZT, Altenbach C, Hubbell WL. Spin labeled cysteines as sensors for protein-lipid interaction and conformation in rhodopsin. Photochem Photobiol. 1992;56:1019–33. doi: 10.1111/j.1751-1097.1992.tb09725.x. [DOI] [PubMed] [Google Scholar]
- 53.McHaourab HS, Perozo E. Determination of protein folds and conformational dynamics using spin-labeling EPR spectroscopy. Vol. 19. Springer; 2002. [Google Scholar]
- 54.Batiza AF, Kuo MM, Yoshimura K, Kung C. Gating the bacterial mechanosensitive channel MscL in vivo. Proc Natl Acad Sci U S A. 2002;99:5643–8. doi: 10.1073/pnas.082092599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brooks B, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 56.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–52. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 57.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 58.Stone J. An efficient library for paralel ray tracing and animation. Master Thesis, University of Missouri-Rolla 1998 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.