Abstract
The molecular mechanism of thyroid hormone (TH) effects to fatty acid metabolism in liver is yet to be clear. The carbohydrate response element-binding protein (ChREBP) as well as sterol response element-binding protein (SREBP)-1c plays a pivotal role in hepatic lipogenesis. Both SREBP-1c and ChREBP are target genes of liver X receptors (LXRs). Because LXRs and TH receptors (TRs) cross talk mutually in many aspects of transcription, we examined whether TRs regulate the mouse ChREBP gene expression. In the current study, we demonstrated that TH up-regulated mouse ChREBP mRNA and protein expression in liver. Run-on and luciferase assays showed that TH and TR-β1 positively regulated the ChREBP gene transcription. The mouse ChREBP gene promoter contains two direct repeat-4 sites (LXRE1 and LXRE2) and EMSAs demonstrated that LXR-α and TR-β1 prefer to bind LXRE1 and LXRE2, respectively. The direct repeat-4 deletion and LXRE2 mutants of the promoter deteriorate the positive regulation by TR-β1, indicating that LXRE2 is functionally important for the regulation. We also showed that human ChREBP gene expression and promoter activities were up-regulated by TH. These data suggest that ChREBP mRNA expression is positively regulated by TR-β1 and TH at the transcriptional level in mammals. This novel observation indicates that TH fine-tunes hepatic lipogenesis via regulating SREBP-1c and ChREBP gene expression reciprocally.
Mouse and human carbohydrate response element-binding protein gene expression is positively regulated by thyroid hormone in liver at the transcriptional levels, indicating the hormone fine-tunes hepatic lipogenesis.
Thyroid hormone (TH) gives great impact for lipid homeostasis (1,2,3). The role of TH and TH receptors (TRs) in cholesterol metabolism has been examined intensively for years (1,3,4). However, it still remains unclear how TH affects triglyceride metabolism (5,6,7,8,9). Sterol response element binding protein (SREBP)-1c is a major factor for triglyceride synthesis (10,11,12). We and another group have recently shown that SREBP-1c gene expression is negatively regulated by TH (13,14). The carbohydrate response element-binding protein (ChREBP) is a glucose-responsive basic/helix-loop-helix/leucine zipper transcription factor (15,16), which binds to carbohydrate-responsive element in the glycolytic and lipogenic gene promoter regions (16,17). ChREBP plays a pivotal role to induce liver-pyruvate kinase (L-PK), one of the rate-limiting enzymes of glycolysis, which is exclusively glucose dependent (11,18,19,20). Lipogenic genes such as acetyl-CoA carboxylase (ACC) and fatty acid synthase are regulated by ChREBP and SREBP-1c in response to glucose and insulin, respectively (21). ChREBP is located in the cytosol in low-glucose conditions and enters from the cytosol into the nucleus under high-glucose conditions (22,23). ChREBP is also regulated by glucose at the transcriptional level (24). Therefore, ChREBP, which is a glucose-sensitive transcription factor that is related to convert carbohydrate to lipid in the liver, plays a pivotal role as well as SREBP-1c (25). Intriguingly, ChREBP has been recently identified as a direct target of Liver X receptors (LXRs) (26), which are nuclear receptors that play pivotal roles in the transcriptional control of lipid and carbohydrate metabolism (27,28,29,30,31,32). Thus, both SREBP-1c and ChREBP are regulated by LXRs (26). LXRs and TRs cross talk mutually in many aspects of transcription, sharing the same DNA binding site [direct repeat-4 (DR-4)] with identical geometry and polarity (33,34,35,36,37,38). We recently showed that TR-β1 and LXR-α interact on the mouse cholesterol 7α-hydroxylase gene promoter (39) and that LXR-α gene promoter itself is positively regulated by TH and TR-β1 (40), suggesting cross talk between the two receptors. Therefore, we examined whether TRs regulate the mouse ChREBP gene expression. In the current study, we demonstrated that TH up-regulated mouse ChREBP mRNA and protein expression in liver. We have also shown that mouse ChREBP gene promoter as well as the human promoter is activated by TH.
Materials and Methods
Animals
Four-week-old male C57/BL6 mice were used for the study. All aspects of animal care were approved by the Institutional Animal Care and Use Committee of Gunma University Graduate School of Medicine (Maebashi, Gunma, Japan). Animals were maintained on a 12-h light, 12-h dark schedule (lights on at 0600 h) and fed laboratory chow as indicated and given water ad libitum. The mice were rendered hypothyroid by the inclusion of 0.1% methimazole (MMI) in the drinking water and 1% (wt/wt) propylthiouracil (PTU) in the chow for 21 d (14). To introduce a thyrotoxic status, the mice were injected daily with 10 μg per 100 g body weight of T3 for an additional 5-d period (14). After the treatment, we measured the body weight (24 h) and evaluated the food intake and blood glucose levels (fasting) in each group. The blood glucose levels were measured with Glutest-AceR (Sanwa Kagaku Kenkyusho Co., Ltd., Nagoya, Japan) using mouse tail blood. The number of mice receiving each treatment is indicated in the figure legends. Serum-free T4 levels were determined using a GammaCoat RIA kit (DiaSorin Inc., Stillwater, MN) and free T3 levels were determined using an AMERLEX-MAB kit, and we confirmed the mouse serum TH levels as described in reference (14). The serum TH data were reported in our previous report (14).
Plasmids
The mouse ChREBP gene promoter (−3801/+9bp) plasmid, which contained the region from −3801 to +9 bp of the ChREBP gene, was generated by genomic PCR using 5′-GTGTGTCGACTAGACTGTGTAGATG-3′ as a sense primer and 5′-GTGTGTCGACGGCCACTATTGTCGC-3′ as an antisense primer. A SalI restriction enzyme site was introduced into the primer sequences so that the PCR product could be subcloned into XhoI site in the pGL4-Luc vector (Promega, Madison, WI). The human ChREBP promoter (−2092 to +32 bp) plasmid, which contained the region from −2092 to +32 bp of the human ChREBP gene, was generated by genomic PCR using 5′-GTGTGGATCCACGCTCCAGTGAGGT-3′ as a sense primer and 5′-GTGTAAGCTTAACCGCCTGGTCCCT-3′ as an antisense primer. For the human promoter, BamH1 and HindIII restriction enzyme sites were used for subcloning into the pGL4-Luc vector. We excised the −3801/+9 bp plasmid with Xho1 and EcoR1 restriction enzyme to prepare the DR-4 sites deletion construct (ΔDR-4: −2571 to −2307 bp region was deleted). The −2307 to +9 bp construct of the mouse ChREBP gene were generated from the −3801/+9 bp plasmid with Asp718 and EcoR1 restriction enzyme followed by T4 DNA polymerase reaction to make blunt end. The DR-4 mutants (pGL4-mut1 and pGL4-mut2) were generated with PCR site-directed mutagenesis (40). The details of the mutation were indicated in Fig. 5. All human TR-β1, retinoid X receptor (RXR)-α, and murine LXR-α cDNAs were placed into an Simian virus-40 expression construct, pSG5 (40). All PCR-generated constructs were verified by sequencing the DNA.
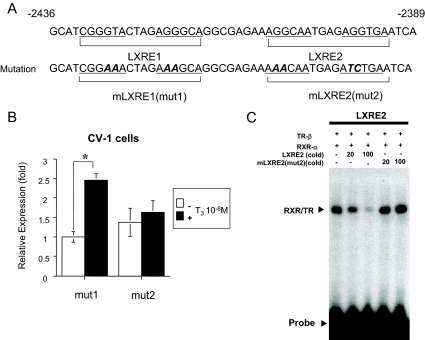
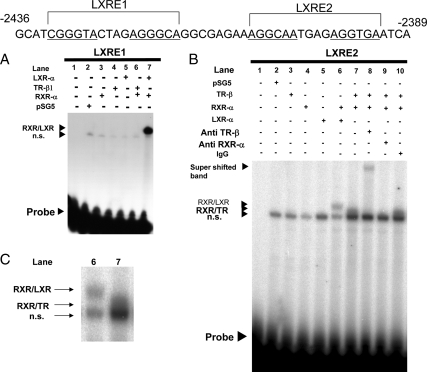
Figure 5.
The LXRE2 is functionally responsible for T3 induction of mouse ChREBP gene promoter. A, Mouse ChREBP gene promoter sequences around the −2436 to −2389 bp region (top panel). The LXRE1 and LXRE2 sequences are as indicated. DR-4 half-site is underlined. The location of the mutation is indicated in bold italic (bottom panel). B, A mouse ChREBP gene promoter (−3801/+9 bp) mutant in which either LXRE1 (mut1) or LXRE2 (mut2) was mutated and coupled to the luciferase reporter construct (pGL4-Luc) was cotransfected into CV-1 cells with an expression vector for wild-type human TR-β1 as indicated in a six-well (w) format together with RXR-α in pSG5 (0.5 μg per well in six-well format). Relative luciferase activities for both mut1 and mut2 and ± T3 groups (mean ± se, n = 3) compared with the light units of mut1 reporter construct in the absence of T3 (10−8 m) (arbitrary light units divided by cellular protein) are shown as relative expression (fold). *, P < 0.01 by t testing. C, EMSAs using cold wild-type (LXRE2) and mutant [mLXRE2(mut2)]competitors. Four microliters of in vitro-translated human TR-β1 and human RXR-α protein were incubated with 32P-radiolabeled DNA probe (LXRE2). For competition experiments, a 20- and 100-fold molar excess of cold oligonucleotides was included as indicated.
Transfections and luciferase assay
For the luciferase assay, we used CV-1 cells. Two micrograms of the reporter plasmid and human RXR-α and human TR-β1 or mouse LXR-α in pSG5 (otherwise indicated) were transfected per well of a six-well plate into CV-1 cells using the calcium-phosphate method. Sixteen hours after transfection, cultures were treated with DMEM containing 10% resin charcoal double-stripped fetal bovine serum for 8 h in the absence or presence of 10−8 m T3. All transfections were equalized for the same total amount of expression vector using an empty vector as needed. We performed β-gal assays to confirm the transfection efficiency of the luciferase assay for each experiment at least once and found no significant difference in transfection efficiency among the plates. Data are presented as fold basal activation expressed as fold induction over vector (pSG5) in the absence of ligand stimulation ± sem otherwise as indicated. Luciferase activity was expressed as arbitrary light units per microgram of cellular protein. All transfection experiments were repeated at least twice with triplicate determinations.
Western blotting
For analysis of the protein expression of ChREBP, 30 μg of whole cell extracts from mouse liver or 20 μg of whole cell extracts from Hepa1-6 cells were subjected to SDS-PAGE. Western blotting was performed using a rabbit anti-ChREBP polyclonal antibody (sc-33764; Santa Cruz Biotechnology, Santa Cruz, CA) and anticyclophilin A (07-313; Upstate, Lake Placid, NY) as a control. The ChREBP detects a specific band at 95 kDa in tissues and cells. The bands were quantitatively measured using Adobe Photoshop CS2 (Adobe Systems Corp., San Jose, CA) and National Institutes of Health Image (Scion Corp., Frederick, MD) and standardized against cyclophilin controls. All Western blotting experiments were repeated at least three times with similar results. ChREBP protein levels are normalized by cyclophilin. Data are presented as fold basal (B) levels ± sem.
RNA preparation and real-time quantitative PCR
Total RNA was extracted from mouse liver, Hepa1-6 cells, and HepG2 cells using ISOGEN (Nippon Gene, Tokyo, Japan). Real-time quantitative PCR assays were performed using an 7700 sequence detector (Applied Biosystems, Foster City, CA) with standard 40 cycles. Briefly, 1 μg of total RNA was reverse transcribed with random hexamers using the Taqman reverse transcription reagent kit (Applied Biosystems) according to the manufacturer’s protocol. Mouse and human ChREBP mRNA expression was analyzed using Taqman probes (Mm02342723_m1 and Hs00263027_m1, respectively; Applied Biosystems). We set up standard curve for each real-time PCR and confirmed that all PCR products were on the standard curve as we previously described (40). The PCR results were normalized to mouse/human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression using a probe and primers from previously developed assays for GAPDH (Applied Biosystems). The number of samples is indicated in the figure legends.
Gel-shift assays
EMSAs (gel-shift assays) were performed as described previously (41). Mouse LXR-α, human TR-β1, and wild-type and human RXR-α recombinant proteins were synthesized from constructs in the pSG5 expression vector, using the TNT T7 quick-coupled transcription/translation system (Promega). Binding reactions contained 20 mm HEPES (pH 7.6), 50 mm KCl, 12% glycerol, 1 mm dithiothreitol, 1 μg of polydeoxyinosine-deoxycytosine, and 4 μl of each of the synthesized nuclear receptors or unprogrammed reticulocyte lysates. Double-stranded oligonucleotides (LXRE1; 5′-AGCTTCGGGTACTAGAGGGCA-3′, LXRE2; 5′-AGCTTAGGCAATGAGAGGTGA-3′) were labeled with [α-32P] deoxy-CTP by a fill-in reaction using a Klenow fragment of DNA polymerase. Binding reactions were performed at room temperature for 30 min. For competition experiments, a 20- and 100-fold molar excess of cold oligonucleotides (wild-type LXRE2 or mutant [mLXRE2 (mut2)]; 5′-AGCTTAAACAATGAGATCTGA-3′) was included as indicated in Fig. 5. For supershift experiments, 3 μl of rabbit anti-TR-β1 polyclonal antibody (06-539; Upstate) were added, and the mixture was incubated for an additional 30 min at room temperature. The protein-DNA complexes were resolved on a 5% polyacrylamide gel in 0.5× 45 mm Tris-base and 1 mm EDTA. T3 was dissolved in 20 mm NaOH as a 1 mm stock solution and diluted to the indicated concentration in 20 mm Tris (pH 7.5). All gel-shift assays were repeated at least three times with similar results and a representative result is shown.
Nuclear run-on assays
Nuclear run-on assays were performed as described previously with minor modification (42,43,44). HepG2 cells (10 cm dish) were treated with 10−8 m T3 for 16 h. Then cells were washed twice with 10 ml of PBS and harvested using trypsine and collected with DMEM containing 10% resin charcoal double-stripped fetal bovine serum. Then 4 ml lysis buffer [0.5% IGEPAL CA630, 10 mm NaCl, 3 mm MgCl2, 10 mm Tris-HCl (pH7.4), 150 mm sucrose] were added per plate, and cell suspensions were incubated on ice for 5 min. After centrifugation, the nuclei were washed once with 2ml of lysis buffer without IGEPAL CA630 and then centrifuged again. Finally nuclei were resuspended in 100 μl of freezing buffer [50 mm Tris-HCl (pH8.3), 40% glycerol, 5 mm MgCl2, 0.1 ml EDTA] and kept frozen at −80 C.
For transcription, 20 μl of transcription buffer (2 times) [200 mm KCL, 20 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 200 mm sucrose, 20% glycerol, 4 mm dithiothreitol], 4 μl of 10× biotin RNA labeling mix (Roche Diagnostics, Penzberg, Germany) was added to 40 μl of the nuclei, and the reaction was incubated. at 29 C for 30 min. Then RNA was isolated using ISOGEN reagent (Nippon Gene). The final RNA was dissolved in 50 μl of diethylpyrocarbonate water. Streptavidin-conjugated magnetic beads (Dynabeads M-280 streptavidin, 50 μl; Invitrogen, Carlsbad, CA) resuspended in 50 μl of binding buffer [10 mm Tris-HCl (pH 7.5), 1 mm EDTA, 2M NaCl] were added to 50 μl of RNA, and the mix was incubated at room temperature for 2 h. Beads were separated using magnetic apparatus and the supernatant was kept for extracting steady-state RNA. The beads were washed twice with 500 μl of 15% formamide and 2× standard sodium citrate for 15 min and once in 1 ml of 2× standard sodium citrate for 5 min. Beads were finally resuspended in 12 μl of diethylpyrocarbonate water. The run-on and steady-state RNA were reverse transcribed, and the real-time quantitative PCR was carried out to detect human ChREBP mRNA. The Taqman probe used for the nuclear run-on assays were the same as that used for gene expression experiments.
Statistical analyses
Statistical analysis was performed using InStat version 2.0 (GraphPad Software, La Jolla, CA). Values are expressed as the mean ± sem. The significance of differences between the mean values was evaluated using the unpaired Student’s t test after ANOVA.
Results
ChREBP gene expression and protein levels are induced by TH in mouse liver
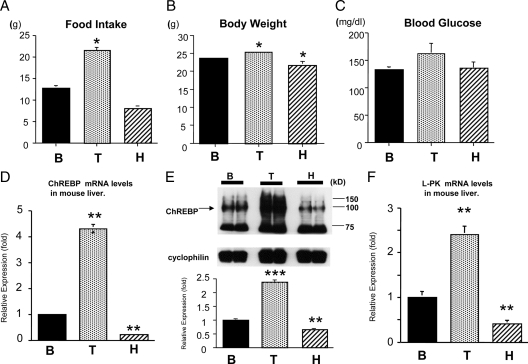
To examine whether TH regulates mouse ChREBP gene expression, we performed real-time RT-PCR using mouse liver steady-state total RNA. For this purpose, we first rendered the mice in a hypothyroid state with an MMI/PTU diet and then injected them with T3 ip to make them thyrotoxic. (Fig. 1A-C). We measured the amount of food intake, body weight and blood glucose levels. As shown in Fig. 1A, the amount of food intake was significantly increased in thyrotoxic animals compared with that of the basal status. Thyrotoxic and hypothyroid mice gained and lost weight, respectively, compared with the basal status (Fig. 1B). However, there were no significant differences in blood glucose levels among the treatment (Fig. 1C). As shown in Fig. 1D, TH induced the mouse ChREBP gene expression by about 4.3-fold and the hypothyroid state decreased the expression to 22% compared with the B level. Western blot analysis using mouse liver whole-cell extract demonstrated that thyrotoxic treatment increased the ChREBP protein levels by about 2.34-fold and hypothyroid state reduced the levels by 36% compared with the B level (Fig. 1E). In addition to an increase in hepatic ChREBP mRNA and protein levels, there is a concomitant increase in L-PK mRNA by TH and a significant decrease in the hypothyroid state. These data suggest that TH increases active ChREBP because L-PK gene expression is largely regulated by ChREBP (19,20) (Fig. 1F).
Figure 1.
The amount of food intake, body weight, and blood glucose levels in mice in different thyroid status (panels A–C). TH induced ChREBP mRNA and protein expression in the mouse liver (D–F). C57/B6 mice (4 wk old male) were first rendered hypothyroid (H) with MMI and PTU diet and then were treated with T3 (T). B was control. Each treatment involved six mice. *, Difference compared with B is significant at a confidence level of P < 0.05 by t testing. Panels D and F, Liver total RNA was isolated, and 1 μg of total RNA was subjected to RT. Real-time RT-PCR analysis for ChREBP (panel D) or L-PK (panel F) was performed using mouse liver cDNA. Relative values (mean ± se, n = 6) normalized by GAPDH mRNA levels compared with B are shown as relative expression (fold). **, Difference compared with B is significant at a confidence level of P < 0.001 by t testing. Panel E, Western blot analysis using mouse liver whole-cell extract. Representative Western blots for ChREBP are shown. Relative ODs (mean ± se, n = 6) normalized by cyclophilin levels compared with B are shown as relative expression (fold). **, Difference compared with B is significant at a confidence level of P < 0.001; ***, P < 0.0001 by t testing.
ChREBP gene expression and protein levels in the nucleus are induced by TH in Hepa1-6 cells
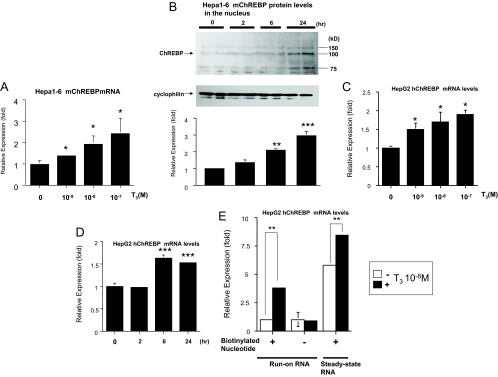
We treated Hepa1-6 cells with T3 and then subjected them to real-time RT-PCR to examine mouse ChREBP gene expression. As shown in Fig. 2A, ChREBP mRNA expression was increased in a T3 dose-dependent manner. Because ChREBP enters from the cytosol into the nucleus and exerts its function (22,23), we evaluate ChREBP protein levels in the nucleus. Nuclear ChREBP protein levels were increased in time-dependent manner under T3 treatment (Fig. 2B).
Figure 2.
T3 up-regulated ChREBP gene and protein expression at the transcriptional level. A, Hepa1-6 cells were cultured in 10-cm-diameter dishes and T3 was added to the medium as indicated. Real-time RT-PCR analysis was performed using Hepa1-6 cDNA. Results (mean ± se, n = 3) are normalized by GAPDH mRNA levels and showed as fold expression correlated to the level in the absence of T3, which was set to 1. *, Difference compared with the level in the absence of T3 is significant at a confidence level of P < 0.01 by t testing. B, Western blot analysis using the nuclear protein in Hepa1-6 cells. Representative Western blots for ChREBP are shown. Relative OD (mean ± se, n = 6) normalized by cyclophilin levels compared with the level in the absence of T3 (10−8 m) are shown as relative expression (fold). **, Difference compared with the level in the absence of T3 (10−8 m) is significant at a confidence level of P < 0.001; ***, P < 0.0001 by t testing. C and D, HepG2 cells were cultured in 10-cm-diameter dishes and T3 was added to the medium as indicated D, T3 (10−8 m). Real-time RT-PCR analysis was performed using HepG2 cellular cDNA. Results (mean ± se, n = 3) are normalized by GAPDH mRNA levels and showed as fold expression correlated to the level in the absence of T3, which was set to 1. *, Difference compared with the level in the absence of T3 is significant at a confidence level of P < 0.01; *** P < 0.0001 by t testing. E, Nuclear run-on assays were performed on nuclei isolated from HepG2 cells after treatment with T3 (10−8 m) for 16 h. Results (mean ± se, n = 3) are normalized by GAPDH mRNA levels and shown as fold expression correlated to the level in the absence of T3 with biotinylated nucleotide, which was set to 1. P < 0.001 (**) by t testing.
TH increased ChREBP gene expression in HepG2 cells by inducing the gene transcription
To examine whether species difference exist in ChREBP gene regulation by TH, we used HepG2 cells. We treated HepG2 cells with T3 and then subjected them to real-time RT-PCR to examine human ChREBP gene expression. As shown in Fig. 2C, ChREBP mRNA expression was increased in a T3 dose-dependent manner in HepG2 cells. We also confirmed that ChREBP gene expression was induced in a time-dependent manner under the T3 treatment (Fig. 2D). To examine whether TH stimulate ChREBP gene transcription, we performed nuclear run-on assays using biotinylated nucleotides and magnetic beads in the nuclei of HepG2 cells. This new method enabled us to evaluate the run-on and steady-state RNAs individually (42,43,44). As shown in Fig. 2E, T3 treatment significantly induced the run-on RNA in addition to steady-state RNA of ChREBP, indicating that TH stimulates ChREBP gene transcription. We also performed run-on assays with Hepa1-6 cells and obtained the similar results (data not shown).
Mouse ChREBP gene promoter is activated by TR-β1 via DR-4 sites
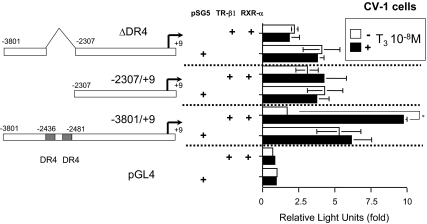
We also subcloned the mouse ChREBP gene promoter (−3801 to +9 bp) and ligated it into pGL4 luciferase reporter plasmid. We used CV-1 cells for luciferase assays. As shown in Fig. 3, T3 significantly induced the mouse ChREBP promoter activity (about 1.8-fold), indicating that TH positively regulates the mouse ChREBP gene expression at the transcriptional level.
Figure 3.
T3 induced the mouse ChREBP gene promoter activity via DR-4 sites. The mouse ChREBP gene promoter (−3801/+9 bp) coupled to the luciferase reporter construct (pGL4) or the deletion mutant reporters were cotransfected into CV-1 cells in the presence of an expression vector (pSG5) for RXR-α and TR-β1 (0.5 μg/well in a six well format). Relative luciferase activities (mean ± se, n = 3) compared with the light unit of pGL4 reporter construct in the absence of T3 (arbitrary light units divided by cellular protein) are shown as relative light units (fold). Vehicle for T3 was distilled water buffered with 100 mm HEPES (pH 7.5). *, P < 0.01 by t testing.
Next, we prepared deletion constructs of the mouse ChREBP gene promoter, which were subjected to transfection into CV-1 cells with these reporters together with TR-β1 and RXR-α. As shown in Fig. 3, neither the ΔDR-4 nor −2307/+9 reporter showed induction by T3, suggesting that the DR-4 sites in the mouse ChREBP promoter are responsible for induction by TH.
RXR-α/TR-β1 heterodimer prefers to bind to LXRE2 in the mouse ChREBP gene promoter
Based on the reporter assay data (Fig. 3), we hypothesized that TR-β1 could bind to the DR-4 sites (LXRE1 and LXRE2 as indicated in Fig. 4, top panel) in the mouse ChREBP gene promoter. In the recent report, it was revealed that LXR-α prefers to bind to the LXRE1 (26); therefore, we examined whether TR-β1 has the preference to bind either LXRE1 or LXRE2. For this purpose, we performed gel-shift assays using double-stranded oligonucleotides for the LXRE1 and LXRE2 of the mouse ChREBP gene promoter. As shown in Fig. 4A, RXR/LXR clearly bound to LXRE1, whereas RXR/TR did not bind to LXRE1. Instead, RXR/TR heterodimer bound to LXRE2 (Fig. 4, B and C). We detected a supershifted band of the RXR/TR heterodimer on LXRE2 probe using anti-TR-β1 antibody (Fig. 4, B and C, lane 8) and an anti-RXR-α antibody clearly pulled out the heterodimer band on LXRE2 probe (Fig. 4B, lane 9). In contrast, mouse normal IgG did not affect the heterodimerization (Fig. 4B, lane 10). These data indicated its specific binding to LXRE2.
Figure 4.
A and B, RXR-α/LXR-α and RXR-α/TR-β1 prefers to bind to LXRE1 and LXRE2, respectively. Mouse ChREBP gene promoter sequences around the −2436 to −2389 bp region (top panel). The LXRE1 and LXRE2 sequences are as indicated. DR-4 half-site is underlined. Four microliters of in vitro-translated human TR-β1 and/or human RXR-α and/or mouse LXR-α protein or unprogrammed rabbit reticulocyte lysates were incubated with 32P-radiolabeled DNA probes (LXRE1, LXRE2). C, The magnified region of RXR/LXR and RXR/TR heterodimer bands (Fig 5B, lanes 6 and 7). n.s., Nonspecific bands; probe, free radiolabeled probes.
LXRE2 in the mouse ChREBP gene promoter is responsible for the regulation by TH
To confirm whether LXRE2 was functionally important, we mutated LXRE1 or LXRE2 in the −3801/+9 pGL4 reporter (Fig. 5A). As shown in Fig. 5B, the −3801/+9 reporter harboring mutated LXRE1 (mut1-pGL4) was activated by TH, whereas mut2-pGL4, which is LXRE2 mutant, was not. To examine whether RXR/TR heterodimer binds to mutant LXRE2, we performed EMSA with LXRE2 radiolabeled probe using cold competitors. As shown in Fig. 5C, cold competitors (LXRE2) deteriorated the heterodimerization, whereas mutant cold competitors [mLXRE2(mut2)] did not. These data indicated that RXR/TR heterodimer binding to LXRE2 is functionally important for the promoter activation by T3.
Human ChREBP gene promoter is also up-regulated by TH
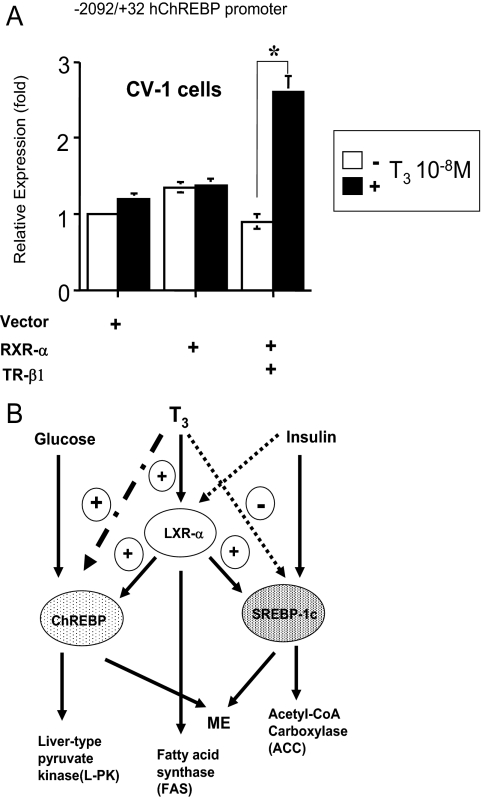
We subcloned the human ChREBP gene promoter (−2092 to +32 bp) and ligated it into pGL4 Luciferase reporter plasmid. As shown in Fig. 6A, T3 significantly induced the human ChREBP promoter activity (about 2.8-fold), indicating that TH positively regulates the human ChREBP gene expression at the transcriptional level. Thus, we concluded that the human ChREBP gene expression is positively regulated by TH.
Figure 6.
A, The human ChREBP promoter (−2092/+32 bp) coupled to the luciferase reporter construct (pGL4) was cotransfected into CV-1 cells in the presence of an expression vector for TR-β1 and/or RXR-α (0.5 μg/well in a six well format) as indicated in the figure. Vector, pSG5. Relative luciferase activities (for both ± T3 groups) (mean ± se, n = 3) compared with the light units of the reporter construct in the absence of T3 (10−8 m) (arbitrary light units divided by cellular protein) are shown as relative expression (fold). *, P < 0.01 by t testing. B, The contribution of T3 to the hepatic lipid metabolism. Schematic diagram illustrates the relationship between T3 and key factors in hepatic lipid metabolism. T3 induces LXR-α gene expression and represses SREBP-1c mRNA levels, which promotes ACC and malicenzyme (ME) upon insulin signaling. Insulin represses LXR-α gene expression. LXR-α positively regulates both ChREBP and SREBP-1c gene expression and stimulates fatty acid synthase. T3 induces ChREBP gene expression and ChREBP promotes L-PK and ME by glucose.
Discussion
In the current study, we demonstrated that mouse ChREBP mRNA and protein expression levels in the mouse liver were up-regulated by TH and also showed that the gene promoter was activated by T3. First, we found that thyrotoxic treatment increased the amount of food intake and body weight, whereas hypothyroid state decreased the body weight compared with the group fed normal chow. However, blood glucose levels were not significantly different among the groups. Because ChREBP is regulated by glucose at the transcriptional level (24), we speculated that these factors such as the amount of food intake and body weight did not necessarily affect the ChREBP gene expression in this study. We and another group (13,14) recently reported that SREBP-1c gene expression is negatively regulated by TH. The physiological relevance of which ChREBP gene expression is up-regulated by T3 remains unclear. However, because it has been reported the interaction between TH and carbohydrate (glucose) to induce lipogenic enzymes (45,46,47,48,49), the current finding could be a help to account for this phenomenon. In terms of the interaction, further study should be required. Cha and Repa (26) recently reported that ChREBP gene promoter is regulated by LXR. They concluded that LXR-RXR heterodimer prefers to bind to LXRE1 but not LXRE2 in the promoter. In this study, we confirmed their data, and in addition, we found that RXR-TR heterodimer prefers to bind to LXRE2. Moreover, the functional data using luciferase assay supported the binding study; LXRE2 mutant promoter was not activated by TH. To date, there has been no evidence that TH directly regulates the L-PK gene expression; therefore, the increase of L-PK mRNA in the thyrotoxic mouse liver indicated that ChREBP protein up-regulated by TH is functionally active. Thus, we concluded that ChREBP is a novel target gene of TH. We also demonstrated human ChREBP gene promoter is positively regulated by TH. First, we searched the mouse LXRE1 or LXRE2 sequence in the human promoter and found no complete LXRE1 or LXRE2 sites. We found only the half-site of mouse LXRE1 or LXRE2 in the human promoter.
The human ChREBP gene promoter contains two discrete DR-4 sites between −2970 and −2937 bp; however, the −2092/+32 construct clearly showed the up-regulation by TH, suggesting that the upstream DR-4 sites is not necessary for the positive gene regulation by TH. Because the −2092/+32 construct possesses only the half-site of canonical DR-4 site, there could be novel TH response elements in the construct, which needs further examination. Both ChREBP and SREBP-1c are the key factors of triglyceride synthesis in the liver (21,50). The function of TH in triglyceride metabolism has been unclear. In fact, serum triglyceride levels are unstable in thyrotoxic and hypothyroid patients (4,8,51,52,53). The up-regulation of ChREBP and down-regulation of SREBP-1c by TH may account for this instability. Furthermore, because we recently reported that LXR-α gene expression is positively regulated by TH (40), we speculated that TH would fine-tune the triglyceride synthesis (Fig. 6B). Considering recent reports, LXR plays essential roles to regulate lipogenic enzymes and transcription factors to promote glucose conversion to lipid; however, because TR regulates LXR, SREBP-1c, and ChREBP gene expression, TR could serve as a master transcriptional regulator for hepatic lipogenesis (Fig. 6B). Indeed, a very recent study reported that ChREBP but not LXRs is required for the induction of glucose-related genes such as L-PK and ACC (50), indicating that another master transcriptional regulator other than LXR could exist in liver. These findings including the current study will shed new light on the role of TH in the lipid metabolism.
Footnotes
Disclosure Summary: K.H., E.I., S.M., S.O., M.Y., T.S., T.M., and M.M. have nothing to declare.
This work was supported by a grant from the Japan Intractable Disease Research Foundation, Yamaguchi Endocrine Disease Research Foundation, and Kowa Life Foundation (to K.H.).
First Published Online March 26, 2009
Abbreviations: ACC, Acetyl-CoA carboxylase; B, basal; ChREBP, carbohydrate response element-binding protein; DR-4, direct repeat-4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; L-PK, liver-pyruvate kinase; LXR, liver X receptor; MMI, methimazole; PTU, propylthiouracil; RXR, retinoid X receptor; SREBP, sterol response element-binding protein; TH, thyroid hormone; TR, TH receptor.
References
- Oppenheimer JH, Schwartz HL, Mariash CN, Kinlaw WB, Wong NC, Freake HC 1987 Advances in our understanding of thyroid hormone action at the cellular level. Endocr Rev 8:288–308 [DOI] [PubMed] [Google Scholar]
- Lazar MA 1993 Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev 14:184–193 [DOI] [PubMed] [Google Scholar]
- Silva JE 2005 Intermediary metabolism and the sympathoadrenal system in hypothyroidism. 9th ed. Philadelphia: Lippincott Williams and Wilkins [Google Scholar]
- Duntas LH 2002 Thyroid disease and lipids. Thyroid 12:287–293 [DOI] [PubMed] [Google Scholar]
- O'Brien T, Katz K, Hodge D, Nguyen TT, Kottke BA, Hay ID 1997 The effect of the treatment of hypothyroidism and hyperthyroidism on plasma lipids and apolipoproteins AI, AII and E. Clin Endocrinol (Oxf) 46:17–20 [DOI] [PubMed] [Google Scholar]
- Nishitani H, Okamura K, Noguchi S, Inoue K, Morotomi Y, Fujishima M 1990 Serum lipid levels in thyroid dysfunction with special reference to transient elevation during treatment in hyperthyroid Graves’ disease. Horm Metab Res 22:490–493 [DOI] [PubMed] [Google Scholar]
- Miura S, Iitaka M, Yoshimura H, Kitahama S, Fukasawa N, Kawakami Y, Sakurai S, Urabe M, Sakatsume Y, Ito K, Ishii J 1994 Disturbed lipid metabolism in patients with subclinical hypothyroidism: effect of l-thyroxine therapy. Intern Med 33:413–417 [DOI] [PubMed] [Google Scholar]
- Pearce EN 2004 Hypothyroidism and dyslipidemia: modern concepts and approaches. Curr Cardiol Rep 6:451–456 [DOI] [PubMed] [Google Scholar]
- Pearce EN, Wilson PW, Yang Q, Vasan RS, Braverman LE 2008 Thyroid function and lipid subparticle sizes in patients with short-term hypothyroidism and a population-based cohort. J Clin Endocrinol Metab 93:888–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckman AK, Towle HC 2002 The role of SREBP-1c in nutritional regulation of lipogenic enzyme gene expression. J Biol Chem 277:27029–27035 [DOI] [PubMed] [Google Scholar]
- Uyeda K, Repa JJ 2006 Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab 4:107–110 [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS 2002 SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109:1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viguerie N, Millet L, Avizou S, Vidal H, Larrouy D, Langin D 2002 Regulation of human adipocyte gene expression by thyroid hormone. J Clin Endocrinol Metab 87:630–634 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Yamada M, Matsumoto S, Monden T, Satoh T, Mori M 2006 Mouse sterol response element binding protein-1c gene expression is negatively regulated by thyroid hormone. Endocrinology 147:4292–4302 [DOI] [PubMed] [Google Scholar]
- Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, Shillinglaw W, Arnot D, Uyeda K 2001 A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci USA 98:9116–9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka K, Horikawa Y 2008 ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr J 55:617–624 [DOI] [PubMed] [Google Scholar]
- Ishii S, Iizuka K, Miller BC, Uyeda K 2004 Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc Natl Acad Sci USA 101:15597–15602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaux JF, Antoine B, Kahn A 1989 Regulation of the expression of the L-type pyruvate kinase gene in adult rat hepatocytes in primary culture. J Biol Chem 264:11584–11590 [PubMed] [Google Scholar]
- Thompson KS, Towle HC 1991 Localization of the carbohydrate response element of the rat L-type pyruvate kinase gene. J Biol Chem 266:8679–8682 [PubMed] [Google Scholar]
- da Silva Xavier G, Rutter GA, Diraison F, Andreolas C, Leclerc I 2006 ChREBP binding to fatty acid synthase and L-type pyruvate kinase genes is stimulated by glucose in pancreatic β-cells. J Lipid Res 47:2482–2491 [DOI] [PubMed] [Google Scholar]
- Dentin R, Girard J, Postic C 2005 Carbohydrate responsive element binding protein (ChREBP) and sterol regulatory element binding protein-1c (SREBP-1c): two key regulators of glucose metabolism and lipid synthesis in liver. Biochimie (Paris) 87:81–86 [DOI] [PubMed] [Google Scholar]
- Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K 2001 Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc Natl Acad Sci USA 98:13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentin R, Benhamed F, Pégorier JP, Foufelle F, Viollet B, Vaulont S, Girard J, Postic C 2005 Polyunsaturated fatty acids suppress glycolytic and lipogenic genes through the inhibition of ChREBP nuclear protein translocation. J Clin Invest 115:2843–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentin R, Peéorier JP, Benhamed F, Foufelle F, Ferré P, Fauveau V, Magnuson MA, Girard J, Postic C 2004 Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J Biol Chem 279:20314–20326 [DOI] [PubMed] [Google Scholar]
- Uyeda K, Yamashita H, Kawaguchi T 2002 Carbohydrate responsive element-binding protein (ChREBP): a key regulator of glucose metabolism and fat storage. Biochem Pharmacol 63:2075–2080 [DOI] [PubMed] [Google Scholar]
- Cha JY, Repa JJ 2007 The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J Biol Chem 282:743–751 [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ 2001 Nuclear receptors and lipid physiology: opening the X-files. Science 294:1866–1870 [DOI] [PubMed] [Google Scholar]
- Stulnig TM, Steffensen KR, Gao H, Reimers M, Dahlman-Wright K, Schuster GU, Gustafsson JA 2002 Novel roles of liver X receptors exposed by gene expression profiling in liver and adipose tissue. Mol Pharmacol 62:1299–1305 [DOI] [PubMed] [Google Scholar]
- Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P 2003 Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med 9:213–219 [DOI] [PubMed] [Google Scholar]
- Berkenstam A, Gustafsson JA 2005 Nuclear receptors and their relevance to diseases related to lipid metabolism. Curr Opin Pharmacol 5:171–176 [DOI] [PubMed] [Google Scholar]
- Zelcer N, Tontonoz P 2006 Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest 116:607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitro N, Mak PA, Vargas L, Godio C, Hampton E, Molteni V, Kreusch A, Saez E 2007 The nuclear receptor LXR is a glucose sensor. Nature 445:219–223 [DOI] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM 1991 Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65:1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfel R, Benbrook D, Lernhardt E, Ortiz MA, Salbert G, Pfahl M 1994 A novel orphan receptor specific for a subset of thyroid hormone-responsive elements and its interaction with the retinoid/thyroid hormone receptor subfamily. Mol Cell Biol 14:7025–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teboul M, Enmark E, Li Q, Wikström AC, Pelto-Huikko M, Gustafsson JA 1995 OR-1, a member of the nuclear receptor superfamily that interacts with the 9-cis-retinoic acid receptor. Proc Natl Acad Sci USA 92:2096–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ 1995 LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev 9:1033–1045 [DOI] [PubMed] [Google Scholar]
- Quack M, Frank C, Carlberg C 2002 Differential nuclear receptor signalling from DR4-type response elements. J Cell Biochem 86:601–612 [DOI] [PubMed] [Google Scholar]
- Berkenstam A, Färnegårdh M, Gustafsson JA 2004 Convergence of lipid homeostasis through liver X and thyroid hormone receptors. Mech Ageing Dev 125:707–717 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Cohen RN, Yamada M, Markan KR, Monden T, Satoh T, Mori M, Wondisford FE 2006 Cross-talk between thyroid hormone receptor and liver X receptor regulatory pathways is revealed in a thyroid hormone resistance mouse model. J Biol Chem 281:295–302 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Matsumoto S, Yamada M, Satoh T, Mori M 2007 Liver X receptor-α gene expression is positively regulated by thyroid hormone. Endocrinology 148:4667–4675 [DOI] [PubMed] [Google Scholar]
- Ren Y, Satoh T, Yamada M, Hashimoto K, Konaka S, Iwasaki T, Mori M 1998 Stimulation of the preprothyrotropin-releasing hormone gene by epidermal growth factor. Endocrinology 139:195–203 [DOI] [PubMed] [Google Scholar]
- Patrone G, Puppo F, Cusano R, Scaranari M, Ceccherini I, Puliti A, Revazzolo R 2000 Nuclear run-on assay using biotin labeling, magnetic bead capture and analysis by fluorescence-based RT-PCR. Biotechniques 29:1012–1014, 1016–1017 [DOI] [PubMed] [Google Scholar]
- Schbeler D, Bode J 1997 A sensitive transcription assay based on a simplified nuclear runon protocol. Elsevier Technical Tips Online 2:140–142 [Google Scholar]
- Madak-Erdogan Z, Kieser KJ, Kim SH, Komm B, Katzenellenbogen JA, Katzenellenbogen BS 2008 Nuclear and extranuclear pathway inputs in the regulation of global gene expression by estrogen receptors. Mol Endocrinol 22:2116–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariash CN, Kaiser FE, Schwartz HL, Towle HC, Oppenheimer JH 1980 Synergism of thyroid hormone and high carbohydrate diet in the induction of lipogenic enzymes in the rat. Mechanisms and implications. J Clin Invest 65:1126–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forciea MA, Schwartz HL, Towle HC, Mariash CN, Kaiser FE, Oppenheimer JH 1981 Thyroid hormone-carbohydrate interaction in the rat: correlation between age-related reductions in the inducibility of hepatic malic enzyme by triiodo-l-thyronine and a high carbohydrate, fat-free diet. J Clin Invest 67:1739–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennemann B, Moon YK, Freake HC 1992 Tissue-specific regulation of fatty acid synthesis by thyroid hormone. Endocrinology 130:637–643 [DOI] [PubMed] [Google Scholar]
- Brown SB, Maloney M, Kinlaw WB 1997 “Spot 14” protein functions at the pretranslational level in the regulation of hepatic metabolism by thyroid hormone and glucose. J Biol Chem 272:2163–2166 [PubMed] [Google Scholar]
- Koo SH, Towle HC 2000 Glucose regulation of mouse S(14) gene expression in hepatocytes. Involvement of a novel transcription factor complex. J Biol Chem 275:5200–5207 [DOI] [PubMed] [Google Scholar]
- Denechaud PD, Dentin R, Girard J, Postic C 2008 Role of ChREBP in hepatic steatosis and insulin resistance. FEBS Lett 582:68–73 [DOI] [PubMed] [Google Scholar]
- Heimberg M, Olubadewo JO, Wilcox HG 1985 Plasma lipoproteins and regulation of hepatic metabolism of fatty acids in altered thyroid states. Endocr Rev 6:590–607 [DOI] [PubMed] [Google Scholar]
- Abbas JM, Chakraborty J, Akanji AO, Doi SA 2008 Hypothyroidism results in small dense LDL independent of IRS traits and hypertriglyceridemia. Endocr J 55:381–389 [DOI] [PubMed] [Google Scholar]
- Teixeira Pde F, Reuters VS, Ferreira MM, Almeida CP, Reis FA, Buescu A, Costa AJ, Vaisman M 2008 Lipid profile in different degrees of hypothyroidism and effects of levothyroxine replacement in mild thyroid failure. Transl Res 151:224–231 [DOI] [PubMed] [Google Scholar]