Abstract
We have conducted an in silico analysis of progesterone response elements (PRE) in progesterone receptor (PR) up-regulated promoters. Imperfect inverted repeats, direct repeats, and half-site PRE are widespread, not only in PR-regulated, but also in non-PR-regulated and random promoters. Few resemble the commonly used palindromic PRE with three nucleotide (nt) spacers. We speculated that PRE may be necessary but insufficient to control endogenous PR-dependent transcription. A search for PRE partners identified a highly conserved 234-nt sequence invariably located within 1–2 kb of transcription start sites. It resembles ALU repeats and contains binding sites for 11 transcription factors. The 234-nt sequence of the PR-regulated 8-oxoguanine DNA glycosylase promoter was cloned in the forward or reverse orientation in front of zero, one, or two inverted repeat PRE, and one or tandem PRE half-sites, driving luciferase. Under these conditions the 234-nt sequence functions as a co-response element (coRE). From the PRE or tandem half-sites, the reverse coRE is a strong activator of PR and glucocorticoid receptor-dependent transcription. The forward coRE is a powerful repressor. The prevalence of PRE half-sites in natural promoters suggested that PR monomers regulate transcription. Indeed, dimerization-domain mutant PR monomers were stronger transactivators than wild-type PR on PRE or tandem half-sites. This was repressed by the forward coRE. We propose that in natural promoters the coRE functions as a composite response element with imperfect PRE and half-sites to present variable, orientation-dependent transcription factors for interaction with nearby PR.
ALU repeats are co-response elements (coRE) that enhance or repress transcription mediated by dimeric and monomeric progesterone receptors from imperfect PREs or PRE half sites.
Progesterone receptors (PR) are thought to regulate genes by binding DNA at progesterone response elements (PRE); palindromic inverted repeats consisting of two hexamers separated by three nucleotides. This model holds that upon hormone activation, PR dimerize, bind the PRE, and interact with coregulatory proteins. One problem with this is that the hormone response elements (HRE) recognized by PR are also recognized by glucocorticoid receptors (GR), androgen receptors (AR), and mineralocorticoid receptors. For historical reasons, the consensus HRE was defined using GR. In the TRANSFAC database (1) there are two binding site matrices for GR. The consensus glucocorticoid response element (GRE) (V$GRE_C), derived from a position weight matrix generated by ConsIndex (2), uses 10 functionally confirmed HRE from rat tyrosine amino transferase (TAT); rat tryptophan oxidase; human metallothionein IIA; murine sarcoma virus; human GH gene; and the mouse mammary tumor virus long terminal repeat (MMTV-LTR). The second GRE matrix (V$GR_Q6) stems from 38 binding sequences the identities of which are unavailable.
Transcription experiments tend to use the same two high-affinity palindromic HRE. The first model, derived from the rat TAT promoter, was identified as a GRE (3) but also confers strong PR-mediated transcription when it is cloned in tandem upstream of a minimal promoter (4). Subsequently, AR and mineralocorticoid receptors were shown to bind and stimulate transcription from the same isolated TAT PRE2. The second model uses the MMTV-LTR. Unlike the TAT PRE2, which is PR responsive in isolation, for PR-dependent transcription the imperfect palindromic PRE of the MMTV-LTR requires, in addition, at least two of three proximate PRE half-sites (5).
The sequence of an optimal PRE has been assessed (5). Point mutations in the palindromic PRE of MMTV-LTR coupled to in vitro transcription studies pointed to specific nucleotides needed for optimal PR activity. The 3′-TGTTCT hexamer tolerated few mutations but the 5′-TTTACA hexamer required only the cytosine (5). Fewer mutations were tolerated when gel mobility shift assays were used to assess DNA binding (5). However, differences among constructs, such as inclusion of two MMTV half-sites to the palindrome for the transcription but not gel shift studies, makes comparisons between transcription and DNA binding difficult. With regard to MMTV-LTR there are differences between PR- and GR-dependent transcription with the palindrome more heavily required by GR, and the half-sites more important for PR. Footprinting and methylation protection studies of MMTV-LTR with PR and GR show subtle differences, especially over the half-sites, and PR covers approximately 10 additional base pairs just upstream of the half-sites, not seen with GR (6). Thus whereas PR bind to and are functional on a synthetic GRE, these sequences may not reflect the authentic endogenous PRE for PR, which may rely more heavily on half-sites.
Historically PR were believed to bind DNA as preformed dimers. Recent thermodynamic studies dispute this. Using the PRE of TAT and MMTV-LTR, Bain and co-workers (7) demonstrate that PR bind DNA as monomers, especially on half-sites, and that PR binding to the MMTV-LTR is highly cooperative (7,8). Thermodynamic analysis shows that solution dimer formation is 1000-fold weaker than biochemical studies suggest, indicating that few or no dimers exist at the low PR concentrations adequate for DNA binding (8). PR are bound as head-to-head dimers in crystals of the PR coRE DNA-binding domain, and a simulated TGTTCT palindrome with the trinucleotide spacer of MMTV (9). However, by mobility shift assays, PR bind half-sites as monomers (9) with a dissociation constant (Kd) similar to that of PR binding to a palindromic PRE.
Other steroid receptors function as monomers. GR dimerization mutants maintain transcriptional activity on some genes (10). Indeed, knockout of wild-type GR is embryonic lethal but mice expressing dimerization-deficient GR are viable (11), suggesting that GR monomers are sufficient for development. Analysis of endogenous GR binding sites demonstrates that the majority are composite elements containing imperfect palindromes plus binding sites for other transcription factors (12). Endogenous androgen response elements in immortalized prostate epithelial and cancer cells are largely noncanonical half-sites, suggesting that AR too bind DNA as monomers (13,14).
Endogenous binding sites for GR and AR have been identified (12,13,14) but chromatin immunoprecipitation (ChIP) on chip studies of in vivo PR binding sites have not been reported. However, several proximal progesterone-responsive promoters have been mapped (15,16,17). They either contain nonconsensus PRE or lack them altogether (15,16,17). Further complexity results from the fact that PR regulate transcription without PRE by tethering to DNA-bound proteins like SP1 and AP1 (17,18). A genome-wide in silico sequence analysis to locate perfect palindromic steroid receptor HRE identified 565 perfect inverted repeats, of which only 26 were within 10 kb of a transcription start site. By ChIP analysis four of these were significantly bound by liganded PR in T47D breast cancer cells (19). One would have to conclude either that the majority of PR binding sites are not contained within 10 kb of transcription start sites or that PR binding sites are not the perfect inverted repeats for which the authors were searching.
We recently published several studies (20,21,22) that identified endogenous genes regulated by liganded or unliganded PR or by PRA vs. PRB isoforms in breast cancer cells. Here we use these datasets for an in silico analysis of endogenous PR-regulated promoters. We show that PRE are widespread even in genes that are not PR regulated. The PRE represent a mixture of imperfect inverted repeats, direct repeats, and half-sites. In close association to PRE are highly conserved 234-nt transcription factor-binding sites commonly associated with ALU repeats. When cloned together with PRE or half-sites, these 234-nt sequences function as co-response elements (coRE). The coRE repress or enhance PR-mediated transcription, depending on their orientation. We also engineered and tested PR dimerization domain mutants. The PR monomers regulate transcription more strongly than wild-type PR from a variety of PRE including half-sites, and this is also modulated by the coRE. We conclude that endogenous PR binding sites do not resemble canonical inverted repeats. Rather, PR commonly use half-sites, with transcription aided or repressed by neighboring DNA-bound factors.
Results
PRE in human promoters
In vitro transcription assays use high-affinity consensus PRE consisting of palindromic hexamers separated by three nonconserved spacers. They are usually derived from the rat TAT promoter (3), or the mouse MMTV-LTR which, in addition to an imperfect palindrome, has three proximal half-sites (5). How these rat and mouse viral PRE compare with human PRE is unclear. We defined PR-regulated genes in human breast cancer cells and used this dataset to identify PRE in natural promoters by an in silico approach (Fig. 1, A and B). EZ-retrieve (23) yielded 2000 bp of proximal promoters that were analyzed by the Silico Regulation Management System (SRMS) program. SRMS employs TRANSFAC GRE matrices to identify DNA-binding sites by two methods: a consensus PRE/GRE (V$GRE_C: 5′-GGTACAANNTGTYCTK-3′; Y = C or T; K = G or T); and a PRE/GRE derived from 38 GR-binding sites (V$GR_Q6: 5′-NNNNNNCNNTNTGTNCTNN-3′).
Figure 1.
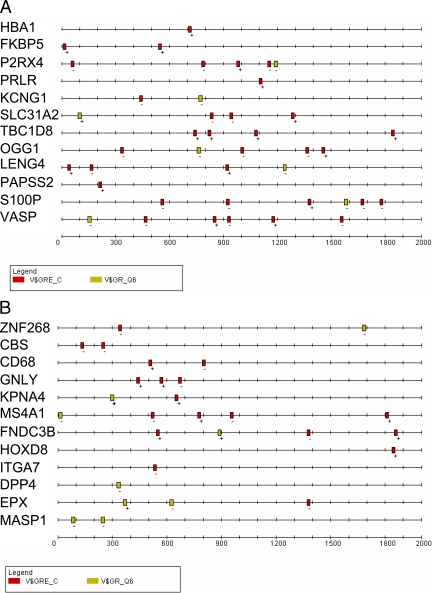
Position and spacing of putative PRE in 2000-nt proximal promoters of PR-up-regulated or random genes. A, 12 PR up-regulated (20,21,22) and B) 12 random genes were searched for 2000-nt promoter sequences using the EZ retrieve database (23). SRMS was used to define 16-nt conserved motifs and patterns within them. The MEME database was used for large-scale sequence alignment (27). The TRANSFAC database was specifically searched for consensus PRE using two matrices: V$GRE_C (red boxes) and V$GR_Q6 (yellow boxes). Note: the nucleotide numbering on the abscissa is the reverse of conventional promoter positions due to the default numbering in SRMS with 2000 at the putative transcription start site, and ″0″ designates -2000.
The 3′-hexamer (TGTNCT) is conserved between the two matrices but the 5′-hexamer is variable. Both (V$GRE_C, red boxes; V$GR_Q6, yellow boxes) were used for the initial search of PR up-regulated (Fig. 1A), and random promoters (Fig. 1B) using the same dataset (20,21). Twelve representative PR up-regulated promoters show widely varying numbers of PRE, ranging from one in the hemoglobin α chain 1 (HBA1) gene to six in the vasodilator phosphoprotein gene (Fig. 1A). However, the random promoters also contained multiple PRE (Fig. 1B). None were perfect inverted repeats. They were direct repeats or more often, conserved 3′-hexamer half-sites downstream of nonconserved 5′-hexamer half-sites. Also identifiable were simple TGTTCT half-site hexamers at additional sites (data not shown).
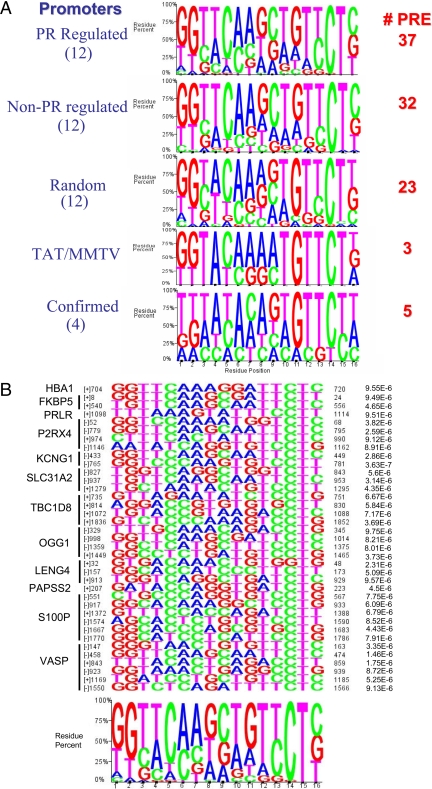
Nucleotide sequence pileups of PRE from different datasets are shown in Fig. 2A. They are derived from: 12 PR up-regulated promoters containing 37 PRE; 12 experimentally determined non-PR-regulated promoters containing 32 PRE; 12 promoters selected at random containing 23 PRE. (An additional 42 random promoters contained at least one PRE each.) The non-PR-regulated promoters were selected from our datasets because random promoters have a negligible (<3%) possibility of being PR regulated. Also shown in Fig. 2A is the compiled logo for the two consensus PRE of the TAT promoter, the imperfect palindrome of MMTV-LTR, and five PRE found in four promoters the PR responsiveness of which has been experimentally confirmed (16,24,25,26). None of the PRE differ significantly. In each set, the 3′-hexamer is better conserved than the 5′-hexamer, as previously reported (5). The experimentally confirmed vs. computer-defined PRE are similar and demonstrate the accuracy of the SRMS program.
Figure 2.
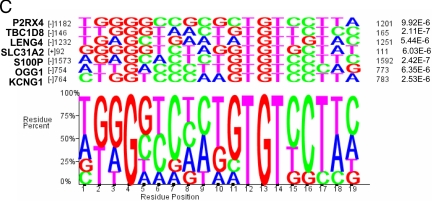
Composition of PRE in PR-regulated and random promoters, the TAT promoter and MMTV-LTR, and four experimentally confirmed PR-regulated promoters. A, SRMS was used to search 12 PR-up-regulated, 12 non-PR-regulated, and 12 random promoters for the presence of PRE using the TRANSFAC database and the 16-nt consensus PRE/GRE 5′-GGTACAANNTGTYCTK-3′, where Y = C or T; K = G or T. A total of 37 PR-regulated, 32 non-PR-regulated, and 23 random PRE were used to construct a logo illustrating sequence conservation at each of the 16 positions. Letter heights represent the degree of base conservation at that position. Four promoters containing five experimentally confirmed PRE (16,24,25,26) and the three PRE of MMTV-LTR and TAT were also analyzed. B, The sequences of 37 PRE in 12 PR-up-regulated promoters from the TRANSFAC database 16-nt V$GRE_C matrix are shown in the pileup above the derived logo. The statistical significance (P values) for each sequence is compared with the matrix sequence. The nucleotide location of each PRE and its presence on the sense (+) or antisense (−) strand is indicated on the left. Examples of PRE sequences include 1) imperfect indirect repeats (KCNG1, site 765–781); 2) direct repeats (SLC31A2, site 827–843); 3) perfect half-sites (vasodilator phosphoprotein, site 147–163); and 4) imperfect half-sites (P2RX4, site 52–68). C, In the same genes, the more stringent 19-nt V$GR_Q6 matrix identified seven PRE the pileup and summary logo of which are shown together with their statistical significance (P values) compared with the matrix sequence.
Figure 2B is a detailed pileup of 37 PRE in promoters of 12 PR up-regulated genes using the V$GRE_C matrix (red boxes in Fig. 1A). They are highly statistically significant as shown by E-values on the right, assigned by SRMS. Conservation of the 3′-hexamer is evident, especially the CT at positions 14 and 15. In some cases the 5′-hexamer is more highly conserved as seen for FKBP5, KCNG1, and SLC31A2. No PRE is a perfect palindrome (see figure legend for details).
Figure 2C shows a pileup of PRE in the same 12 PR-up-regulated promoters with the V$GR_Q6 matrix (yellow boxes in Fig. 1A). Because this matrix uses 19 nt instead of the 16 nt used by V$GRE_C, their numbering is not directly comparable. Because of the greater stringency, fewer PRE were identified. Here the TGT at position 12–14 in the 3′-hexamer is particularly well conserved. Interestingly, the G at position 4 in the 5′-hexamer is also highly conserved. It may be an important contact point for tandemly bound PR.
Conserved ALU repeats near PRE
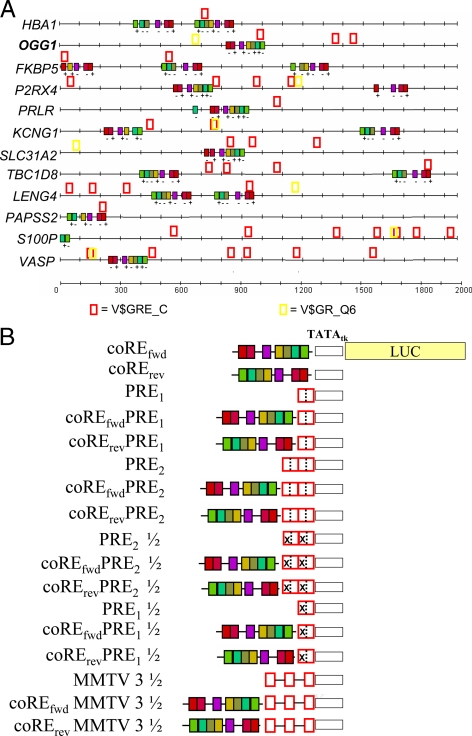
Because the PRE of PR-up-regulated and random genes did not differ, we postulated that a PRE may be necessary, but not sufficient, to control PR-dependent transcription. The 2000-nt proximal promoters of the 12 PR-regulated genes were therefore searched for additional approximately 16-nt motifs using the MEME database (27) and SRMS, an interface for multiple databases. Ten of the 12 contain a highly conserved pattern of seven linked approximately 16-nt motifs spanning about 234 nt. Each colored box in Fig. 3A represents one such 16-nt motif. The 234 nt are CpG rich and resemble an ALU repeat. Some promoters contain the ALU in one orientation and others in the opposite orientation. Some promoters, including HBA1, FKBP5, KCNG1, TBC1D8, and LENG4, contain two or three copies of the ALU repeat. The two remaining promoters, S100P and PAPSS2, contain a partial ALU with the remainder apparently upstream of the 2-kb cutoff. If 3 kb of 5′-flanking sequence are searched, they generally contain at least one ALU repeat (data not shown). Figure 3A also shows the location of the ALU on the sense (+) or antisense (−) strand, and its position relative to the PRE as defined in Fig. 1 (open red or yellow boxes). In general, the ALU are located approximately 1 kb from the putative transcription start site and are flanked by PRE. Interestingly, of the 12 promoters analyzed, none contains a TATA sequence by in silico analysis. Two promoters, 8-oxoguanine DNA glycosylase (OGG1) and FKBP5, have been cloned and characterized, and neither contains a TATA or CAATT box (28,29). Note that CpG-rich promoters often contain a TFIIB-recognition element to launch transcription, thereby bypassing the need for a TATA box (30).
Figure 3.
Location of PRE and coRE on 12 PR-up-regulated promoters and other promoter constructs. A, SRMS was used to locate 2000-nt proximal promoters of 12 PR-regulated genes, which were searched for PRE using the TRANSFAC database 16-nt consensus PRE V$GRE_C matrix (open red boxes) or the 19-nt experimental GRE V$Gr_Q6 matrix (open yellow boxes). A highly conserved pattern of seven motifs, each approximately 16 nt in length (each colored box), spanning about 234 bp total, was found on 11 of the 12 promoters. The 234-nt sequence is termed a “coRE”. Location of each coRE on the sense (+) and antisense (−) strand is indicated. The numbering on the abscissa is the reverse of standard promoter nomenclature with 2000 at the transcription start site, and 0 at −2000 bp. B, Luciferase reporters driven by promoters containing PRE and the coRE of the OGG1 promoter, cloned in the forward or reverse orientation as it aligns in OGG1, were made as described in Materials and Methods. Open red boxes denote ½, one, or two palindromic PRE derived from the TAT promoter with each half-site separated by a dashed line. Mutated sites are designated by X. Also shown are constructs containing the three half-sites of the MMTV-LTR. Open black box is the TATA box of the minimal tk promoter.
ALU repeats are coresponse elements (coRE): wild-type PR
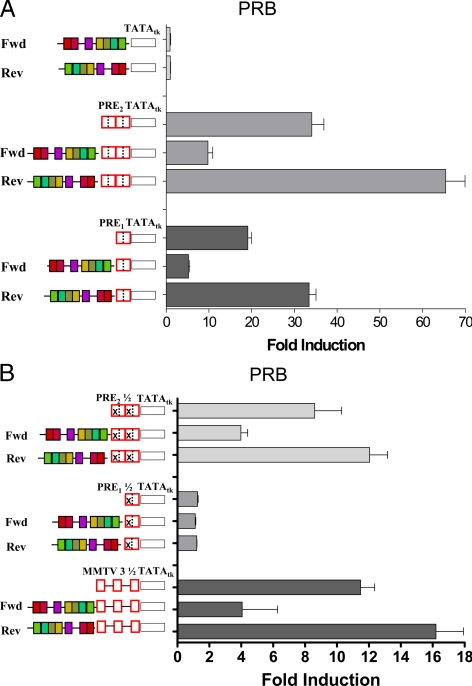
To determine whether these highly conserved 234-nt sequences influence PR-mediated transcription, a series of promoter-driven luciferase constructs were cloned containing PRE plus the 234-nt ALU derived from the OGG1 promoter (Fig. 3B). The approximately 234-nt construct was cloned upstream of no PRE; a single PRE (PRE1); tandem PRE (PRE2); two PRE half-sites (PRE2 ½); or one PRE half-site (PRE1 ½), placed upstream of the minimal thymidine kinase (tk) promoter and luciferase reporter. Constructs containing half-sites were created by mutating the 5′-hexamer of single or tandem PRE, leaving the 3′-hexamer intact. Because the ALU is found in two orientations in natural promoters, it was cloned in both the forward (fwd; its natural position in the OGG1 promoter) and reverse (rev) orientation, in the various constructs. Because of their regulatory functions (below) we term the 234-nt sequence a “coRE.”
The constructs described in Fig. 3B were transiently transfected together with a PRB expression vector into PR-negative HeLa cells (Fig. 4A). Without a PRE, neither the forward nor reverse coRE is transcriptionally active. Basal transcription levels are 2-fold higher in the reverse than the forward orientation (data not shown). The middle set shows that whereas PRE2-tk-LUC alone confers strong transcription under PRB control (∼28-fold from basal) addition of the coRE has opposing effects depending on its orientation. The forward coRE is a potent repressor, reducing transcription 3- to 4-fold; the reverse coRE doubles transcription levels (Fig. 4A). The same results were observed using PR-negative T47D-Y breast cancer cells (data not shown). Figure 4A also shows transcription from a PRE1, on which PRB are weaker transactivators (∼17-fold from basal). Here again, the forward coRE was a repressor and the reverse coRE an enhancer, generating transcription levels equal to those achieved by PR on a PRE2.
Figure 4.
Enhancement or suppression of PRB-mediated transcription is dependent on the orientation of the coRE. A, The coRE of the OGG1 promoter in the fwd or rev orientation was cloned upstream of PRE2-tk-LUC, PRE1-tk-LUC, or tk-LUC. HeLa cells were transiently transfected with the reporters, an expression vector encoding PRB, and an simian virus 40-driven Renilla luciferase control vector. Cells were treated with 10 nm R5020 or ethanol for 24 h. Relative firefly luciferase units were corrected for transfection efficiency based on Renilla activity, and fold changes between control and hormone-driven activity were calculated. Error bars represent the sem of three independent studies. B, The coRE in the fwd or rev orientation was cloned upstream of one or two PRE half-sites or the three half-sites of the MMTV-LTR in front of tk-LUC. HeLa cells were cotransfected with the reporters, an expression vector encoding PRB, and a Renilla-LUC control, and treated with R5020 or ethanol. Fold transcription was calculated as above.
Effects of the coRE on PRA were also tested (supplemental Fig. S1A published as supplemental data on The Endocrine Society’s Journals Online web site at http://endojournals.mend. org). The relatively weak transcription by PRA on PRE2 was somewhat repressed by the forward coRE or enhanced by the reverse coRE. Interestingly, PRA stimulate transcription as well from a PRE1 as they do from a PRE2 (∼2.6-fold; supplemental Fig. S1A). On the PRE1, transcriptional enhancement by the reverse coRE was especially evident.
To determine whether the coRE modifies PR-dependent transcription from half-sites, a series of constructs were created in PRE1 or PRE2 leaving the 3′-hexamer intact but mutating the 5′-hexamer. The reporters were transiently transfected into HeLa cells together with PRB (Fig. 4B) or PRA (supplemental Fig. S1B). PRB increased transcription from tandem half-sites approximately 10-fold, which was repressed by the forward coRE, but unaffected by the reverse coRE. The forward coRE was ineffective with PRA, whereas the reverse coRE was an enhancer. Thus the coRE has different effects on PRB vs. PRA from tandem half-sites. Single half-sites were transcriptionally inactive and not affected by the coRE with either receptor.
We also assessed the three natural half-sites of MMTV-LTR (Fig. 4B). PRB-dependent transcription on these was similar to that on the artificial tandem half-sites with repression by the forward coRE and minimal effects by the reverse coRE. PRA is a good transactivator of the three MMTV half-sites, which were unaffected by the coRE (supplemental Fig. S1B).
Other steroid receptors and effects of coRE
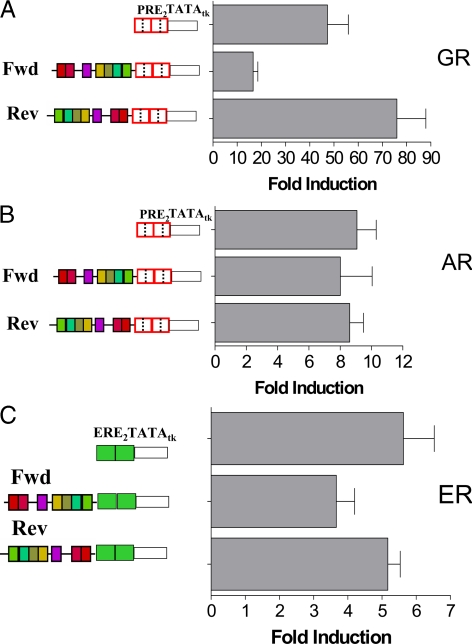
Because GR also transactivate from PRE, effects of the coRE constructs were tested in dexamethasone-treated HeLa cells (Fig. 5A). The endogenous liganded GR generated high luciferase levels that were suppressed by the forward coRE and increased by the reverse coRE, akin to the effects of PR. In contrast, the coRE had no effects on AR (Fig. 5B). Parenthetically, AR have their highest transcriptional activity on tandem half-sites (18-fold), but the coRE had no effect on these either (data not shown). To determine effects of the coRE on ER, HeLa cells were transiently transfected with ERα expression vectors and forward or reverse coRE cloned upstream of two estrogen response elements (ERE) derived from the vitellogenin A2 promoter. Treatment with 17β-estradiol increased transcription (Fig. 5C), which was weakly suppressed by the forward coRE but unaffected by the reverse coRE. Taken together the data suggest that effects of the coRE vary depending on the receptors and DNA binding sites.
Figure 5.
Effects of the coRE on transcription by other nuclear receptors. A, The PRE2-tk-LUC reporter without or with the coRE in the forward or reverse orientation was transiently transfected into HeLa cells together with a Renilla-LUC control, and cells were treated 24 h with 10 nm dexamethasone. B, The same reporters were transiently transfected into HeLa cells together with an expression plasmid encoding AR and the Renilla-LUC control. Cells were treated 24 h with 10 nm of the androgen R1881. C, The coRE in either orientation was cloned upstream of an ERE2-tk-LUC reporter, HeLa cells were transiently transfected with the reporter, an ERα expression vector, and Renilla-LUC and treated 24 h with 10 nm 17β-estradiol. Fold changes are shown for hormone treated vs. untreated transcription, corrected for the Renilla control. Error bars represent the sem of three independent studies.
The coRE of the natural OGG promoter
The natural orientation of the coRE in the human OGG1 promoter was defined as the forward case (Fig. 3A). The data in Figs. 4 and 5 demonstrate that in this orientation the coRE is a repressor. This was confirmed with T47D cells stably transfected with 2 kb of the wild-type OGG promoter or a coRE deletion mutant linked to luciferase. Low basal levels of luciferase activity were observed with the wild-type promoter. Deletion of the coRE increased basal transcription approximately 10-fold (data not shown). These data indicate that the forward coRE is a potent repressor in the context of the natural promoter; repressor properties that can be transferred to synthetic promoters.
Monomeric PR and transcriptional regulation
Analysis of PR-regulated promoters (Fig. 2, B and C) indicates that unlike experimental PRE, natural PRE are not palindromic. Rather, they are mainly direct repeats and half-sites. We therefore asked whether PR monomers could regulate transcription through these noncanonical sites. For this, a single amino acid mutation (R606A; supplemental Fig. S2A) was introduced into the D-box dimerization domain of the second zinc finger of PRA (to generate DAX) and PRB (to generate DBX). This mutation blocks dimerization in GR and MR (31,32). Unlike wild-type PR, DAX and DBX bind poorly to a palindromic PRE, as assessed by EMSA (supplemental Fig. S2B).
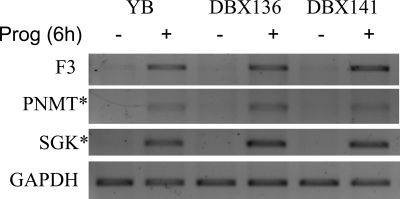
We first asked whether PR monomers regulate endogenous genes. To test this, the DBX and DAX mutants were stably transfected into PR-negative T47D-Y breast cancer cells, and several independent clonal cell lines were propagated. The ability of progesterone to regulate transcription of select, PR-regulated genes (20,21,22) in these cells was assessed by RT-PCR. Figure 6 shows transcript expression levels without and with 6 h progesterone of three PRB-regulated genes—tissue factor (F3), phenylethanolamine N-methyltransferase (PNMT), and serum/glucocorticoid-regulated kinase (SGK)—in cells expressing wild-type PRB (YB), or two DBX-expressing lines (DBX136; DBX141). Liganded wild-type PRB up-regulated all three transcripts, as did monomeric DBX in both independent cell lines. This suggests that PR monomers are capable of regulating at least some endogenous genes. In this regard, the PNMT promoter contains multiple half-sites (32), and PNMT and SGK are regulated by monomeric GR (10). The PRA dimerization mutant was also tested on endogenous genes (supplemental Fig. S3), and regulation by PRA mutants was observed on TSC22 domain family, member 3 (TSC22D3), and BCL2-like 1 (BCL2L1) genes at levels similar to, or higher than, those observed with wild-type PRA.
Figure 6.
A PRB dimerization mutant monomer up-regulates transcription of endogenous genes. T47D breast cancer cell lines that stably express wild-type PRB (YB) or PRB dimerization mutants (DBX136; DBX141), were treated 6 h without (−) or with (+) 10 nm progesterone and harvested and RNA was isolated. RT-PCR was performed with primers specific for tissue factor (F3), PNMT, or SGK. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is shown as a loading control. Prog, Progesterone.
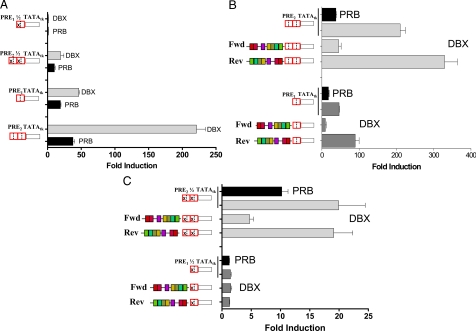
We next tested PR monomers on the single or tandem PRE or half-site reporters (Fig. 7A). Remarkably, on palindromic PRE and on tandem half-sites, DBX monomers were much stronger transactivators (light gray bars) than their wild-type PRB counterparts (black bars). Similar data for wild-type PRA vs. monomeric DAX can be found in supplemental Fig. S4A.
Figure 7.
Compared with wild-type PRB, DBX monomers strongly stimulate PRE-dependent transcription, which is modified by the coRE. A, HeLa cells were transiently transfected with the single or tandem PRE half-site tk-LUC, or single or tandem palindromic PRE tk-LUC, plus wild-type PRB or DBX dimerization mutants, and a Renilla-LUC vector. Cells were treated 24 h without or with 10 nm R5020. Luciferase activity was corrected for the Renilla control, and fold regulation by liganded vs. unliganded receptors was quantified. B, HeLa cells were transiently transfected with the single or tandem palindromic PRE tk-LUC without or with the coRE in the fwd or rev orientation. They were cotransfected with wild-type PRB or DBX expression vectors, and a Renilla-LUC vector, treated 24 h without or with 10 nm R5020, and quantified as above. C, HeLa cells were transiently transfected with the single or tandem PRE half-site tk-LUC without or with the coRE in the fwd or rev orientation. They were cotransfected with PRB or DBX expression vectors and a Renilla-LUC vector and treated 24 h without or with 10 nm R5020. Transcription was quantified as above. Error bars represent the sem of three independent studies.
Figure 7B shows that R5020-dependent transcription by the DBX monomer on PRE2 was much stronger (∼200-fold, light gray bar), than transcription by wild-type PRB (∼40-fold, black bar; see also Fig. 4A). Transcription by DBX monomers was markedly suppressed by the forward coRE and enhanced by the reverse coRE (Fig. 7B). The same pattern was observed on a PRE1, where the DBX monomer activated transcription much more effectively (∼50-fold) than did wild-type PRB (∼20-fold, black bar; and Fig. 7A). Here too transcription by monomeric DBX was suppressed by the forward coRE and enhanced by the reverse coRE. Similarly, the DAX dimerization mutant was a stronger transactivator than wild-type PRA (supplemental Fig. S4A), and the coRE repressed or enhanced transcription depending on its orientation (supplemental Fig. S4B).
The PR dimerization mutants were also tested on PRE half-sites with or without the coRE (Fig. 7C). As with wild-type PRB, single half-sites were inactive. However, on tandem half-sites the DBX mutant exhibited remarkably strong transcriptional activity (∼20-fold; gray bar, Fig. 7C); much stronger than that seen with wild-type PRB (∼10-fold; black bar). The forward coRE was a strong repressor of tandem half-sites, but the reverse coRE had no effect. PRA DAX monomer data on half-sites are shown in supplemental Fig. S4C. In sum, PR monomers are exceptionally strong transactivators even on PRE half-sites where the major effect of the coRE is repressive. On palindromic PRE the coRE represses or enhances the strong transcription exhibited by PR monomers, depending on its orientation.
Discussion
PRE structure
Classical transcription studies with PR demonstrate minimal activity on a single palindromic PRE and synergistic activity with two or more PRE (33,34). We show here, however, as previously reported (35), that a single PRE strongly mediates transcription (Fig. 4A). The reported need for two or more PRE may reflect differences in basal factor-binding sites among minimal promoters; in characteristics of artificial start sites; in the stability and sensitivity of reporter genes; and/or in the plasmid vector backbones used. For example, many transcription studies use the pGL family of vectors that contain an AP1-binding site and are progesterone responsive even in the absence of a promoter (our unpublished data). Here we use the pA3Luc vector, which is completely free of AP1 binding sites, and the minimal tk promoter, which exhibits negligible basal transcription levels. We observe good transcriptional activity from a single PRE, and the increase with two PRE is additive, not synergistic (Fig. 4A). We also demonstrate strong transcriptional activity on tandem PRE half-sites (Fig. 4B).
Palindromic PRE are thought to be important for PR-dependent transcription. We sought to define the PRE of endogenous PR-up-regulated promoters and discovered that they lack the expected structure. Our analysis demonstrates that PRE are scattered throughout promoters, not just PR-regulated ones (Figs. 1 and 2). Of course, any non-PR-regulated PRE-containing promoters could be subject to regulation by other steroid receptors. If so, what, for instance, are the rules that confer GR regulation but not PR regulation? Our data also indicate that the classical requirement for a palindromic PRE appears to be oversimplified. Perfect palindromic PRE were not found in the subset of promoters we examined (Fig. 2, B and C) and imperfect palindromes were rare. On the other hand, direct repeats and tandem and single half-sites were widespread. Indeed, there are too many PRE for these sequences, alone, to be dispositive.
Two different binding site matrices from the TRANSFAC database that allowed flexibility and recognized nonconsensus PRE were used in the search. Both showed strong conservation of the 3′-hexamer structure TGTNCT. Detailed studies may determine the relationship between these specific sequence constraints and in vitro or in vivo PR-regulatory specificity. Our data support previous findings based on mutagenesis of the MMTV palindrome that the 3′-hexamer tolerates fewer mutations and is functionally more important than the 5′-hexamer (5). Receptor/DNA crystal studies (9), chemical modification of DNA bases (36,37), and DNA footprinting (6) using either the imperfect palindrome of MMTV or an optimized PRE also indicate that PR interact more strongly with nucleotides in the 3′- than the 5′-hexamer.
Monomers and half-sites
It appears that the DNA sequences used for exogenous transcription studies or structural analyses may not be representative of endogenous PRE in human genes, which are, for the most part, half-sites and imperfect direct or inverted repeats (Fig. 2B). The prevalence of half-sites suggested to us that PR function as monomers, and we found this to be the case on both endogenous (Fig. 6) and exogenous (Fig. 7) promoters. This is not surprising in retrospect, because GR dimerization mutants are functional and regulate endogenous genes like SGK (10), or the multiple half-sites of the PNMT promoter (32). Dimerization domain mutants of MR can also be transcriptionally active (31), and AR-binding sites are often composed of half-sites (14).
That PR function as monomers is supported by mapping studies of the MMTV-LTR. They show that the three half-sites are more important than the palindrome for regulation by progesterone (38). This has been confirmed recently by the elegant thermodynamic studies of Bain and co-workers (7) who show that whereas a solution dimer binds at the palindrome, PR monomers assemble at the half-sites where they interact cooperatively. Sedimentation equilibrium confirms a binding stoichiometry of one monomer per half-site. Interestingly, monomeric PR only cooperatively engage receptors bound at the palindrome after the half-sites are completely saturated. Bain and colleagues (7) also argue that although PR can self-associate as dimers, the energetics of dimerization are so weak that few, if any, dimers are present at protein concentrations that initiate DNA binding. This raises the possibility that, even at palindromes, sequential PR monomer assembly loads the DNA. Indeed, the extensive transcription we observe on a PRE palindrome with dimerization domain mutants (Fig. 7B) suggests that sequential monomer assembly may be transcriptionally more efficient than binding of preformed dimers. Because the DNA binding affinity of monomers is weaker than of dimers (39), and if the kinetics of binding/assembly on DNA are the same for monomers and dimmers, a rate only limited by diffusion (D. Bain, personal communication), then the off rate of monomers from DNA would be faster than of dimers. If so, the more efficient release of monomers from DNA would accelerate their ligand-dependent down-regulation, perhaps explaining their strong transcription (40).
PR and other transcription factors
PR binding to PRE can be bypassed by tethering the receptors to DNA-bound proteins like SP1 in the p21 promoter (17) or AP1 in IL-2 and other promoters (18). The spacing between PRE and flanking transcription factor-binding sites like SP1, NF1, OTF, and CP1 may also be important (34). Much remains to be learned about this because most studies that address PR-dependent transcription use artificial constructs with optimized, isolated, receptor-binding sites that are an oversimplification of the intricate structure and length of endogenous promoters. In addition to the role of PRE, we address the role of flanking sequences on the direction and strength of transcription by PR. We describe a highly conserved 234-nt sequence composed of seven motifs of approximately 16 nt each. It is always found near PRE in proximal promoters. Depending on its orientation this 234-nt sequence is either an enhancer or repressor. We dub this a “co-response element” based on these properties. These findings were based on a series of studies using the 234-nt sequence derived from the OGG1 promoter.
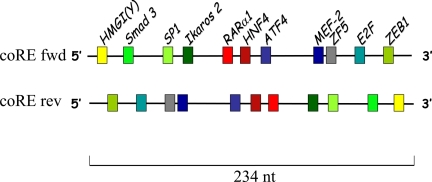
Putative transcription factor-binding sites in the 234-nt coRE were searched using the TRANSFAC database (Fig. 8). Eleven were identified: HMGI(Y), Smad3, SP1, Ikaros 2, RARα1, HNF4, ATF4, MEF-2, ZF5, E2F, and ZEB1. Several are regulated by and/or interact with PR. For example, ZEB1 and Ikaros 2 are up-regulated by progesterone (20,41); ATF4 is up-regulated by the selective PR modulator asoprisnil (42); the inhibitory effect of ΤGFβ1 on PR is dependent on Smad3 signaling (43), and importantly, Smad3 is a known PR coactivator (43); the ability of SP1 to tether PR to DNA is well known (17). Of course, the order of binding sites is flipped, and their distance to neighboring PRE is altered, when the coRE is reversed. We speculate that depending on their position relative to PRE, transcription factors bound at the coRE enhance or repress transcription. Interestingly, identity and characterization of endogenous GRE by ChIP chip showed an enrichment of motifs near GRE resembling AP1-, HNF4-, C/EBP-, ETS-, and SP1-binding sites (12) suggesting that the compound coRE/PRE promoter elements we report here are analogous to previously described composite GRE (12) made up of binding sites for GR together with multiple nonreceptor-regulatory factors. Indeed, as we show in Fig. 5A, the coRE modifies not only PR- but also GR-dependent transcription.
Figure 8.
The coRE of the OGG1 promoter contains 11 putative transcription factor-binding sites. Factor binding sites shown are variable in size and were identified using the TRANSFAC database.
GR and PR regulate some genes in common, but they also regulate unique gene subsets (44). This may be due to preferential receptor interaction with different neighboring transcription factors. For example, the MMTV-LTR, which has an NF1-binding site 3′ of the half-sites, is regulated by GR in an NF1-dependent manner and by PR in an NF1-independent manner (38). PR-specific regulation at sites not regulated by GR has also been reported. An example are tandem PRE in Intron E of the FKBP5 gene, one of which is solely inducible by progesterone whereas the other is inducible by both progesterone and glucocorticoids (45). This suggests that flanking sequences play a role in receptor specificity.
The coRE, CpG islands, and ALU repeats
Many promoters that lack canonical TATA or CCAAT-box sequences contain GC-rich regions, or so-called CpG islands (30). There are two major subtypes: 1) a region of at least 200 nt with G+C content greater than 50% and observed/expected CpG ratio greater than 0.6 (46); or 2) a region longer than 500 nt, G+C content greater than 55%, and an observed/expected CpG ratio greater than 0.65 (47). The second excludes ALU repeats, which are highly repetitive short interspersed elements with an approximately 280-bp consensus GC-rich sequence. By this definition, and by alignment analysis with the Repbase Update database (48,49), the coRE is an ALU repeat that conforms to subtype 1.
ALU are highly conserved and present in approximately 1 million copies per haploid genome (47). There are 31 ALU subfamilies in the human database (48,49) with 12 consensus subclasses, although a more recent analysis subdivides them into 213 subclasses (50). These conserved sequences sometimes overlap with CpG islands in promoters, and the distinction between the two is not always clear. Many ALU repeats are located within introns or the 3′-untranslated region of genes and, through retrotransposition and/or ALU-ALU mediated recombination, they disrupt RNA splicing, alter patterns of gene expression, and silence genes (51). It is estimated that more than 1000 human genes are controlled by ALU (52). One recent study finds that ALU sequences are underrepresented in proximal promoters (i.e. between −1000 to +200 nt) compared with introns. Within our 12 PR up-regulated dataset (Fig. 3A), three contain ALU repeats between positions −1000 and 0. If 2000 nt are queried, 11 contain at least one ALU and the twelfth contains a partial one.
ALU repeats contain binding sites for basal transcription factors and sometimes for nuclear receptors (52,53) although GRE are rare (54). They play important roles in transcription: 1) The ERα promoter has an ALU repeat that contains an ER-dependent enhancer between −1416 to −1703 (55). 2) The ALU repeat in the BRCA1 promoter contains an estrogen-responsive region (53) and is a transcriptional enhancer, but the effect is specific to subclasses of ALU repeats. Subtle variations in ERE within ALU are enough to eliminate ER-mediated regulation (53). 3) ALU repeats confer tissue specificity to GR-dependent transcription (63). 4) They function as transcriptional activators or repressors depending on their orientation (56). 5) They control micro-RNA expression (57). 6) In the promoters of CETP, nAChR, and BRCA2 they function as silencers (58,59,60). 7) ALU repeats play a role in human genetic diseases including familial hypercholesterolemia, Lesch-Nyhan syndrome, β-thalassemia, and Tay-Sachs disease (61). 8) The PROGINS PR variant contains an ALU insertion in intron G, generating two-point mutations in PR exons that increase PR transcriptional activity (62) and increase risk of sporadic ovarian and breast cancers after 3 or more years of combined hormone replacement therapy.
We show here that ALU repeats in proximity to PRE enhance or repress transcription in an orientation-dependent manner. We propose that these sequences function as coresponse elements in partnership with PRE. Here we tested only a single coRE. However, natural promoters can contain multiple such sites. For example, the FKBP5 promoter has three reverse coRE. It is the most strongly PR-regulated gene in our dataset. Whether the number and orientation of coRE controls the extent of endogenous transcription, and even whether a gene is up- or down-regulated by PR depending on the number and orientation of coRE, requires more study.
In summary, although PRE are present throughout the genome, not all can be functional PR-binding sites. Canonical palindromic PRE are almost nonexistent. More common are half-sites on which PR monomers are active. We speculate that other factors control PR-dependent transcription and demonstrate that sequences we call “coRE” partner with PRE to accomplish this. The orientation of the coRE relative to palindromic and half-site PRE may variably present transcription factors for binding to PR. Such factors may not only modify transcription by PR, but could confer unique gene-regulatory properties to GR, AR, and MR, all of which bind apparently similar PRE. The strong transcription generated by the reverse coRE on a single repeat or half-site—sequences commonly found in proximal promoters—is equivalent to levels seen with tandem palindromic experimental PRE that are rarely found naturally. Cooperativity between PR binding to suboptimal PRE and adjacent transcription factors would be sufficient to confer extensive regulatory control.
Materials and Methods
Promoter profiling
Sets of PR-regulated genes that we previously defined (1,2,3) were selected based on the availability of 2000 bp of putative proximal promoter sequences in the EZ retrieve (23) database. These included genes up-regulated by both PR isoforms, known non-PR-regulated genes, and random genes. Retrospectively, none of the random genes appeared on the PR-regulated gene lists. The Silico Regulation Management System (SRMS) program version 1.3.5 (H. Patel, Silico Informatics, Santa Clara, CA) was used to define 16-nt conserved motifs and patterns within the 2000-bp promoters, and the MEME database (version 3.0) was used for large-scale sequence alignments (27). The TRANSFAC database (version 7.2) was also searched for conserved motifs and putative binding sites of known transcription factors (64), and specifically, for consensus PRE using the TRANSFAC GRE matrix. This database lists two GREs: A consensus GRE (V$GRE_C: 5′-GGTACAANNTGTYCTK-3′) and a GRE derived from 38 GR-binding sites (V$GR_Q6: 5′-NNNNNNCNNTNTGTNCTNN-3′). The 3′-hexamer is conserved between these two sites, whereas the 5′-hexamer is variable. From the GRE/PRE sequences, logos were generated by SRMS using the position-specific probability matrix derived from the MEME algorithm (27). The dominant residues at each position in a predicted binding site are indicated by letter size. Independently, SRMS was used to generate logos of the palindromic PRE in the MMTV-LTR, two PRE of the rat TAT promoter, and five endogenous PRE previously defined experimentally as PR regulated in human promoters (16,24,25,26).
Reporter constructs
PRE2-tk-LUC (65) contains two identical PRE from site II of the rat TAT gene (3). They are spaced 25 nt apart, and the 3′-PRE is 40 nt from the TATA box of the minimal 67-nt tk promoter [+145 to +211; (66)], which is cloned upstream of the luciferase reporter gene in the pA3LUC backbone (a gift of Dr. William Wood, UCHSC, Aurora, CO). To generate a single PRE, the PRE2 vector was digested with SalI and HindIII to release the minimal tk promoter, which was amplified by PCR, using specific primers: tk bottom, 5′-CCCaagcttATCGATTTCGAACCCGGGTCGCTCGG-3′; and tk top, 5′-acgcgtcgacagtcggggcggcgcggtccgaggtcc-3′. The fragment was digested with SalI and HindIII and subcloned into the pA3LUC vector to generate a tk-LUC minimal construct. This was linearized with SalI, filled in, and dephosphorylated. A single double-stranded PRE, 5′-AAAGTCTGTACAGGATGTTCTGATCAA-3′, was subcloned 45 nt upstream of the minimal promoter, and clones were sequenced to determine orientation. The ERE2-tk-LUC vector was previously described (67).
PRE2 ½-tk-LUC contains two nucleotide mutations in each 5′-hexamer resulting in two half-sites. Two primers were used for PCR amplification: 5′-CCCCGGTCGACTCTAGATtTAaAGGATGTTCTGATCCCGGGCTAGACCTAGGGGATCTtTAaAGGATGTTCT-3′ (PRE shown in bold, mutations in lowercase), and the reverse primer to the tk promoter: 5′-CCCaagcttATCGATTTCGAACCCGGGTCGCTCGG-3′ (HindIII site shown in lowercase). Fragments were PCR amplified, digested with SalI and HindIII and subcloned into the pA3LUC vector.
The 2-kb hOGG1/Luc promoter (29) was kindly provided by J. P. Radicella (Commissariat à l’Energie Atomique, France). It was digested with PstI and SmaI to release the 234-nt coRE. The fragment was filled in and subcloned 10 nt upstream of the 5′-PRE in PRE2-tk-LUC or 40 nt upstream of the 5′-ERE in ERE2-tk-LUC in pA3LUC (65). Constructs with the coRE and PRE ½ sites were created by PCR amplification of the 234-bp coRE fragment, using the 2-kb promoter as template, and the following primers containing NcoI digestion sites: NcoI, −1181 OGG1 forward (fwd) 5′-CATGCCATGGCTGCAGATGTTGCTCTTCAAGAATTTGCAG-3′; NcoI, −943 OGG1 reverse (rev) 5′-CATGCCATGGGGGTTCAAGCGATTCTCCTGCCTCAGCCTCCC-3′. After amplification the PCR products were subcloned into the PRE2 half-site vectors that had been digested with NcoI and dephosphorylated. The spacing between the two PRE half-sites and the spacing of the proximal half-site to the TATA box is the same as that of the PRE2 construct. They were sequenced to determine orientation: the forward (fwd) coRE is sense in the OGG1 promoter; the reverse (rev) coRE is antisense.
Transfection and transcription
HeLa cells were transfected by CaPO4 precipitation with 1 μg of firefly luciferase reporter plasmids, 10 ng of a Renilla luciferase plasmid, and 20 ng PRB, PRA, GR, AR, or ERα expression plasmids as described elsewhere (65). Transfected cells were treated 24 h with ethanol vehicle, or 10 nm of the progestin R5020, the glucocorticoid dexamethasone, the androgen R1881, or 17β-estradiol, before harvesting and quantitation of luciferase activity (65) corrected for the Renilla control. Absolute luciferase activity levels and their fold regulation in ethanol vs. hormone were calculated. Error bars represent the sem of three independent studies.
PR dimerization mutants
A single amino acid mutation (R606A) was introduced in the second zinc finger via PCR mutagenesis using the oligonucleotide: 5-TTATGTGCTGGAgcAAATGACTGCATC-3′ (mutated bases shown in lowercase) with wild-type PR as template. Fragments generated were digested with MluI and BclI and subcloned into the pSG5 expression vector containing wild-type PRB or PRA (68) digested with MluI and BclI to create DBX and DAX, respectively. Plasmids were sequenced for confirmation.
Stable cell lines
The pSG5-DBX or DAX mutants were transfected into PR-negative T47D-Y breast cancer cells together with a neo resistance vector (69). Several independent stable cell lines expressing each PR mutant were selected in G418, and PR expression was confirmed by immunoblotting with the 1294 anti-PR antibody (DAKO Corp., Carpinteria, CA).
EMSA
Nuclear extracts were prepared from T47D cells stably expressing PRA, PRB, DAX, or DBX treated with progesterone for 30 min. A double-stranded TAT PRE sequence: 5′-CCAAAGTCTGTACAGGATGTTCTGATCAAAG-3′ was radiolabeled, and gel shifts were performed as previously described (70). Anti-PR 1294 was added to confirm the presence of PR in the shifted complexes.
Semiquantitative RT-PCR
Cells were plated and treated, and RT-PCR was performed as previously described (20). Primers for BCL2L1, F3, and glyceraldehyde-3-phosphate dehydrogenase were previously reported (20). Other primers are: SGK fwd, 5′-GACACTTGCAGGACACTACAACGTG-3′, SGK rev, 5′-GTTATAACCCAATGTACAGACGTTC-3′; PNMT fwd, 5′-GGTGAAACGGGTCCTGCCCATCGAC-3′, PNMT rev, 5′-GACCTTGTAGCCACTACGCACCAGG-3′; TSC22D3 fwd, 5′-CGTGAGAACACCCTGTTGAAGACCC-3′, TSC22D3 rev, 5′-CTTACACCGCAGAACCACCAGGGGC-3′.
Supplementary Material
Acknowledgments
We thank J. P. Radicella and W. M. Wood for reagents, the Sequencing Core of the University of Colorado Cancer Center, and Robert Burke and David Bain (University of Colorado Denver) for helpful discussions.
Footnotes
This work was supported by National Institutes of Health Grant CA26869-30, the National Foundation for Cancer Research, the Breast Cancer Research Foundation, and the Avon Foundation.
Disclosure Summary: The authors have nothing to declare.
First Published Online April 16, 2009
Abbreviations: AR, Androgen receptors; ARE, androgen response elements; ChIP, chromatin immunoprecipitation; CoRE, coresponse elements; ER, estrogen receptor; ERE, estrogen response elements; fwd, forward; GR, glucocorticoid receptor; GRE, glucocorticoid response elements; HRE, hormone response elements; MMTV-LTR, mammary tumor virus long terminal repeat; nt, nucleotide; OGG1, 8-oxoguanine DNA glycosylase; PNMT, phenylethanolamine N-methyltransferase; PRE, progesterone response elements; rev, reverse; SGK, serum/glucocorticoid-regulated kinase; SRMS, Silico Regulation Management System; TAT, tyrosine amino transferase; tk, thymidine kinase.
References
- Wingender E, Kel AE, Kel OV, Karas H, Heinemeyer T, Dietze P, Knüppel R, Romaschenko AG, Kolchanov NA 1997 TRANSFAC, TRRD and COMPEL: towards a federated database system on transcriptional regulation. Nucleic Acids Res 25:265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech K, Herrmann G, Werner T 1993 Computer-assisted prediction, classification, and delimitation of protein binding sites in nucleic acids. Nucleic Acids Res 21:1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen HM, Strähle U, Gloss B, Stewart F, Schmid W, Boshart M, Miksicek R, Schütz G 1987 Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell 49:29–38 [DOI] [PubMed] [Google Scholar]
- Strähle U, Klock G, Schütz G 1987 A DNA sequence of 15 base pairs is sufficient to mediate both glucocorticoid and progesterone induction of gene expression. Proc Natl Acad Sci USA 84:7871–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman BA, Bona BJ, Edwards DP, Nordeen SK 1993 The constitution of a progesterone response element. Mol Endocrinol 7:515–527 [DOI] [PubMed] [Google Scholar]
- Chalepakis G, Arnemann J, Slater E, Brüller HJ, Gross B, Beato M 1988 Differential gene activation by glucocorticoids and progestins through the hormone regulatory element of mouse mammary tumor virus. Cell 53:371–382 [DOI] [PubMed] [Google Scholar]
- Connaghan-Jones KD, Heneghan AF, Miura MT, Bain DL 2008 Thermodynamic dissection of progesterone receptor interactions at the mouse mammary tumor virus promoter: monomer binding and strong cooperativity dominate the assembly reaction. J Mol Biol 377:1144–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connaghan-Jones KD, Bain DL 2009 Using thermodynamics to understand progesterone receptor function: method and theory. Methods Enzymol 455:41–70 [DOI] [PubMed] [Google Scholar]
- Roemer SC, Donham DC, Sherman L, Pon VH, Edwards DP, Churchill ME 2006 Structure of the progesterone receptor-deoxyribonucleic acid complex: novel interactions required for binding to half-site response elements. Mol Endocrinol 20:3042–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I, Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq CM, Darimont BD, Garabedian MJ, Yamamoto KR 2003 Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci USA 100:13845–13850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schütz G 1998 DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93:531–541 [DOI] [PubMed] [Google Scholar]
- So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR 2007 Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet 3:e94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR 2007 Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev 21:2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M 2007 A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaratanakornkit V, Strong DD, Mohan S, Baylink DJ, Beck CA, Linkhart TA 1999 Progesterone stimulation of human insulin-like growth factor-binding protein-5 gene transcription in human osteoblasts is mediated by a CACCC sequence in the proximal promoter. J Biol Chem 274:26431–26438 [DOI] [PubMed] [Google Scholar]
- Gao J, Mazella J, Tang M, Tseng L 2000 Ligand-activated progesterone receptor isoform hPR-A is a stronger transactivator than hPR-B for the expression of IGFBP-1 (insulin-like growth factor binding protein-1) in human endometrial stromal cells. Mol Endocrinol 14:1954–1961 [DOI] [PubMed] [Google Scholar]
- Owen GI, Richer JK, Tung L, Takimoto G, Horwitz KB 1998 Progesterone regulates transcription of the p21(WAF1) cyclin-dependent kinase inhibitor gene through Sp1 and CBP/p300. J Biol Chem 273:10696–10701 [DOI] [PubMed] [Google Scholar]
- Bamberger AM, Bamberger CM, Gellersen B, Schulte HM 1996 Modulation of AP-1 activity by the human progesterone receptor in endometrial adenocarcinoma cells. Proc Natl Acad Sci USA 93:6169–6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie-Inoue K, Takayama K, Bono HU, Ouchi Y, Okazaki Y, Inoue S 2006 Identification of novel steroid target genes through the combination of bioinformatics and functional analysis of hormone response elements. Biochem Biophys Res Commun 339:99–106 [DOI] [PubMed] [Google Scholar]
- Richer JK, Jacobsen BM, Manning NG, Abel MG, Wolf DM, Horwitz KB 2002 Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J Biol Chem 277:5209–5218 [DOI] [PubMed] [Google Scholar]
- Jacobsen BM, Schittone SA, Richer JK, Horwitz KB 2005 Progesterone-independent effects of human progesterone receptors (PR) in estrogen receptor-positive breast cancer: PR isoform-specific gene regulation and tumor biology. Mol Endocrinol 19:574–587 [DOI] [PubMed] [Google Scholar]
- Ghatge RP, Jacobsen BM, Schittone SA, Horwitz KB 2005 The progestational and androgenic properties of medroxyprogesterone acetate: gene regulatory overlap with dihydrotestosterone in breast cancer cells. Breast Cancer Res 7:R1036–R1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ramanathan Y, Soteropoulos P, Recce ML, Tolias PP 2002 EZ-Retrieve: a web-server for batch retrieval of coordinate-specified human DNA sequences and underscoring putative transcription factor-binding sites. Nucleic Acids Res 30:e121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PA, Chaparro O 1999 The 5′-flanking sequence and regulatory elements of the cystatin S gene. Biochem Biophys Res Commun 261:705–711 [DOI] [PubMed] [Google Scholar]
- Cheng KW, Cheng CK, Leung PC 2001 Differential role of PR-A and -B isoforms in transcription regulation of human GnRH receptor gene. Mol Endocrinol 15:2078–2092 [DOI] [PubMed] [Google Scholar]
- Balbín M, Lopez-Otín C 1996 Hormonal regulation of the human pepsinogen C gene in breast cancer cells. Identification of a cis-acting element mediating its induction by androgens, glucocorticoids, and progesterone. J Biol Chem 271:15175–15181 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994 Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36 [PubMed] [Google Scholar]
- Hubler TR, Denny WB, Valentine DL, Cheung-Flynn J, Smith DF, Scammell JG 2003 The FK506-binding immunophilin FKBP51 is transcriptionally regulated by progestin and attenuates progestin responsiveness. Endocrinology 144:2380–2387 [DOI] [PubMed] [Google Scholar]
- Dhénaut A, Boiteux S, Radicella JP 2000 Characterization of the hOGG1 promoter and its expression during the cell cycle. Mutat Res 461:109–118 [DOI] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR 2006 Transcriptional regulatory elements in the human genome. Annu Rev Genom Hum Genet 7:29–59 [DOI] [PubMed] [Google Scholar]
- Liu W, Wang J, Sauter NK, Pearce D 1995 Steroid receptor heterodimerization demonstrated in vitro and in vivo. Proc Natl Acad Sci USA 92:12480–12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M, Meijer OC, Wang J, Bhargava A, Pearce D 2003 Homodimerization of the glucocorticoid receptor is not essential for response element binding: activation of the phenylethanolamine N-methyltransferase gene by dimerization-defective mutants. Mol Endocrinol 17:2583–2592 [DOI] [PubMed] [Google Scholar]
- Tsai SY, Tsai MJ, O'Malley BW 1989 Cooperative binding of steroid hormone receptors contributes to transcriptional synergism at target enhancer elements. Cell 57:443–448 [DOI] [PubMed] [Google Scholar]
- Schüle R, Muller M, Kaltschmidt C, Renkawitz R 1988 Many transcription factors interact synergistically with steroid receptors. Science 242:1418–1420 [DOI] [PubMed] [Google Scholar]
- Nordeen SK, Ogden CA, Taraseviciene L, Lieberman BA 1998 Extreme position dependence of a canonical hormone response element. Mol Endocrinol 12:891–898 [DOI] [PubMed] [Google Scholar]
- Truss M, Chalepakis G, Beato M 1990 Contacts between steroid hormone receptors and thymines in DNA: an interference method. Proc Natl Acad Sci USA 87:7180–7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G, Beato M 1989 Hydroxyl radical interference: a new method for the study of protein-DNA interactions. Nucleic Acids Res 17:1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowland PL, Buetti E 1989 Mutations in the hormone regulatory element of mouse mammary tumor virus differentially affect the response to progestins, androgens, and glucocorticoids. Mol Cell Biol 9:3999–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connaghan-Jones KD, Heneghan AF, Miura MT, Bain DL 2007 Thermodynamic analysis of progesterone receptor-promoter interactions reveals a molecular model for isoform-specific function. Proc Natl Acad Sci USA 104:2187–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T, Horwitz KB, Lange CA 2001 Transcriptional hyperactivity of human progesterone receptors is coupled to their ligand-dependent down-regulation by mitogen-activated protein kinase-dependent phosphorylation of serine 294. Mol Cell Biol 21:6122–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarrone M, Bari M, Di Rienzo M, Finazzi-Agrò A, Rossi A 2003 Progesterone activates fatty acid amide hydrolase (FAAH) promoter in human T lymphocytes through the transcription factor Ikaros. Evidence for a synergistic effect of leptin. J Biol Chem 278:32726–32732 [DOI] [PubMed] [Google Scholar]
- Xu Q, Ohara N, Liu J, Nakabayashi K, DeManno D, Chwalisz K, Yoshida S, Maruo T 2007 Selective progesterone receptor modulator asoprisnil induces endoplasmic reticulum stress in cultured human uterine leiomyoma cells. Am J Physiol Endocrinol Metab 293:E1002–E1011 [DOI] [PubMed] [Google Scholar]
- Kang HY, Lin HK, Hu YC, Yeh S, Huang KE, Chang C 2001 From transforming growth factor-β signaling to androgen action: identification of Smad3 as an androgen receptor coregulator in prostate cancer cells. Proc Natl Acad Sci USA 98:3018–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Nordeen SK 2002 Overlapping but distinct gene regulation profiles by glucocorticoids and progestins in human breast cancer cells. Mol Endocrinol 16:1204–1214 [DOI] [PubMed] [Google Scholar]
- Hubler TR, Scammell JG 2004 Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones 9:243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M 1987 CpG islands in vertebrate genomes. J Mol Biol 196:261–282 [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA 2002 Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA 99:3740–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J 1998 Repeats in genomic DNA: mining and meaning. Curr Opin Struct Biol 8:333–337 [DOI] [PubMed] [Google Scholar]
- Jurka J 2000 Repbase update: a database and an electronic journal of repetitive elements. Trends Genet 16:418–420 [DOI] [PubMed] [Google Scholar]
- Price AL, Eskin E, Pevzner PA 2004 Whole-genome analysis of ALU repeat elements reveals complex evolutionary history. Genome Res 14:2245–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan PA, Wang J, Herke SW, Garber RK, Liang P, Batzer MA 2005 ALU retrotransposition-mediated deletion. J Mol Biol 348:791–800 [DOI] [PubMed] [Google Scholar]
- Oei SL, Babich VS, Kazakov VI, Usmanova NM, Kropotov AV, Tomilin NV 2004 Clusters of regulatory signals for RNA polymerase II transcription associated with ALU family repeats and CpG islands in human promoters. Genomics 83:873–882 [DOI] [PubMed] [Google Scholar]
- Norris J, Fan D, Aleman C, Marks JR, Futreal PA, Wiseman RW, Iglehart JD, Deininger PL, McDonnell DP 1995 Identification of a new subclass of ALU DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J Biol Chem 270:22777–22782 [DOI] [PubMed] [Google Scholar]
- Babich V, Aksenov N, Alexeenko V, Oei SL, Buchlow G, Tomilin N 1999 Association of some potential hormone response elements in human genes with the ALU family repeats. Gene 239:341–349 [DOI] [PubMed] [Google Scholar]
- Li LC, Yeh CC, Nojima D, Dahiya R 2000 Cloning and characterization of human estrogen receptor β promoter. Biochem Biophys Res Commun 275:682–689 [DOI] [PubMed] [Google Scholar]
- Rowold DJ, Herrera RJ 2000 ALU elements and the human genome. Genetica 108:57–72 [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI 2006 ALU elements within human mRNAs are probable microRNA targets. Trends Genet 22:532–536 [DOI] [PubMed] [Google Scholar]
- Le Goff W, Guerin M, Petit L, Chapman MJ, Thillet J 2003 Regulation of human CETP gene expression: role of SP1 and SP3 transcription factors at promoter sites −690, −629, and −37. J Lipid Res 44:1322–1331 [DOI] [PubMed] [Google Scholar]
- Ebihara M, Ohba H, Ohno SI, Yoshikawa T 2002 Genomic organization and promoter analysis of the human nicotinic acetylcholine receptor α6 subunit (CHNRA6) gene: ALU and other elements direct transcriptional repression. Gene 298:101–108 [DOI] [PubMed] [Google Scholar]
- Sharan C, Hamilton NM, Parl AK, Singh PK, Chaudhuri G 1999 Identification and characterization of a transcriptional silencer upstream of the human BRCA2 gene. Biochem Biophys Res Commun 265:285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA 1999 ALU repeats and human disease. Mol Genet Metab 67:183–193 [DOI] [PubMed] [Google Scholar]
- Agoulnik IU, Tong XW, Fischer DC, Körner K, Atkinson NE, Edwards DP, Headon DR, Weigel NL, Kieback DG 2004 A germline variation in the progesterone receptor gene increases transcriptional activity and may modify ovarian cancer risk. J Clin Endocrinol Metab 89:6340–6347 [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Troxel AB, Norman S, Bunin G, DeMichele A, Schinnar R, Berlin JA, Strom BL 2007 Pharmacogenetic modulation of combined hormone replacement therapy by progesterone-metabolism genotypes in postmenopausal breast cancer risk. Am J Epidemiol [Erratum (2008) 167:888] 166:1392–1399 [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T 1995 MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23:4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung L, Shen T, Abel MG, Powell RL, Takimoto GS, Sartorius CA, Horwitz KB 2001 Mapping the unique activation function 3 in the progesterone B-receptor upstream segment. Two LXXLL motifs and a tryptophan residue are required for activity. J Biol Chem 276:39843–39851 [DOI] [PubMed] [Google Scholar]
- McKnight SL 1980 The nucleotide sequence and transcript map of the herpes simplex virus thymidine kinase gene. Nucleic Acids Res 8:5949–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Hafiz H, Takimoto GS, Tung L, Horwitz KB 2002 The inhibitory function in human progesterone receptor N termini binds SUMO-1 protein to regulate autoinhibition and transrepression. J Biol Chem 277:33950–33956 [DOI] [PubMed] [Google Scholar]
- Tung L, Mohamed MK, Hoeffler JP, Takimoto GS, Horwitz KB 1993 Antagonist-occupied human progesterone B-receptors activate transcription without binding to progesterone response elements and are dominantly inhibited by A-receptors. Mol Endocrinol [Erratum (1993) 7:1378] 7:1256–1265 [DOI] [PubMed] [Google Scholar]
- Sartorius CA, Groshong SD, Miller LA, Powell RL, Tung L, Takimoto GS, Horwitz KB 1994 New T47D breast cancer cell lines for the independent study of progesterone B- and A-receptors: only antiprogestin-occupied B-receptors are switched to transcriptional agonists by cAMP. Cancer Res 54:3868–3877 [PubMed] [Google Scholar]
- Jacobsen BM, Skalnik DG 1999 YY1 binds five cis-elements and trans-activates the myeloid cell-restricted gp91(phox) promoter. J Biol Chem 274:29984–29993 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.