Abstract
The adapter protein SH2B1 (SH2-B, PSM) is recruited to multiple ligand-activated receptor tyrosine kinases, including the receptors for nerve growth factor (NGF), insulin, and IGF-I as well as the cytokine receptor-associated Janus kinase family kinases. In this study, we examine SH2B1’s function in NGF signaling. We show that depleting endogenous SH2B1 using short hairpin RNA against SH2B1 inhibits NGF-dependent neurite outgrowth, but not NGF-mediated phosphorylation of Akt or ERKs 1/2. SH2B1 has been hypothesized to localize and function at the plasma membrane. We identify a nuclear localization signal within SH2B1 and show that it is required for nuclear translocation of SH2B1β. Mutation of the nuclear localization signal has no effect on NGF-induced activation of TrkA and ERKs 1/2 but prevents SH2B1β from enhancing NGF-induced neurite outgrowth. Disruption of SH2B1β nuclear import also prevents SH2B1β from enhancing NGF-induced transcription of genes important for neuronal differentiation, including those encoding urokinase plasminogen activator receptor, and matrix metalloproteinases 3 and 10. Disruption of SH2B1β nuclear export by mutation of its nuclear export sequence similarly prevents SH2B1β enhancement of NGF-induced transcription of those genes. Nuclear translocation of the highly homologous family member SH2B2(APS) was not observed. Together, these data suggest that rather than simply acting as an adapter protein linking signaling proteins to the activated TrkA receptor at the plasma membrane, SH2B1β must shuttle between the plasma membrane and nucleus to function as a critical component of NGF-induced gene expression and neuronal differentiation.
SH2B1β requires access to both cytoplasmic and nuclear compartments to enhance NGF-induced gene transcription and neurite outgrowth.
The adapter protein SH2B1 (SH2-B, PSM) has been shown to be recruited to multiple ligand-activated receptor tyrosine kinases, including the receptors for nerve growth factor (NGF) (TrkA) (1,2), insulin (3), IGF-I (4), and the cytokine receptor-associated Janus kinase (JAK) family kinases (e.g. receptors for GH and leptin) (5,6,7,8). Yet, little is understood about the exact cellular consequences of these interactions. In this paper, we delve more deeply into the physiological function of SH2B1 within the context of NGF. SH2B1 belongs to a family of adapter proteins that includes SH2B2 (APS) and SH2B3 (Lnk) (9,10,11,12). Each of the family members contains at least one N-terminal dimerization domain (DD), a pleckstrin homology (PH) domain, a C-terminal Src homology (SH2) domain (13), and several proline-rich regions. Splice variation leads to the formation of four SH2B1 isoforms, α, β, γ, and δ, which differ only in their C termini after the SH2 domain (9,14). We and others have shown that NGF promotes the rapid association of SH2B1 with TrkA via its SH2 domain and subsequent phosphorylation of SH2B1 on tyrosines as well as serines/threonines (1,2,15).
Overexpression of SH2B1 has been shown to enhance NGF-dependent neurite outgrowth in PC12 cells. In contrast, the overexpression of dominant-negative SH2B1(R555E), which lacks a critical arginine in its SH2 domain required for binding to phosphorylated tyrosines, inhibits NGF-induced neurite outgrowth (2,15). Furthermore, antibodies to SH2B1 introduced into dissociated primary sympathetic neurons grown in NGF-containing medium exhibit a reduced rate of survival. Similarly, axonal processes are nearly eliminated when an SH2B1 mutant that blocks SH2B1-mediated signaling is introduced within explants of sympathetic ganglia grown in the presence of NGF. In contrast, NGF-treated neurons into which wild-type (WT) SH2B1 is introduced thrive and have elaborate long-branching axonal processes (2). These results led to the hypothesis that SH2B1 is required for maximal NGF-induced neurite outgrowth and maintenance of the differentiated state. However, a role for endogenous SH2B1 in NGF-induced neurite outgrowth remains to be demonstrated. In addition, the molecular mechanism by which SH2B1 contributes to enhanced neurite outgrowth remains unclear. Previous efforts to elucidate this mechanism have largely centered on the interaction between SH2B1 with TrkA. The apparent membrane localization of SH2B1β (1) and its recruitment to NGF-activated TrkA (15) support a mechanism in which SH2B1 enhances the kinase activity of TrkA and/or recruits essential NGF effector molecules to the activated receptor complex (2,16), thereby driving the prolonged activation of ERKs required for NGF-dependent differentiation (17). However, expression of SH2B1β(R555E) blocks NGF-dependent neurite outgrowth without affecting NGF-induced phosphorylation of TrkA, phospholipase C (PLC)-γ, Akt, ERK1, or ERK2 (15,18). These findings indicate that the dominant-negative effect of overexpressed SH2B1β(R555E) on NGF-dependent neurite outgrowth is unlikely to be a result of reduced TrkA activation or a diminished ability of TrkA to phosphorylate the substrates responsible for activating the ERK1/2, Akt, or PLCγ pathways. Rather, these results raised the possibility that SH2B1β might facilitate NGF-dependent differentiation through a novel pathway at a point downstream of or parallel to ERKs 1 and 2.
Here, we provide the first direct evidence that endogenous SH2B1β is required for NGF-dependent neurite outgrowth but is nonessential for NGF activation of Akt and ERKs 1 and 2. We use deletion and mutational analysis to identify in SH2B1 a monopartite nuclear localization signal (NLS). We show that the adjacent motifs of the NLS and a previously identified nuclear export sequence (NES) direct the cycling of SH2B1β between the nucleus and cytoplasm. Similar nucleocytoplasmic shuttling was not observed for the highly homologous family member SH2B2. Mutation of its NLS prevented SH2B1β from enhancing NGF induction of neurite outgrowth and transcription of the NGF-dependent genes encoding urokinase plasminogen activator receptor (uPAR), matrix metalloproteinase (MMP)-3, and MMP10. Mutation of the NES also significantly decreased the ability of SH2B1β to enhance NGF-induced transcription of these three genes. Mutation of the SH2 domain of SH2B1β, which we have shown previously to cause SH2B1 to act as a dominant negative and block NGF-induced neurite outgrowth, was shown to render SH2B1β unable to translocate to the nucleus. These results provide the first characterization of a nuclear role for SH2B1 and strong evidence in support of a paradigm shift in our thinking about the roles of membrane receptor-binding adapter proteins in growth factor function. Our data suggest that rather than simply acting as an adapter protein linking signaling proteins to the activated TrkA receptor at the plasma membrane, SH2B1 must both bind to activated TrkA and shuttle between the plasma membrane and nucleus to carry out its role as a critical component in NGF-induced gene expression and neuronal differentiation. The data also provide additional evidence that SH2B1 and SH2B2 have distinct cellular functions.
Results
SH2B1 is required for NGF-dependent differentiation of PC12 cells
Previous results using overexpression of WT and dominant-negative forms of SH2B1 indirectly implicated SH2B1 as contributing to NGF-induced neurite outgrowth. To provide more direct evidence for a critical role of endogenous SH2B1 in NGF-induced neurite outgrowth, we used an RNA interference approach to stably reduce endogenous levels of SH2B1 in PC12 cells. PC12 cells were chosen as a model system because they have provided much, if not most, of our current understanding about NGF-induced (dependent) neuronal differentiation and the responsible signaling pathways. Derived from a rat adrenal pheochromocytoma, PC12 cells cease proliferation in response to NGF, exhibit somatic hypertrophy, and acquire neurites (19). The resulting NGF-dependent morphological and biochemical changes of the PC12 cell are highly analogous to those of a true sympathetic neuron. NGF-treated PC12 cells express neuronal-specific genes (20) and are capable of forming synapses with primary neurons from rat cortex (21). Additionally, en route to NGF-mediated differentiation, the PC12 cells develop a dependence on NGF for survival as has been documented for both sympathetic and sensory neurons (22). As demonstrated previously (23) and in Fig. 2, A and B, endogenous SH2B1 levels in pooled cell lines stably expressing a 21-nucleotide long small hairpin (sh)RNA targeted against all isoforms of SH2B1 (shSH2B1) were reduced by about 50–70% (n = 3). We compared the ability of the shControl and shSH2B1 PC12 cell lines to differentiate in the presence of NGF. The shControl and shSH2B1 cell lines were incubated in serum-free medium overnight before treatment with 50 ng/ml NGF for 4 d. Cells were assessed for differentiation on each of the 4 d. They were considered differentiated if they had neurites at least twice the length of their cell diameter. As shown in Fig. 1A, the rate of NGF-induced differentiation in the SH2B1 knockdown cells was reduced by about 50% compared with control cells. Furthermore, NGF-dependent differentiation was restored in the shSH2B1 PC12 cell line upon transfection of a green fluorescent protein (GFP)-SH2B1β cDNA construct harboring three silent mutations within the shSH2B1 target sequence [GFP-SH2B1β(res)] (Fig. 1B) These results suggest that endogenous SH2B1 is critical for NGF-induced neurite outgrowth in PC12 cells.
Figure 2.
Effect of shRNA-mediated knockdown of endogenous SH2B1 on NGF-induced phosphorylation of Akt and ERKs 1 and 2. PC12 cells stably expressing control shRNA (shControl) or shRNA targeted against SH2B1 (shSH2B1) were incubated in serum-free medium for 14 h before treatment with 50 ng/ml NGF for 0, 15, 120, 240, or 360 min. Proteins in cell lysates were separated by SDS-PAGE and immunoblotted with αSH2B1, αpAkt(Ser473), αpERK, αErk, and α-αTubulin as a loading control. The relative signal intensity of the bands corresponding to endogenous SH2B1β (B), pErk (C), and pAkt (D) was quantified.
Figure 1.
Endogenous SH2B1 is required for NGF-dependent neurite outgrowth. A, PC12 cells stably expressing control shRNA (shControl) or shRNA targeted against SH2B1 (shSH2B1) were incubated in serum-free medium for 14 h and then treated with 50 ng/ml NGF for 4 d. The percentage of differentiated cells (y-axis) was calculated on d 1, 2, 3, and 4 of the 4-d assay (x-axis). B, shSH2B1 PC12 cells were transfected with either cDNA encoding GFP or GFP-SH2B1β(res) (a construct containing silent mutations within the shRNA target sequence). The cells were incubated in serum-free medium for 14 h and then treated with 50 ng/ml NGF for 2 d, after which differentiation was assessed in GFP-positive cells and expressed as percent differentiation relative to shControl+GFP cells. The values are means ± sem from three different experiments. Asterisks represent P values of <0.05 using a one-tailed, paired Student’s t test. At least 250 cells were counted for each condition in A and at least 50 cells in B.
We next addressed the question of whether endogenous SH2B1 is required for NGF-dependent activation of ERKs 1 and 2, because ERK1/2 activation is critical for NGF-induced neurite outgrowth of PC12 cells (24,25,26). Both shControl and shSH2B1 PC12 cells were treated with NGF (50 ng/ml) for 0, 15, 120, 240, and 360 min. As mentioned above, Fig. 2A, top panel, reveals a greater than 50% reduction of endogenous SH2B1β in the shSH2B1 vs. shControl cells. The relative signal intensity is quantified in Fig. 2B. Figure 2A, top panel, reveals that the mobility of endogenous SH2B1β is dramatically reduced after 15 min treatment with NGF. Previous results attributed the decreased mobility to phosphorylation on tyrosines and serines/threonines (1). To assess the NGF-dependent activation of ERKs 1 and 2, we performed Western blotting of cell lysates with αpERK1/2 that recognizes only the doubly phosphorylated, activated form of ERKs 1 and 2 as well as an antibody against total ERK1/2 (αERK1/2). The extent and duration of phosphorylation of ERKs 1 and 2 in response to NGF was similar (data not shown) or even greater (Fig. 2A, middle panel, and 2C) in the shSH2B1 cells compared with the shControl cells, when normalized to levels of total ERKs 1 and 2. The membrane was reblotted with an antibody to pAkt (αpAkt) that recognizes phosphorylated Ser 473. Akt, which lies downstream of phosphatidylinositol-3-kinase, requires phosphorylation on Ser 473 to be active (27). We observed no significant difference in NGF-induced phosphorylation of Akt in the shSH2B1 line compared with the shControl line (Fig. 2A, second panel, and 2D). These results suggest that SH2B1’s ability to enhance NGF-induced neurite outgrowth is not secondary to enhanced NGF activation of Akt or ERKs 1 and 2.
Identification of a region within SH2B1 required for nuclear translocation
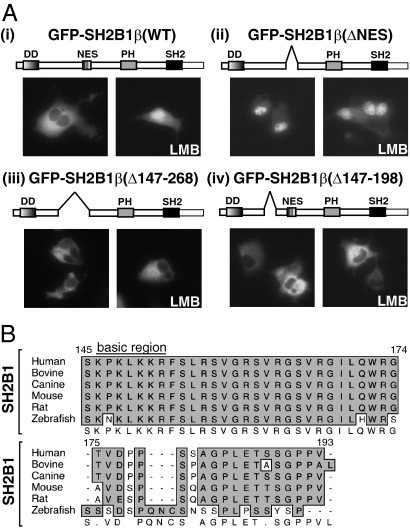
We have shown previously (28) that endogenous SH2B1, as well as overexpressed SH2B1β, accumulates in the nucleus of cells incubated in the presence of a nuclear export inhibitor, leptomycin B (LMB) (28), indicating that SH2B1β undergoes nucleocytoplasmic shuttling. Additionally, the deletion of amino acids 198-268 results in the nuclear accumulation of SH2B1β, an otherwise membrane/cytoplasmic localized protein (15). Close examination of the sequence contained within amino acids 198-268 revealed a match (28) to the NES motif (LX2–3LX2–4LXL) (29,30,31). Indeed, point mutations of two key leucine residues (L231 and L233) in this sequence rendered SH2B1β defective in nuclear export, resulting in SH2B1β’s nuclear accumulation (28). The dramatic localization shift observed with these SH2B1β mutants suggested that SH2B1β undergoes rapid and continual shuttling between the nucleus and cytoplasm, with the rate of nuclear export greatly exceeding the rate of nuclear import, giving rise to a steady-state membrane/cytoplasmic appearance. However, because there is substantial mixing of cytoplasmic and nuclear proteins after nuclear envelope breakdown during mitosis (32), the presence of an NES alone within a protein is not sufficient evidence to ascribe a nuclear role for that protein. In fact, an NES in the absence of an NLS may function to assure that a protein stays out of the nucleus. Thus, to determine whether nuclear localization of SH2B1 is important for its function, it was critical to identify a nuclear localization sequence in SH2B1 that we could then mutate to prevent SH2B1 from entering the nucleus. As the readout of a screen to identify the regions necessary for SH2B1β’s nuclear import, we used the dramatic nuclear accumulation of GFP-SH2B1β(Δ198-268) lacking the NES [henceforth designated as GFP-SH2B1β(ΔNES)] (Fig. 3A, ii) or GFP-SH2B1β(WT) treated with the nuclear export inhibitor LMB (Fig. 3A, i), In this screen in which nuclear export is blocked, nuclear accumulation of SH2B1β represents functional nuclear import of SH2B1β, whereas nuclear exclusion represents defective nuclear import of SH2B1β. Initially, a series of deletion and point mutations were made on top of GFP-SH2B1β(ΔNES). The modified forms of SH2B1 were transiently expressed in COS-7 cells. COS-7 cells were used for the initial screen because of their high transfection efficiency and flat morphology, which lends itself to rapid and unambiguous identification of subcellular localization of the fluorescent proteins. We made the same series of mutations in a GFP-tagged WT SH2B1β and incubated the cells expressing those mutants in the absence or presence of LMB.
Figure 3.
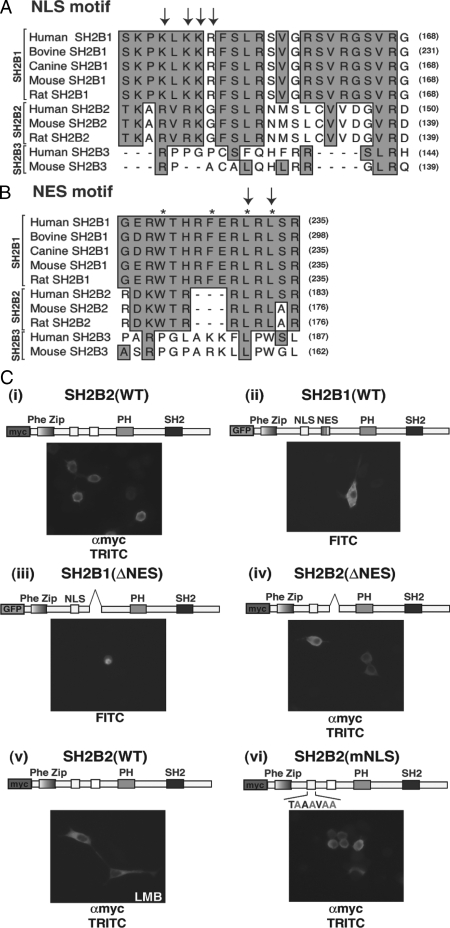
The region between amino acids 147 and 198 is essential for nuclear translocation. A, COS-7 cells were transiently transfected with cDNA encoding GFP-SH2B1β(WT) (i), GFP-SH2B1β(ΔNES) (ii), GFP-SH2B1β(Δ147-268) (iii), or GFP-SH2B1β(Δ147-198) (iv). Fourteen hours after transfection, the cells were either mock treated with vehicle (left panels) or treated with 20 nm LMB (right panels) for 7 h. Cells were then fixed and visualized using epifluorescence microscopy. The schematic representations of SH2B1β indicate the positions of internal deletions and/or previously defined domains within SH2B1, including the DD, NES, PH, and SH2. B, MacVector ClustalW sequence alignment of multiple species of SH2B1 between residues 145 and 193.
As shown previously (28), overexpressed GFP-SH2B1β(WT) appears to have a membrane and cytoplasmic localization (Fig. 3A, i, left panel). After incubation with LMB, GFP-SH2B1β(WT) was found to accumulate in the nucleus (Fig. 3A, i, right panel). The GFP-SH2B1β(ΔNES) mutant, which retains the ability to translocate to the nucleus but is unable to cycle to the cytoplasm without the signal responsible for nuclear export, accumulates in the nucleus even in the absence of LMB (Fig. 3A, ii). Another mutant of SH2B1β [SH2B1β(Δ147-268)] that contains a slightly larger region of deletion than SH2B1β(ΔNES) (Δ198-268) was observed in the cytoplasm with or without LMB (Fig. 3A, iii). This suggests that the region required for nuclear import of SH2B1 lies within amino acids 147-198. As predicted, GFP-SH2B1β(Δ147-198) failed to accumulate in the nuclei of COS-7 cells in the absence or presence of LMB (Fig. 3A, iv). Examination of the amino acid sequence between 147 and 198 in SH2B1β (Fig. 3B) revealed a small stretch of highly basic residues that resembles the classical nuclear localization sequence motif (PKKKRKV) of the SV40 large T antigen (33,34). This putative NLS is almost perfectly conserved in all known vertebrate SH2B1 homologs from zebrafish to human (Fig. 3B). The basic region identified in SH2B1 spans from amino acids 146–152. Thus, four basic residues likely to be critical for nuclear import are missing from the nuclear import defective SH2B1β(Δ147-198).
SH2B1 requires an NLS and a functional SH2 domain for nuclear translocation in PC12 cells
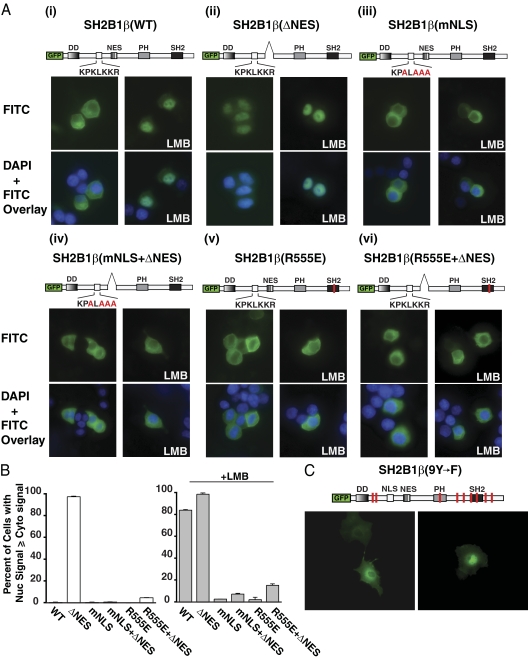
To confirm that this basic region found between amino acids 146–152 is responsible for nuclear import of SH2B1β in PC12 cells, a series of GFP-SH2B1β constructs encoding point mutations and/or deletions in this region were expressed in PC12 cells in the presence or absence of LMB. As shown previously (1,28), GFP-SH2B1β(WT) appears to localize exclusively to the membrane and cytoplasm of PC12 cells (Fig. 4A, i, top left panel) and incubation with LMB results in its nuclear accumulation (Fig. 4A, top right panel). These results are quantified in Fig. 4B and expressed as the percentage of cells in which the intensity of GFP-SH2B1β epifluorescence in the nucleus is greater than or equal to its intensity in the cytoplasm. GFP-SH2B1β(ΔNES) is found in the nucleus whether or not the cells are incubated with LMB (Fig. 4A, ii). Site-directed mutagenesis was used to change the putative NLS from K148/K150/K151/R152 to A148/A150/A151/A152 in SH2B1β. This mutant SH2B1β will be referred to as SH2B1β(mNLS). The mutation of the putative NLS resulted in a failure of GFP-SH2B1β to localize to the nucleus even in the presence of LMB (Fig. 4A, iii, and 4B) or when combined with deletion of the NES (Fig. 4A, iv, and 4B). We therefore concluded that the basic residues K148/K150/K151/R152 are required for the nuclear localization of SH2B1β and constitute a previously unrecognized monopartite NLS present within all isoforms of SH2B1.
Figure 4.
Point mutations in the putative NLS motif inhibit nuclear accumulation of SH2B1β. A and B, PC12 cells were transiently transfected with cDNAs encoding GFP-SH2B1β(WT) (i), GFP-SH2B1β(ΔNES) (ii), GFP-SH2B1β(mNLS) (iii), GFP-SH2B1β(mNLS+ΔNES) (iv), GFP-SH2B1β(R555E) (v), and GFP-SH2B1β(R555E+ΔNES) (vi). Fourteen hours after transfection, the cells were either mock treated (left panels) or treated with 20 nm LMB for 7 h (right panels). A, Cells were fixed and stained with 4′,6-diamidino-2-phenylindole to visualize nuclei (blue color in overlay image, lower panels) and then visualized using epifluorescence microscopy. B, In a blinded experiment, at least 100 cells from each condition were assessed for a nuclear fluorescence signal greater than or equal to the cytoplasmic fluorescence. The results are expressed as the percentage of cells exhibiting nuclear fluorescence out of the total number of cells counted. Means and range calculated from two independent experiments are shown. C, COS-7 cells were transiently transfected with cDNA encoding GFP-SH2B1β(9Y-F), a mutant lacking all nine tyrosines within SH2B1β. Fourteen hours aftert transfection, the cells were either mock treated (left panel) or treated with 20 nm LMB for 7 h (right panel). Cells were then fixed and visualized using epifluorescence microscopy. The schematic representations of SH2B1β indicate the positions of internal deletions, mutations, and/or previously defined domains within SH2B1, including the DD, NES, PH, and SH2. The open white box represents the putative NLS motif within SH2B1, and the corresponding sequence is indicated below. Single basic amino acid substitutions are indicated in red.
Figure 4A, v and vi, illustrates that GFP-SH2B1β(R555E) also does not accumulate in the nucleus when treated with LMB (Fig. 4A, v) or when the NES is deleted (Fig. 4A, vi). Mutation of the critical arginine within the SH2 domain to a glutamate [SH2B1β(R555E)] abolishes the phosphotyrosine binding ability of SH2B1, and the expression of SH2B1β(R555E) in PC12 cells results in the dominant-negative inhibition of NGF-dependent neurite outgrowth (35). The failure of SH2B1β(R555E) to associate with activated TrkA also results in a loss of SH2B1β(R555E) tyrosyl phosphorylation (15). To test whether SH2B1β requires tyrosine phosphorylation for nuclear translocation, we mutated all nine tyrosines within SH2B1β to phenylalanines [SH2B1β(9Y→F)] using site-directed mutagenesis. COS-7 cells were transiently transfected with GFP-SH2B1β(9Y→F) and incubated with or without LMB for 7 h. As shown in Fig. 4C, GFP-SH2B1β(9Y→F) accumulates in the nucleus after LMB treatment, indicating that, like SH2B1β(WT), GFP-SH2B1β(9Y→F) undergoes nucleocytoplasmic shuttling. Thus, phosphorylated tyrosines within SH2B1β are not required for its nuclear translocation.
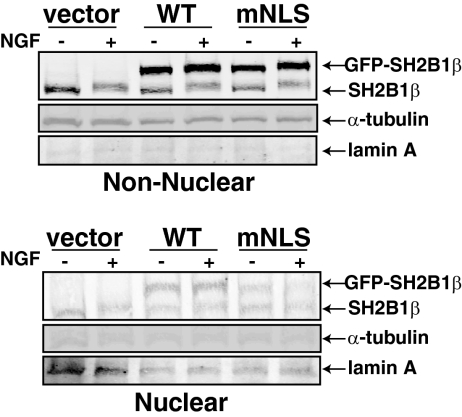
To assess whether or not NGF stimulation affects the amount of SH2B1β in the nucleus, we used subcellular fractionation of PC12 cells transiently expressing GFP, GFP-SH2B1β(WT), or GFP-SH2B1β(mNLS). PC12 cells were incubated in serum-free medium before NGF treatment for 0 or 30 min. Consistent with the subcellular localization of GFP-SH2B1β observed in Figs. 3A and 4A, the majority of endogenous and overexpressed SH2B1β appeared to reside in the cytoplasm/plasma membrane (nonnuclear) fraction (Fig. 5). However, both endogenous SH2B1β and overexpressed GFP-SH2B1β(WT) were present in purified nuclei devoid of cytoplasmic contamination, as assessed by the absence of α-tubulin. Consistent with impaired nuclear import, levels of GFP-SH2B1β(mNLS) in the nuclear fraction were significantly reduced compared with WT SH2B1β. NGF did not detectably affect the levels of endogenous SH2B1 or ectopically expressed GFP-SH2B1β in the nuclear fraction. However, an NGF-dependent upward shift in the migration of endogenous SH2B1, shown previously to be a consequence of phosphorylation (1), was observable for both nuclear and nonnuclear SH2B1β. Together these results suggest that both endogenous and overexpressed SH2B1β constitutively shuttle between the cytoplasm and nucleus. Furthermore, both the highly phosphorylated form of SH2B1 seen after NGF treatment and the more minimally phosphorylated form of SH2B1 seen after serum deprivation appears capable of crossing the nuclear membrane.
Figure 5.
Endogenous and overexpressed SH2B1β constitutively shuttles between the cytoplasm and nucleus. PC12 cells were transiently transfected with cDNAs encoding GFP, GFP-SH2B1β(WT), and GFP-SH2B1β(mNLS). The cells were incubated in serum-free medium for 14 h before treatment with 100 ng/ml NGF for 0 or 30 min. Proteins (25 μg) in nonnuclear and nuclear lysates were separated by SDS-PAGE and immunoblotted with αSH2B1 (to visualize endogenous and overexpressed SH2B1β), α-αtubulin (nonnuclear), or αlamin (nuclear).
The nuclear import of SH2B1β is disrupted in energy-depleted cells
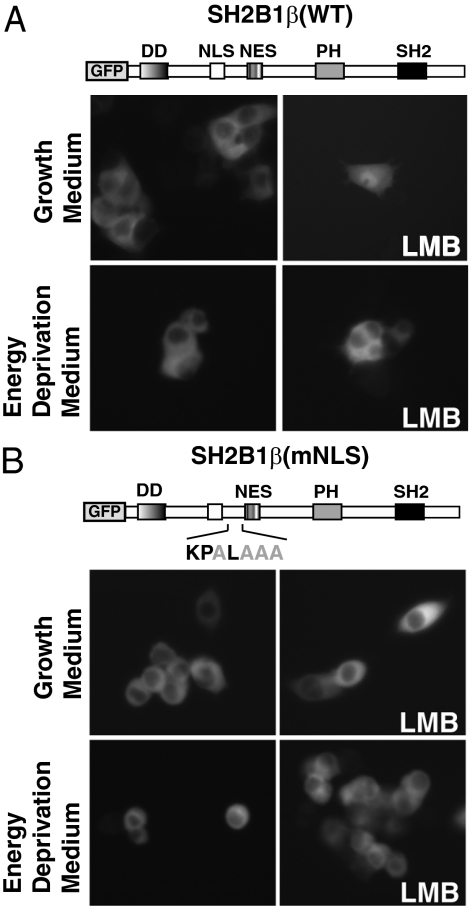
To address whether the nuclear translocation of SH2B1β is an energy-dependent mechanism, we depleted ATP levels within living cells and assessed the nucleocytoplasmic shuttling of SH2B1β. ATP was depleted from cells by incubation in glucose-free medium supplemented with 10 mm sodium azide and 10 mm 2-deoxy-d-glucose (energy depletion medium). This method has been used to reversibly inhibit classical Ran-dependent nuclear transport by limiting GTP-bound Ran (36,37). PC12 cells stably overexpressing GFP-SH2B1β(WT) and GFP-SH2B1β(mNLS) were incubated in normal growth medium or energy depletion medium for 1 h before the addition of 20 nm LMB for 4 h to inhibit nuclear export. As a result of the continuous nucleocytoplasmic shuttling, the steady-state localization of SH2B1β(WT) favors a plasma membrane/cytoplasmic appearance when cells are incubated in normal growth medium but shifts to a predominately nuclear localization after addition of LMB (Fig. 6A, top panels). However, preexposure of the cells to the energy depletion medium blocked LMB-dependent nuclear accumulation of GFP-SH2B1β(WT) (Fig. 6A, bottom panels). As expected, stably overexpressed GFP-SH2B1β(mNLS) is localized at the plasma membrane/cytoplasm with or without LMB treatment and does not change upon preexposure to energy depletion medium (Fig. 6B). These results suggest that the presence of SH2B1β(WT) in the nucleus is not a result of passive diffusion but rather an energy-dependent process.
Figure 6.
The nuclear import of SH2B1β is disrupted in energy-depleted cells. PC12 cells stably expressing GFP-SH2B1β(WT) (A) or GFP-SH2B1β(mNLS) (B) were incubated in normal growth medium or energy depletion medium for 1 h before treatment with or without 20 nm LMB for 4 h, as indicated.
SH2B2 does not undergo nucleocytoplasmic shuttling
The finding that SH2B1 contains functional NES and NLS raises the question as to whether other SH2B gene family members [SH2B2 (APS), SH2B3 (Lnk)] can also undergo nucleocytoplasmic shuttling. The ClustalW multiple sequence alignment of several species of SH2B1 family members revealed that SH2B2, but not SH2B3, shares a high degree of homology with both the NLS (Fig. 7A) and NES (Fig. 7B) of SH2B1. The alignment also revealed that three of the four basic residues required for SH2B1 nuclear localization (R119, R121, and K122, indicated with arrows) are conserved in rat, mouse, and human SH2B2; SH2B2 contains a putative NES between residues 168 and 174, based on the loosely defined NES motif, LX2–3LX2–4LXL (29,30,31). The putative NES of SH2B2 contains three of the four hydrophobic residues, including the amino acid motif LXL (Fig. 7B), that have been shown to be required for nuclear export of SH2B1 (28) as well as of 14-3-3 (30).
Figure 7.
Subcellular distribution of internal deletion and point mutants of SH2B2 in PC12 cells. MacVector ClustalW multiple sequence alignment was used to compare several species of SH2B1, SH2B2, and SH2B3. A, Interspecies conservation among SH2B family members within the region corresponding to the NLS of SH2B1. Arrows designate the basic residues required for nuclear import. B, Interspecies conservation among SH2B family members within the region corresponding to the NES of SH2B1. Asterisks denote the general NES motif, and arrows designate the hydrophobic residues that have been shown to be required for SH2B1β nuclear export. C, PC12 cells were transiently transfected with cDNAs encoding myc-SH2B2(WT) (i), GFP-SH2B1β(WT) (ii), GFP-SH2B1β(ΔNES) (iii), myc-SH2B2(ΔNES) (iv), myc-SH2B2(WT) (v), and myc-SH2B2(mNLS) (vi). Fourteen hours after transfection, the cells were treated for 7 h with either vehicle (methanol) (i, ii, iii, iv, and vi) or 20 nm LMB (v). Cells transiently expressing myc-tagged proteins were stained with αmyc and antimouse IgG Alexa 594 and visualized in the tetramethyl rhodamine isothiocyanate (TRITC) channel (i, iv, v, and vi). GFP-SH2B1 expression was visualized in the fluorescein isothiocyanate (FITC) channel (ii and iii). Images were taken using epifluorescence microscopy.
In agreement with previous findings (38,39), SH2B2(WT) appears to localize at the plasma membrane as well as in the cytoplasm, as shown in Fig. 7C, i. The subcellular localization of myc-SH2B2 resembles closely the localization of GFP-SH2B1β (Fig. 7C, ii). However, although GFP-SH2B1β(ΔNES) localizes to the nucleus (Fig. 7B, iii), myc-tagged SH2B2 mutant carrying a deletion of the putative NES (Δ167-184) [SH2B2(ΔNES)] does not accumulate in the nucleus (Fig. 7C, iv). To confirm the inability of SH2B2 to undergo nucleocytoplasmic shuttling, we incubated cells expressing myc-tagged SH2B2(WT) with LMB for up to 24 h. As shown in Fig. 7C, v, even a 24-h treatment with LMB does not cause myc-SH2B2(WT) to accumulate in the nucleus. Mutating the putative NLS (K117A, R119A, R121A, and K122A) [SH2B2(mNLS)] also did not alter the subcellular localization of SH2B2 (Fig. 7C, vi). To address the possibility that SH2B2 can translocate to the nucleus but only in response to NGF stimulation or possibly serum deprivation, myc-SH2B2(ΔNES)-expressing cells were incubated in serum-free medium for 14 h and incubated in the absence or presence of NGF for 5 min, 30 min, 1 h, and 6 h. None of the conditions tested resulted in detectable nuclear accumulation of myc-SH2B2(ΔNES) (data not shown). The failure to detect even the slightest nuclear localization of SH2B2 after inhibiting nuclear export with LMB or removing its putative NES suggests that the cytoplasmic appearance of SH2B2 reflects a more-or-less static localization. This is in contrast to SH2B1β(WT) that is seemingly in constitutive flux between the plasma membrane/cytoplasm and nucleus. Consequently, we have concluded that despite the high degree of sequence similarity between family members SH2B1 and SH2B2, the ability to undergo nucleocytoplasmic shuttling is unique to SH2B1.
Ability of nuclear localization-defective SH2B1β mutants to regulate NGF-induced differentiation
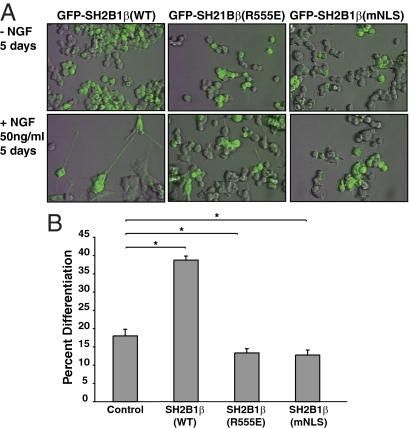
The identification of an NLS in SH2B1 enabled us to examine whether nuclear SH2B1 serves a function. We first examined whether nuclear localization of SH2B1β is required for NGF-dependent differentiation of PC12 cells. We stably expressed WT SH2B1β or the nuclear localization-defective forms of SH2B1β [SH2B1β(mNLS) and SH2B1β(R555E)] in PC12 cells. Pooled cell lines were created by combining the top 5% GFP-positive cells, using fluorescence-activated cell sorting after 4 wk of selection in G418-containing growth medium. The resulting pooled stable PC12 cell lines display a spectrum of GFP expression levels. In Fig. 8A, only PC12 cells expressing high levels of GFP can be seen after merging the fluorescent and bright-field images, although all cells were GFP positive and were counted as such. PC12 cells stably expressing SH2B1β(WT), SH2B1β(R555E), or SH2B1β(mNLS) were incubated in differentiation medium in the absence or presence of 50 ng/ml NGF for a total of 5 d. As shown in Fig. 8A, we observed NGF-dependent neurite outgrowth in PC12 cells overexpressing SH2B1β(WT), but not in PC12 cells overexpressing SH2B1β(R555E) or SH2B1β (mNLS). In Fig. 8B, we quantified the ability of NGF to induce differentiation of PC12 cells overexpressing SH2B1β(WT), SH2B1β(R555E), or SH2B1β(mNLS) in comparison with PC12 cells overexpressing vector control. In agreement with previous findings (28), stable overexpression of SH2B1β(WT) resulted in more than a doubling of the number of cells exhibiting NGF-dependent neurite outgrowth (38 ± 1% compared with 18 ± 2% for control cells). Also in agreement with previous findings, the stable expression of the dominant-negative form of SH2B1β, SH2B1β(R555E), resulted in inhibition of NGF-dependent neurite outgrowth (13 ± 1% of cells) compared with GFP alone (18 ± 2% of cells). Strikingly, mutation of the NLS prevented SH2B1β from enhancing NGF-induced neurite outgrowth, suggesting that SH2B1β must enter the nucleus for it to promote NGF-induced neuronal differentiation.
Figure 8.
Stable expression of the SH2B1β nuclear import mutant inhibits NGF-dependent neurite outgrowth of PC12 cells. A, PC12 cells stably expressing GFP-SH2B1β(WT), GFP-SH2B1β(R555E), or GFP-SH2B1B(mNLS) were incubated in serum-free medium overnight before incubation in the absence (top panels) or presence (bottom panels) of 50 ng/ml NGF for 5 d. Representative pictures of each condition were imaged by overlaying the FITC (GFP) on top of standard bright-field illumination. B, The percentage of differentiated cells was scored on d 5. Means ± sem from six different experiments are shown. Asterisks represent P values of <0.05 using a one-tailed, paired Student’s t test. A combined total of at least 400 cells were counted for each condition.
Effect of overexpression of SH2B1β(mNLS) on NGF-induced tyrosyl phosphorylation of TrkA and activation of Erks 1 and 2
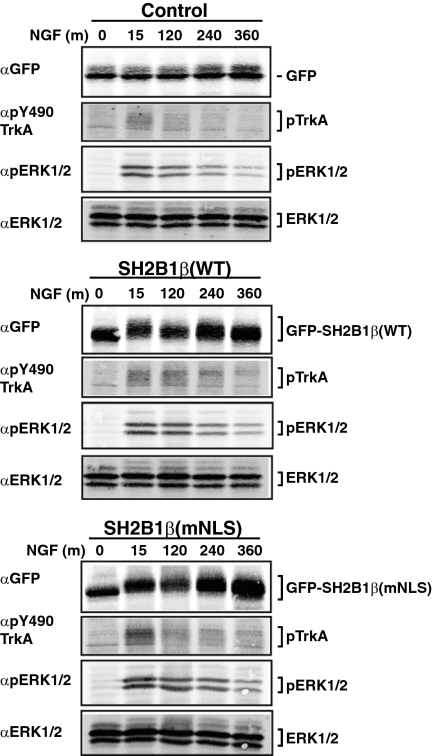
In PC12 cells, prolonged activation of ERK is thought to mediate differentiation, whereas transient activation of ERK is thought to promote proliferation (17). Because overexpression of SH2B1β(mNLS) inhibits NGF-dependent neurite outgrowth, it was of interest to establish whether the overexpression of SH2B1β(mNLS) affects the kinetics of NGF-dependent activation of TrkA and ERKs 1 and 2. PC12 cell lines stably expressing GFP, GFP-SH2B1β(WT), or GFP-SH2B1β(mNLS) were incubated in serum-free medium and then stimulated with NGF for 0, 15, 120, 240, and 360 min. Proteins from cell lysates were resolved by SDS-PAGE and immunoblotted first with αGFP to verify the presence of GFP-SH2B1β(WT) and GFP-SH2B1β(mNLS). Importantly, Fig. 9 reveals that expression levels of GFP-SH2B1β(WT) and GFP-SH2B1β(mNLS) are similar, therefore eliminating the potential for SH2B1β dosage-dependent differences. Figure 9 also reveals similar upward shifts in migration of GFP-SH2B1β(WT) and GFP-SH2B1β(mNLS), consistent with the two forms of SH2B1β undergoing similar degrees of phosphorylation in response to NGF. Immunoblotting with an antibody that recognizes phosphotyrosine 490 (αpY490 TrkA), a primary autophosphorylation site within TrkA, revealed a similar NGF-dependent phosphorylation of TrkA in the SH2B1β(WT)- and SH2B1β(mNLS)-expressing cell lines. Likewise, the extent of phosphorylation of ERKs 1 and 2 after NGF treatment, as well as the duration of phosphorylation, was comparable between SH2B1β(WT)- and SH2B1β(mNLS)-expressing cell lines. The two cell lines also showed similar profiles for NGF-dependent phosphorylation of Akt (data not shown). Collectively, these data strongly suggest that the neurite outgrowth defect observed in the cells expressing SH2B1β(mNLS) is not secondary to a defect in NGF-dependent activation of TrkA or ERKs 1 and 2.
Figure 9.
Effects of SH2B1β(mNLS) expression on NGF-induced phosphorylation of TrkA and Erks 1 and 2. PC12 cells stably expressing GFP, GFP-SH2B1β(WT), or GFP-SH2B1β(mNLS) were incubated in serum-free medium overnight before treatment with 50 ng/ml NGF for 0, 15, 120, 240, and 360 min. Proteins in cell lysates were separated by SDS-PAGE and immunoblotted with αGFP to visualize the relative levels of expression of GFP-SH2B1β(WT) and GFP-SH2B1β(mNLS). The blots were also immunoblotted with αpTrkA(Y490), αpERK1/2, and αERK1/2 as indicated.
Nuclear import and export of SH2B1β are required for NGF-induced enhancement of gene expression required for neuronal differentiation
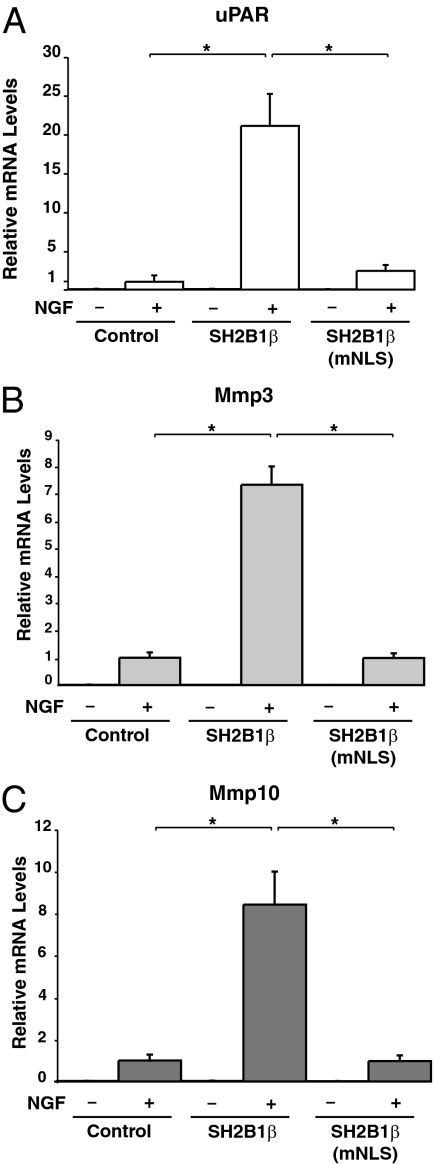
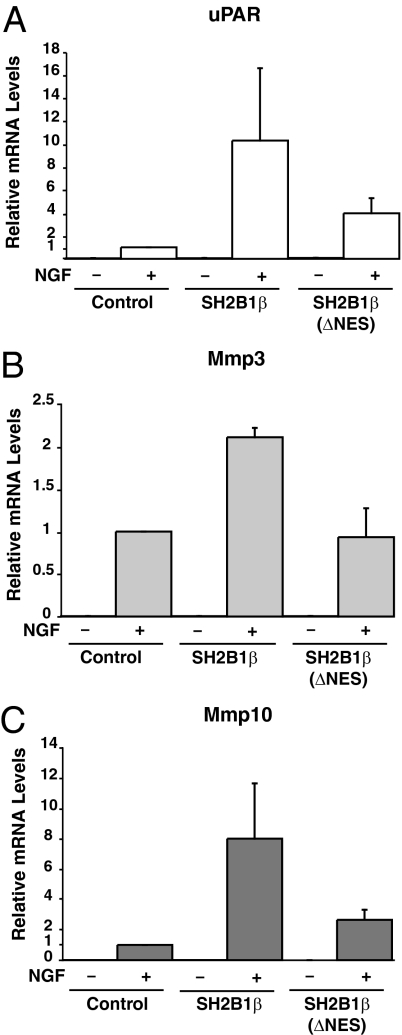
Recently, using microarray analysis of PC12 cells, we established that the overexpression of SH2B1β leads to altered transcription of only a specific subset of NGF-responsive genes, including uPAR (Plaur), Mmp3 (stromelysin-1; transin-1), and Mmp10 whose gene products encode uPAR, MMP3, and MMP10 (23). Interestingly, uPAR present in the plasma membrane of both the cell body and neurites (23) has been shown to be required for NGF-mediated neuronal differentiation (40,41), and MMP3, present in growth cones of NGF-treated PC12 cells, has been implicated in NGF-induced neurite penetration through the extracellular matrix (42). Additionally, uPAR, MMP3, and MMP10 are all situated in the same proteolytic cascade responsible for extracellular matrix degradation required for neurite penetration through the extracellular matrix (reviewed in Ref. 23). Overexpression of the dominant-negative SH2B1β(R555E) blocks both NGF-dependent neurite outgrowth as well as NGF-induced transcription of the genes encoding uPAR, MMP3, and MMP10 (23). Taken together, these results suggested that SH2B1β mediates NGF-induced neuronal differentiation and neurite outgrowth at least in part by enhancing NGF-dependent expression of uPAR, MMP3, and MMP10. In light of these findings, we questioned whether the nucleocytoplasmic shuttling by SH2B1β is required for SH2B1β facilitation of NGF-induced transcription of the genes encoding uPAR, MMP3, and MMP10. To answer this question, we used real-time quantitative PCR (QT-PCR) to assay the NGF-dependent transcription of uPAR, Mmp3, and Mmp10. PC12 cells stably expressing vector control, SH2B1β(WT), and SH2B1β(mNLS) were incubated in serum-free medium for 14 h and then incubated with or without NGF (100 ng/ml) for 0 or 6 h. Untreated PC12 cells displayed very little basal transcription of uPAR, Mmp3, or Mmp10 (Fig. 10). After 6 h NGF treatment, transcription of these genes was dramatically elevated. In agreement with our previous findings (23), NGF-induced transcription of uPAR, Mmp3, and Mmp10 was enhanced in cells stably expressing SH2B1β(WT) (Fig. 10, A–C), whereas the transcription of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was similar in control and SH2B1β-overexpressing cells. However, overexpression of SH2B1β(mNLS) failed to enhance NGF-induced transcription of uPAR, Mmp3, or Mmp10, as shown in Fig. 10, A, B, and C, respectively. The inability of SH2B1β(mNLS) to enhance NGF-induced transcription of uPAR, Mmp3, or Mmp10 suggests that the nuclear translocation of SH2B1β is critical for SH2B1β enhancement of NGF-induced transcription of these genes.
Figure 10.
Stable expression of the SH2B1β nuclear import mutant [SH2B1β(mNLS)] fails to enhance NGF-induced transcription of uPAR, Mmp3, and Mmp10. After incubation in serum-free medium overnight, PC12 cells stably expressing GFP, GFP-SH2B1β(WT), or GFP-SH2B1B(mNLS) were incubated with or without 50 ng/ml NGF for 6 h. The NGF-dependent induction of mRNA for Plaur (uPAR) (A), Mmp3 (B), and Mmp10 (C) was assessed using QT-PCR. Target gene expression was normalized first to levels of GAPDH and then to levels of gene expression seen in GFP control cells treated with NGF. Means ± sem for four to six separate experiments are shown.
We also determined the effects of expressing SH2B1β(ΔNES) on the NGF-dependent transcription of uPAR, Mmp3, and Mmp10. Figure 11 illustrates that the NGF-dependent transcription of uPAR, Mmp3, and Mmp10 in the SH2B1β(ΔNES) cells was also significantly reduced compared with SH2B1β(WT) cells. These findings demonstrate that constitutively nuclear localized SH2B1β is significantly less effective than SH2B1β(WT) at enhancing NGF-dependent transcription of genes required for neurite outgrowth. Collectively, the QT-PCR data indicate that SH2B1β requires both a functional NLS and NES to maximally enhance NGF-dependent transcription of uPAR, Mmp3, and Mmp10.
Figure 11.
Stable expression of the SH2B1β nuclear export mutant [SH2B1β(ΔNES)] fails to enhance NGF-induced transcription of uPAR, Mmp3, and Mmp10. PC12 cells stably expressing GFP, GFP-SH2B1β(WT), or GFP-SH2B1B(ΔNES) were incubated in serum-free medium overnight and then with or without 50 ng/ml NGF for 6 h. The NGF-dependent induction of mRNA for uPAR (A), Mmp3 (B), and Mmp10 (C) was quantified using QT-PCR. Target gene expression was normalized first to levels of GAPDH and then to levels of gene expression seen in GFP control cells treated with NGF. Means and range are shown for two separate experiments.
Discussion
In this work, we provide strong evidence for a critical role of endogenous SH2B1 in NGF-induced neurite outgrowth. We also provide the first evidence of a functional role for nuclear SH2B1: NGF-induced neurite outgrowth and NGF-induced expression of genes vital for NGF-induced neuronal differentiation. Finally, we show that the SH2B1 family member SH2B2 does not appear to localize to the nucleus. Previous studies implicated SH2B1β in NGF-induced neurite outgrowth based upon the ability of overexpressed WT SH2B1β to enhance, and the dominant-negative mutant SH2B1β(R555E) to inhibit, NGF-induced neurite outgrowth (15,16). Here, we more directly demonstrated a critical role for SH2B1 in neuronal differentiation by reducing endogenous levels of SH2B1 in PC12 cells using shRNA targeted against all isoforms of SH2B1. Reduction of endogenous levels of SH2B1 substantially inhibited NGF-dependent neurite outgrowth, whereas reintroduction of SH2B1β restored that neurite outgrowth. Endogenous SH2B1 does not appear to enhance NGF-induced neurite outgrowth as a consequence of enhancing NGF-induced activation of TrkA, Akt, or ERKs 1 and 2, because reduction of endogenous levels of SH2B1 did not reduce the extent or duration of NGF-induced phosphorylation of Akt or ERKs 1 and 2. Recently, SH2B1 has similarly been implicated as a critical factor in glial-cell-line-derived neurotrophic factor (GDNF)-dependent neurite outgrowth (43). GDNF-dependent neurite outgrowth was found to be inhibited when levels of endogenous SH2B1 were reduced by expression of SH2B1β RNA interference and by the overexpression of the dominant-negative SH2B1β(R555E). Similarly, neurite outgrowth did not appear to be a consequence of reduced GDNF signaling because the levels of GDNF-induced phosphorylation of Akt and ERKs 1 and 2 (albeit measured at only one time point) were found to be the same as in control cells (43). Collectively, these data strongly suggest that endogenous SH2B1 is critical for neuronal differentiation induced by both NGF and GDNF and enhances NGF- and GDNF-induced differentiation at a point downstream of or parallel to ERKs 1 and 2 and phosphatidylinositol-3-kinase/Akt.
Although initial analysis of the subcellular localization of SH2B1β suggested that it was located exclusively in the cytoplasm and at the plasma membrane (1), we previously identified an NES present in all isoforms of SH2B1 (28). Mutation or deletion of the NES trapped SH2B1β in the nucleus, providing evidence that the identified NES was indeed functional. Support for endogenous SH2B1β cycling through the nucleus was provided by the finding that pharmacological inhibition of nuclear export using LMB resulted in accumulation of endogenous (as well as ectopically expressed) SH2B1 in the nucleus (28). However, the presence of an NES alone is not sufficient to ascribe a nuclear role to that protein. We report here the identification within SH2B1 of a functional NLS. Specific mutation of four basic amino acids to alanines within the putative NLS [SH2B1β(mNLS)] blocked SH2B1β nuclear translocation in PC12 cells and all other cell lines tested (293T, COS-7, and NIH 3T3, data not shown). The NLS is almost perfectly conserved in all SH2B1 isoforms as well as all known vertebrate SH2B1 homologs from human to zebrafish, suggesting that the NLS bestows an important function to the SH2B1 protein.
Directional transport between the nucleus and the cytoplasm occurs in an energy-dependent manner through the nuclear pore complex. For the vast majority of proteins, crossing the nuclear pore complex from the cytoplasm requires interaction with importinα (Impα) of the Impα/β complex. The NLS endows a protein with a binding motif that is recognized by Impα (32). Consistent with SH2B1β being actively transported into the nucleus via the Impα/β complex, it was unable to translocate into the nucleus of energy-depleted PC12 cells.
Our findings concerning the nuclear localization of SH2B1β are consistent with an increasing amount of recent experimental evidence that signaling molecules, including adapter proteins, actively undergo dynamic translocations and reversible bindings to bring about specific and successful signal transmission (44). Along with SH2B1β, other previously characterized cytoplasmic adapter proteins have been reported to translocate to the nucleus, including insulin receptor substrate-1 (IRS-1) (45,46), Grb4 (Nck2), and APPL (adapter protein containing a PH domain, a phosphotyrosine binding domain, and leucine zipper motif) (47). Together with SH2B1β, these proteins challenge the dogma surrounding the functional roles of cytoplasmic adapter proteins and exemplify a major departure from the classical hardwired signaling concept wherein receptors and their corresponding signaling/adapter proteins stay more or less in place, whereas secondary messengers are actively translocated to the nucleus.
The identification of an NLS in SH2B1 enabled us to start to investigate the function of nuclear SH2B1. Previous findings indicated that the ability of SH2B1β to enhance NGF-dependent neurite outgrowth is lost when the NES is mutated, which suggested that SH2B1β is required either in the cytoplasm or at the plasma membrane to enhance differentiation (28). However, we show here that mutation of the NLS of SH2B1 also results in a loss in SH2B1’s ability to enhance NGF-induced neuronal differentiation. Together these findings suggest that SH2B1 needs access to both extranuclear and nuclear compartments to facilitate NGF-induced neurite outgrowth. This is further supported by our finding that NGF-dependent neurite outgrowth is enhanced by GFP-SH2B1β(9YF) (data not shown), which, like WT SH2B1β, undergoes nuclear-cytoplasmic shuttling but is blocked by SH2B1β(R555E), which appears unable to shuttle.
Mutation of the NLS also prevented SH2B1 from enhancing the NGF-induced transcription of the NGF-responsive genes uPAR, MMP3, and MMP10. These three genes are among those that displayed the most dramatic enhancement among the subset of NGF-responsive genes whose NGF-induced expression was found by microarray analysis to be enhanced by the presence of SH2B1 (23). The protein products of all three of these genes fall within the canonical pathway responsible for extracellular matrix degradation, required during neurite outgrowth. Farias-Eisner et al. (41) demonstrated that NGF-induced transcription of uPAR, in particular, is an essential event for NGF-dependent differentiation of PC12 cells. Both antisense oligonucleotide-mediated knockdown of uPAR transcript as well as functional inhibition of uPAR using an anti-uPAR antibody, blocks NGF-dependent PC12 neurite outgrowth (41). uPAR is an immediate-early gene in neurotrophin-driven neuronal differentiation (41). In PC12 cells, uPAR transcription is specifically induced by NGF, which promotes differentiation, but not epidermal growth factor, which does not promote differentiation (41). Blocking uPAR function inhibited the transcription of a number of NGF-induced secondary response genes including Mmp3 (stromelysin-1; transin-1) (41). The ability of uPAR to initiate a secondary wave of transcription important for differentiation is thought to arise from its capacity to activate intracellular signaling machinery, even though it lacks a cytosolic domain (48). The critical role that SH2B1β plays in mediating or facilitating NGF-induced expression of the genes encoding uPAR, MMP3, and MMP10 is further supported by our finding (23) that even an incomplete reduction in levels of endogenous SH2B1 using shRNA to SH2B1 result in a substantial inhibition of their expression in response to NGF.
Our finding that mutation of the NLS or NES abrogates (NLS) or lessens (NES) the ability of SH2B1β to enhance NGF-induced transcription of uPAR, MMP3, and MMP10 supports the notion that nucleocytoplasmic shuttling of SH2B1β is required for that enhancement. Stable expression of the mutants SH2B1β(mNLS) (Fig. 8) or SH2B1β(ΔNES) (28) did not result in appreciable differences in the NGF-mediated phosphorylation of TrkA Y490, ERKs 1/2, or AKT when compared with the stable expression of SH2B1β(WT). Therefore, we believe that the decreased ability of these mutants, compared with WT SH2B1β, to enhance NGF-induced neurite outgrowth and transcription of NGF-target genes is specific to the inability of SH2B1β to enter and/or exit the nucleus, and not a consequence of their compromising an NGF/TrkA signaling cascade, at least not those involving Akt or ERKs 1 and 2. The precise mechanism by which nuclear SH2B1β facilitates the NGF-dependent induction of specific genes is unknown. However, in a previous study, overexpression of SH2B1β(WT) enhanced the nuclear translocation of FoxO1 to the cytoplasm in response to NGF (18), which raises the possibility that SH2B1β could shuttle activators or repressors of transcription into or out of the nucleus. Interestingly, several previously identified binding partners of SH2B1, including Grb2, Rac1, IRS-1, and JAK2, have each been found to localize and function within the nucleus (45,49,50,51). It remains to be seen whether the aforementioned proteins require interaction with SH2B1 for their nuclear localization and/or function. Alternatively, one could envision SH2B1β serving as a coactivator that recruits specific transcriptional regulators to the promoter of NGF-responsive genes.
The balance between nuclear export and import of proteins such as SH2B1 that contain both an NLS and NES is often subjected to regulation, with regulation via phosphorylation being particularly well documented (52,53). SH2B1 is phosphorylated on serines/threonines in its basal (serum deprivation) state. Furthermore, NGF stimulates phosphorylation of SH2B1 on both tyrosines and serines/threonines. Phosphorylation on tyrosines is presumably by TrkA, whereas NGF-induced phosphorylation on serines/threonines may be carried out by NGF-activated downstream kinases such as ERK1/2 (15). However, in contrast to what we predicted, our data suggest that NGF does not have a major impact on the extranuclear/nuclear distribution of SH2B1β. Although it is possible NGF-induced phosphorylation affects similarly the rate of nuclear import and rate of export, resulting in no detectable change in the extranuclear/nuclear distribution of SH2B1β, we think it more likely that nucleocytoplasmic shuttling of SH2B1 is constitutive and not regulated in a major way by NGF. Consistent with a lack of effect of NGF on nuclear accumulation, SH2B1β lacking all tyrosines is still capable of nuclear import (Fig. 4B). These results suggest that if phosphorylation affects the overall extranuclear/nuclear distribution of SH2B1β, it must occur under basal (non-NGF) conditions and involve serines or threonines rather than a tyrosine. The amino acid sequence surrounding the NLS and NES in SH2B1 makes the NLS a more likely target than the NES for posttranslational phosphorylation due to the presence of multiple flanking putative Ser/Thr phosphorylation sites. It is also possible that phosphorylation of SH2B1 even on Ser/Thr has no impact on the rate of nucleocytoplasmic shuttling. Rather, the phosphorylation of extranuclear SH2B1 that occurs in the presence and/or absence of NGF may alter the proteins that can bind to SH2B1, thereby altering the ability of these proteins to undergo nucleocytoplasmic shuttling or to contribute to a transcriptionally active complex.
The continuous compartmental cycling of SH2B1β we observe even in the absence of NGF is consistent with an emerging theme for a subset of signaling molecules. In contrast to conventional thought that signaling molecules translocate to the nucleus only after their activation by a particular signal, the latent unphosphorylated and inactive signal transducer and activator of transcription 2 (STAT2) is now thought to undergo constant shuttling between the cytoplasm and nucleus (54). Similarly, what was once believed to be a TGF-β-mediated translocation from the cytoplasm to the nucleus of the receptor-regulated Smad (R-Smad) family of proteins has recently been shown to be a continuous cycling process independent of TGF-β stimulation (55,56). A TGF-β-dependent phosphorylation of the R-Smads allows a nuclear R-Smad/Smad4 complex to persist, giving rise to a prolonged and observable nuclear presence (55). Conversely, functional NES have been described for several transcription factors originally thought to localize only in the nucleus. For transcriptional regulators Oct-6, Sox9, and Sox10, disabling their nuclear export reduced their transactivation abilities (57,58,59). It is not clear as to what access to both the cytoplasm/plasma membrane and nucleus affords these latter transcription factors.
The finding that SH2B1β(R555E) does not undergo cytoplasmic-nuclear shuttling was intriguing. SH2B1β(R555E) was previously hypothesized to block neurite outgrowth by competing with endogenous SH2B1β for downstream effectors and sequestering these putative effectors in an inactive state (1). The finding that SH2B1β(R555E) is unable to enter the nucleus provides an alternative or additional explanation for how SH2B1β(R555E) might be acting as a dominant negative. SH2B1β(R555E) may prevent one or more or its binding partners from entering the nucleus, where they function to regulate transcription directly or indirectly. Why an intact SH2 domain is required for nuclear localization of SH2B1 is not known. We think it unlikely that the SH2 domain is directly required for nuclear import. Rather, we favor the hypothesis that the SH2 domain is required for SH2B1 to bind to some protein (e.g. receptor tyrosine kinase) that positions it and/or allows it to be modified appropriately to bind importins.
Based upon the degree of sequence similarity between SH2B1 and SH2B2, we were somewhat surprised to find that SH2B2 did not also cycle between the cytoplasm and nucleus. After the initial identification of SH2B1 and SH2B2 in neurons (2), early conclusions drawn from their characterization indicated a large amount of functional overlap between the two proteins. SH2B1 and SH2B2 were both shown to associate with activated TrkA and enhance NGF-dependent neuritogenesis (2,15). However, functional differences between SH2B1 and SH2B2 have been observed, including phosphotyrosine binding site specificity (60) and unique binding partners [e.g. enigma for SH2B2 but not SH2B1 or SH2B3 (61); c-cbl for SH2B2 and SH2B3 but only the SH2B1α isoform of SH2B1 (60,62,63)]. The ability of SH2B1, but not SH2B2, to cycle between the nucleus and cytoplasm represents another distinction between the two family members. Consistent with this difference, preliminary studies suggest that although overexpression of SH2B2 causes morphological changes in PC12 cells, those changes do not mimic the SH2B1-induced enhancement of NGF-dependent neurite outgrowth.
In summary, we have demonstrated that the adaptor protein SH2B1β is required for NGF-mediated differentiation of PC12 cells in a manner distinct from that of facilitating the activation of ERKs 1 and 2. We identified a functional NLS in SH2B1 that allowed us for the first time to examine the nuclear role of SH2B1. Blocking SH2B1β nuclear localization by mutating the NLS prevented SH2B1 from enhancing both NGF-dependent neurite outgrowth and transcription of the NGF-responsive genes uPAR, Mmp3, and Mmp10. Similarly, blocking nuclear export of SH2B1 by mutation of the NES transcription significantly inhibited the ability of SH2B1β to enhance expression of these genes. These findings suggest that SH2B1β requires access to both the nucleus and cytoplasm to facilitate NGF-mediated morphological and biochemical changes, raising the possibility that SH2B1β directly facilitates transcription of genes required for differentiation. It also raises the possibility that nucleocytoplasmic shuttling of SH2B1β will be important for its actions in the context of the many other receptors, including those for insulin, IGF-I, leptin, and GH, that use SH2B1 as a signaling protein.
Materials and Methods
Antibodies and reagents
Polyclonal antibody to rat SH2B1 (αSH2B1), kind gift of Dr. Liangyou Rui (University of Michigan), was raised against an SH2B1β glutathione S-transferase fusion protein and used at a dilution of 1:1000 for Western blotting (7). Antibodies that recognize the following proteins were used for Western blotting at a dilution of 1:1000: phospho-44/42 MAPK that recognizes both ERK1 and ERK2 that are doubly phosphorylated on T202/Y204 (αpERK; E10), total Erk (αERK), and phospho-Akt (Ser473) (αpAkt-Ser473) from Cell Signaling Technology (Beverly, MA) and phospho-TrkA (Tyr 490) [αpTrkA(Tyr490)] (catalog no. T9691) from Sigma-Aldrich (St. Louis, MO). IRDye800-conjugated, affinity-purified anti-GFP (Rockland Immunochemicals Inc., Gilbertsville, PA) was used at a dilution of 1:20,000 to visualize GFP. IRDye 800- and IRDye 700-conjugated affinity-purified antimouse IgG and antirabbit IgG (Rockland) and Alexa Fluor 680-conjugated anti-rabbit IgG (Invitrogen, Carlsbad, CA) were used at a dilution of 1:20,000. Ascites containing antimouse myc monoclonal antibody, produced by the Michigan Diabetes Research and Training Center Hybridoma Core, was used for immunostaining (dilution of 1:1000). Anti-myc staining was visualized using Alexa Fluor 594-conjugated antimouse IgG (Invitrogen). NGF and rat-tail collagen I were purchased from BD Bioscience (San Diego, CA). Fluorescein was from Bio-Rad (Hercules, CA) and SYBR Green I was from Sigma-Aldrich Chemical Co. (St. Louis, MO). TaqMan RT-PCR kit (catalog item N808-0234) was purchased from Applied Biosystems (Roche, Indianapolis, IN).
Plasmids
All SH2B1β cDNAs were subcloned into pEGFP C1 (Clontech Laboratories, Inc., Palo Alto, CA). cDNAs encoding GFP-tagged SH2B1β, SH2B1β (198–268), and SH2B1β(R555E) have been described previously (15,28). GFP-SH2B1β (147–198) was created from the GFP-SH2B1β(WT) parental plasmid using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Two BamH1 restriction sites (at 147 and 198) were put into SH2B1β(WT) cDNA, flanking the desired region to be deleted. The construct was digested with BamH1, and the resulting insert was omitted during religation. The primers (sense strand, mutations in lowercase) used for generating the deletion in GFP-SH2B1β (147–198) were 5′-CAACGTCCTCAAAGCCGggatcCAAGAAACGCTTCTCCC-3′ for site 147 and 5′-GTTCTAGGTGGAAACggatcCTCCAACTCCTCTGGTGGT for site 198. GFP-SH2B1β (147–268) was created in a manner similar to the previously mentioned GFP-SH2B1β (147–198). The primer (sense strand, mutations in lowercase) used to generate the BamH1 mutation at site 268 was 5′-CCTGACCCAGCAGGAtccGGTCGTGGAGGAGGG-3′. Critical basic residues K148, K150, K151, and R152 were mutated to alanines in the mutant cDNA encoding GFP-SH2B1β(mNLS), which was created from the GFP-SH2B1β(WT) expression vector using site-directed mutagenesis. The primer (sense strand, mutations in lowercase) used for GFP-SH2B1β(mNLS) was 5′-CGTCCTCAAAGCCGgcGCTCgcGgcAgcCTTCTCCCTCCGC-3′. cDNA encoding GFP-SH2B1β(mNLS+ΔNES) was similarly created through site-directed mutagenesis using GFP-SH2-Bβ (198-268) and the primers above. The primer used for the creation of the GFP-SH2B1β(res) (sense strand, silent mutations to the shSH2B1 target sequence are in lowercase) was 5′-TGCCGGGTCCAACAcCTtTGGTTtCAGTCCATTTTCG-3′. cDNA encoding GFP-SH2B1β(9Y-F), in which all nine tyrosines within SH2B1β were changed to phenylalanines, was created using GeneEditor in vitro Site-Directed Mutagenesis System (Promega, Madison, WI). The following primers were used to introduce the nine phenylalanine substitutions:Y48F (5′-CGTTTTCGCCTCTtTCTGGCCTCCCACCC-3′), Y55F (5′-CCCACCCACAATtTGCAGAGCCGGGAGC-3′), Y354F (5′-GGTAGAAGGCCCTtCAGAGTTCATCCTGGAGACAACTG-3′), Y439F (5′-GTCGCAGGGAGCTTtTGGAGGCCTCTCAGACC-3′), Y494F (5′-CCCCTCTCTACCCCGTtCCCTCCCCTGGATAC-3′), Y525F (5′-CCCCTCTCAGGCTtCCCTTGGTTCCACGGC-3′), Y564F (5′-GACGTGGTGAATtTGTCCTCACTTTCAACTTCC-3′), Y624F (5′-CCTTGTCAGCTtTGTGCCCTCCCAGCGG-3′), and Y649F (5′-CGACCGATGCTtCCCCGATGCTTCTTCC-3′). The cDNA encoding myc-tagged rat SH2B2 was kindly provided by Dr. David D. Ginty (Johns Hopkins University). To create myc-SH2B2(ΔNES), two HindIII sites were introduced into myc-SH2B2(WT) at amino acid positions 167 and 184 using site-directed mutagenesis. Primers used to introduce HindIII sites were 5′-GAGCCTCGTGACAAgcttACGCGACGTCTG-3′ and 5′-GCAGCCAAAGTGaAGcTtGTGGACATCCAGCGC-3′, respectively. The resulting myc-SH2B2 mutant contained two HindIII sites flanking the putative NES. The construct was digested with HindIII restriction enzyme (New England Biolabs, Boston, MA), purified, and then religated. The four residues within the putative NLS of myc-SH2B2 were mutated to alanines, using site-directed mutagenesis from Stratagene. The primer used to produce myc-SH2B2(mNLS) was 5′-CACGTGGCTACCgcGGCCgcTGTCgcCgcAGGCTTCTCACTG-3′. All mutations were verified by DNA sequencing.
Stable cell lines and cell culture
The parental PC12 cells were obtained from American Type Culture Collection (Rockville, MD). Pools of PC12 cells stably expressing GFP-SH2B1β(mNLS) and GFP-SH2B1β(R555E) were made as described previously (28). The pooled PC12 cell lines stably expressing GFP, GFP-SH2B1β(WT), GFP-SH2B1β(R555E), and GFP-SH2B1β(NLS) were cultured as described previously (28). PC12 cells were plated on collagen-coated plates (0.1 mg/ml rat tail collagen in 0.02 n acetic acid) and grown at 37 C in 10% CO2 in normal growth medium consisting of DMEM (Invitrogen) containing 10% heat-inactivated horse serum (ICN Biomedicals, Costa Mesa, CA), 5% fetal bovine serum (Invitrogen), 1 mm l-glutamine, and 1 mm antibiotic-antimycotic (Invitrogen). PC12 cells were incubated for 4 h in normal growth medium supplemented with 20 nm leptomycin B (Invitrogen) to inhibit Crm1-dependent nuclear export. For the energy depletion studies, PC12 cells were washed with PBS and incubated in normal growth medium or serum and glucose-free DME (Invitrogen) supplemented with 10 mm sodium azide and 10 mm 2-deoxy-d-glucose. The PC12 cells were incubated in the normal growth medium or energy depletion medium for 1 h before 4 h LMB treatment.
Silencing of SH2B1 gene
SH2B1 siRNA vector was constructed by inserting an oligonucleotide containing the SH2B1 sequence (5′-CATCTGTGGTTCCAGTCCA-3′) corresponding to nucleotides 1771–1789 of rat SH2B1 (GenBank accession no. AF047577) into the pSuper retro vector containing the puromycin resistance gene (pSuper retro puro) (OligoEngine). A pSuper vector containing a nontargeting siRNA with a low sequence similarity to known genes was used as a control. The SH2B1 and control shRNA vectors were transfected into subconfluent PC12 cells using a Bio-Rad Gene Pulser Xcell electroporator (400 V, 500 μF, 0.4 cm cuvette). After 14 h, cells were washed with 1× PBS, pH 7.4, and fresh growth medium was added. Twenty-four hours later, growth medium containing 5 μg/ml puromycin was added to the cells. Selection for pSuper-positive PC12 cells was carried out for 30 d. In the SH2B1β rescue experiment, the shSH2B1 PC12 cells were transfected with either cDNA encoding GFP or GFP-SH2B1β(res) (a construct containing silent mutations within the shRNA target sequence). The cells were incubated in serum-free and puromycin-free medium for 14 h and then treated with 50 ng/ml NGF for 2 d. Neurite outgrowth was assessed in GFP-positive cells and expressed as percent differentiation relative to shControl+GFP cells.
Differentiation of PC12 cells
PC12 cells were plated on six-well, collagen-coated plates. The cells were grown in differentiation medium (DMEM containing 2% horse serum, 1% fetal bovine serum) and 50 or 100 ng/ml NGF was added. The NGF-containing medium was replaced every 2 d until cellular differentiation was assessed. Cells with neurite length at least twice the diameter of the cell body were scored as differentiated cells. The percentage of differentiated cells was determined by dividing the number of morphologically differentiated cells by the total number of cells counted.
Immunolocalization
PC12 cells were transfected using a Gene Pulser Xcell electroporator (400V, 500 μF) (Bio-Rad) in a 0.4-cm cuvette. After 5 h, cells were washed with PBS, and fresh growth medium was added. Cells were incubated for 24 h before they were fixed with 4% paraformaldehyde. Coverslips were mounted onto slides with Prolong (Molecular Probes, Eugene, OR). Cells were incubated with 2 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) for 10 min to visualize nuclei. The subcellular distribution of the various GFP-SH2B1β proteins was determined by GFP fluorescence. The cells were visualized by fluorescence microscopy (Nikon Eclipse TE200) with either a ×20 or ×40 objectives. Images were captured using a CoolSnap HQ digital camera (Roper Scientific, Trenton, NJ) and viewed using MetaVue imaging software (Molecular Devices, Sunnyvale, CA).
RNA preparation and QT-PCR
Total RNA was isolated from control and NGF-treated PC12 cells using Stat60 (Tel-Test, Inc., Friendswood, TX) according to the manufacturer’s instructions. RNA from four experiments was prepared, and the quality of each RNA was checked by OD. cDNA was generated from each RNA preparation using the One-Step RT-PCR Kit with SYBR Green (Bio-Rad; catalog item 170-8892) according to the manufacturer’s instructions. NGF-induced gene expression of uPAR, Mmp3, and Mmp10 was assessed by QT-PCR using SYBR green I and the iCycler system with iCycler iQ Real Time Detection System software (Bio-Rad). Primer sequences were designed using PrimerExpress software (as described in Ref. 23). Amplicons generated from each primer pair were 50–52 bp. The PCR well volume of each sample was normalized with fluorescein. All readings were normalized to the expression of GAPDH. Results are shown as a ratio of gene expression in NGF-treated cells to gene expression in control (no NGF) cells.
Protein preparation and immunoblotting
For whole-cell extract preparation, cells were washed three times with chilled PBSV [10 mm sodium phosphate, 137 mm NaCl, 1 mm Na3VO4 (pH 7.4)] and solubilized in lysis buffer [50 mm Tris, 0.1% Triton X-100, 150 mm NaCl, 2 mm EGTA, 1 mm Na3VO4 (pH 7.5)] containing 1 mm phenylmethylsulfonyl fluoride, 10 mg/ml aprotinin, 10 mg/ml leupeptin, and 25 mm NaF. The solubilized material was centrifuged at 16,750 × g at 4 C for 10 min. Nuclear and nonnuclear extracts were prepared in the following manner. The cells are washed twice in ice-cold PBS and then resuspended in buffer A [10 mm HEPES, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, 1 mm Na3VO4, 10 mg/ml aprotinin, 10 mg/ml leupeptin (pH 7.9)]. After a 15-min incubation on ice, 25 μl buffer B (buffer A with 10% Nonidet P-40) was added, and the cells were vortexed on high for 10 sec. The cells were then pelleted by centrifugation (16,750 × g at 4 C for 30 sec). The cytoplasmic extract (supernatant) was removed and saved, and the pellet was washed twice with buffer A before resuspending in 50 μl buffer C [20 mm HEPES, 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, 1 mm Na3VO4, 10 mg/ml aprotinin, 10 mg/ml leupeptin (pH 7.9)]. The nuclear pellet was sonicated three times (15-sec intervals at level 4) using a Misonix 3000 (Misonix, Inc., Farmingdale, NY) sonicator to reduce the sample viscosity. Protein concentrations of both nuclear and cytoplasmic extracts were determined using Bradford analysis (Sigma) and 25 μg of each were loaded on the SDS-PAGE gels. The lysates from both whole-cell and nuclear/cytoplasmic extractions were boiled for 5 min in a mixture (80:20) of lysis buffer and SDS-PAGE sample buffer [250 mm Tris-HCl, 10% SDS, 10% β-mercaptoethanol, 40% glycerol, 0.01% bromophenol blue (pH 6.8)]. Equal amounts of the solubilized proteins were separated on SDS-PAGE gels, transferred to nitrocellulose, immunoblotted with the indicated antibody, and detected using an Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). Immunoblots were quantified using LiCor Odyssey 2.0 software and normalized to the levels of α-tubulin (Fig. 1) or total ERK (Fig. 7).
Acknowledgments
We thank Dr. Liangyou Rui for his generous gift of the SH2B1 antibody. We appreciate the immunoblotting advice of Dr. Lawrence S. Argetsinger, Dr. Hui Jin, and Nathan J. Lanning. We thank Barbara Hawkins for her help with this manuscript.
Footnotes
This work was supported in part by National Institutes of Health (NIH) Grant RO1-DK54222. The fluorescence cell sorting and DNA sequencing was performed with financial support from the University of Michigan Comprehensive Cancer Center (P30-CA46592). The α-myc antibody was prepared by the Hybridoma Center funded by the Michigan Diabetes Research and Training Center (NIH-5P60-DK20572 from the National Institute of Diabetes and Digestive and Kidney Diseases). T.J.M. was supported by Predoctoral Traineeship in Cellular and Molecular Biology GM-07315 from the NIH, a Cancer Biology Predoctoral Fellowship from the University of Michigan, and a Rackham Predoctoral Fellowship from the University of Michigan.
Current address for L.C.: Institute of Molecular Medicine, National Tsing Hua University, Hsinchu, Taiwan.
Disclosure Summary: The authors have nothing to disclose.
First Published Online April 16, 2009
Abbreviations: DD, Dimerization domain; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GDNF, glial cell line-derived neurotrophic factor; GFP, green fluorescent protein; Impα, importinα; JAK, Janus kinase; LMB, leptomycin B; MMP, matrix metalloproteinase; NES, nuclear export sequence; NGF, nerve growth factor; NLS, nuclear localization signal; PH, pleckstrin homology; PLC, phospholipase C; QT-PCR, real-time quantitative PCR; SH2, Src homology; uPAR, urokinase plasminogen activator receptor; WT, wild type.
References
- Rui L, Herrington J, Carter-Su C 1999 SH2-B, a membrane-associated adapter, is phosphorylated on multiple serines/threonines in response to nerve growth factor by kinases within the MEK/ERK cascade. J Biol Chem 274:26485–26492 [DOI] [PubMed] [Google Scholar]
- Qian X, Riccio A, Zhang Y, Ginty DD 1998 Identification and characterization of novel substrates of Trk receptors in developing neurons. Neuron 21:1017–1029 [DOI] [PubMed] [Google Scholar]
- Riedel H, Wang J, Hansen H, Yousaf N 1997 PSM, an insulin-dependent, pro-rich, PH, SH2 domain containing partner of the insulin receptor. J Biochem 122:1105–1113 [DOI] [PubMed] [Google Scholar]
- Wang J, Riedel H 1998 Insulin-like growth factor-I receptor and insulin receptor association with a Src homology-2 domain-containing putative adapter. J Biol Chem 273:3136–3139 [DOI] [PubMed] [Google Scholar]
- Rui L, Mathews LS, Hotta K, Gustafson TA, Carter-Su C 1997 Identification of SH2-Bβ as a substrate of the tyrosine kinase JAK2 involved in growth hormone signaling. Mol Cell Biol 17:6633–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzer JH, Argetsinger LS, Zhou YJ, Kouadio JL, O'Shea JJ, Carter-Su C 2004 Tyrosine 813 is a site of JAK2 autophosphorylation critical for activation of JAK2 by SH2-Bb. Mol Cell Biol 24:4557–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C, Li M, Rui L 2004 SH2-B promotes insulin receptor substrate 1 (IRS1)- and IRS2-mediated activation of the phosphatidylinositol 3-kinase pathway in response to leptin. J Biol Chem 279:43684–43691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Li M, Duan C, Rui L 2005 Identification of SH2-B as a key regulator of leptin sensitivity, energy balance and body weight in mice. Cell Metab 2:95–104 [DOI] [PubMed] [Google Scholar]
- Yousaf N, Deng Y, Kang Y, Riedel H 2001 Four PSM/SH2-B alternative splice variants and their differential roles in mitogenesis. J Biol Chem 276:40940–40948 [DOI] [PubMed] [Google Scholar]
- Yokouchi M, Suzuki R, Masuhara M, Komiya S, Inoue A, Yoshimura A 1997 Cloning and characterization of APS, an adaptor molecule containing PH and SH2 domains that is tyrosine phosphorylated upon B-cell receptor stimulation. Oncogene 15:7–15 [DOI] [PubMed] [Google Scholar]
- Iseki M, Takaki S, Takatsu K 2000 Molecular cloning of the mouse APS as a member of the Lnk family adaptor proteins. Biochem Biophys Res Commun 272:45–54 [DOI] [PubMed] [Google Scholar]
- Huang X, Li Y, Tanaka K, Moore KG, Hayashi JI 1995 Cloning and characterization of Lnk, a signal transduction protein that links T-cell receptor activation signal to phospholipase C g 1, Grb2, and phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 92:11618–11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maures TJ, Kurzer JH, Carter-Su C 2007 SH2B1 (SH2-B) and JAK2: a multifunctional adaptor protein and kinase made for each other. Trends Endocrinol Metab 18:38–45 [DOI] [PubMed] [Google Scholar]
- Nelms K, O'Neill TJ, Li S, Hubbard SR, Gustafson TA, Paul WE 1999 Alternative splicing, gene localization, and binding of SH2-B to the insulin receptor kinase domain. Mamm Genome 10:1160–1167 [DOI] [PubMed] [Google Scholar]
- Rui L, Herrington J, Carter-Su C 1999 SH2-B is required for nerve growth factor-induced neuronal differentiation. J Biol Chem 274:10590–10594 [DOI] [PubMed] [Google Scholar]
- Qian X, Ginty DD 2001 SH2-B and APS are multimeric adapters that augment TrkA signaling. Mol Cell Biol 21:1613–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ 1995 Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80:179–185 [DOI] [PubMed] [Google Scholar]
- Wang X, Chen L, Maures TJ, Herrington J, Carter-Su C 2004 SH2-B is a positive regulator of nerve growth factor-mediated activation of the Akt/Forkhead pathway in PC12 cells. J Biol Chem 279:133–141 [DOI] [PubMed] [Google Scholar]
- Greene LA, Tischler AS 1976 Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA 73:2424–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Lazarovici P, Guroff G 1989 Regulation of the differentiation of PC12 pheochromocytoma cells. Environ Health Perspect 80:127–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu B, Que H, Lin Q, Lv S, Liu S 2006 Neurons derived from PC12 cells have the potential to develop synapses with primary neurons from rat cortex. Acta Neurobiol Exp (Wars) 66:105–112 [DOI] [PubMed] [Google Scholar]
- Vaudry D, Stork PJ, Lazarovici P, Eiden LE 2002 Signaling pathways for PC12 cell differentiation: making the right connections. Science 296:1648–1649 [DOI] [PubMed] [Google Scholar]
- Chen L, Maures TJ, Jin H, Huo JS, Rabbani SA, Schwartz J, Carter-Su C 2008 SH2B1β (SH2-Bβ) enhances expression of a subset of nerve growth factor-regulated genes important for neuronal differentiation including genes encoding uPAR and MMP3/10. Mol Endocrinol 22:454–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RM, Loeb DM, Copeland TD, Pawson T, Greene LA, Kaplan DR 1994 Trk receptors use redundant signal transduction pathways involving SHC and PLC-γ 1 to mediate NGF responses. Neuron 12:691–705 [DOI] [PubMed] [Google Scholar]
- Pang L, Sawada T, Decker SJ, Saltiel AR 1995 Inhibition of MAP kinase kinase blocks the differentiation of PC-12 cells induced by nerve growth factor. J Biol Chem 270:13585–13588 [DOI] [PubMed] [Google Scholar]
- Cowley S, Paterson H, Kemp P, Marshall CJ 1994 Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell 77:841–852 [DOI] [PubMed] [Google Scholar]
- Meier R, Alessi DR, Cron P, Andjelkoviæ M, Hemmings BA 1997 Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bβ. J Biol Chem 272:30491–30497 [DOI] [PubMed] [Google Scholar]
- Chen L, Carter-Su C 2004 Adapter protein SH2-Bβ undergoes nucleocytoplasmic shuttling: implications for nerve growth factor induction of neuronal differentiation. Mol Cell Biol 24:3633–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR 1996 Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol 16:4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, Frangioni JV, Dalal SN, DeCaprio JA, Greenberg ME, Yaffe MB 2002 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol 156:817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KM, McDonald C, Reich NC 2000 Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J 19:6196–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U 1999 Transport between the cell nucleus and the cytoplasm. Ann Rev Cell Dev Biol 15:607–660 [DOI] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF, Smith AE 1984 Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33–38 [DOI] [PubMed] [Google Scholar]
- Lanford RE, Butel JS 1984 Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell 37:801–813 [DOI] [PubMed] [Google Scholar]
- Rui L, Carter-Su C 1998 Platelet-derived growth factor (PDGF) stimulates the association of SH2-Bβ with PDGF receptor and phosphorylation of SH2-Bβ. J Biol Chem 273:21239–21245 [DOI] [PubMed] [Google Scholar]
- Schwoebel ED, Ho TH, Moore MS 2002 The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J Cell Biol 157:963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marg A, Shan Y, Meyer T, Meissner T, Brandenburg M, Vinkemeier U 2004 Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J Cell Biol 165:823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokouchi M, Wakioka T, Sakamoto H, Yasukawa H, Ohtsuka S, Sasaki A, Ohtsubo M, Valius M, Inoue A, Komiya S, Yoshimura A 1999 APS, an adaptor protein containing PH and SH2 domains, is associated with the PDGF receptor and c-Cbl and inhibits PDGF-induced mitogenesis. Oncogene 18:759–767 [DOI] [PubMed] [Google Scholar]
- Liu J, Kimura A, Baumann CA, Saltiel AR 2002 APS facilitates c-Cbl tyrosine phosphorylation and GLUT4 translocation in response to insulin in 3T3-L1 adipocytes. Mol Cell Biol 22:3599–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias-Eisner R, Vician L, Reddy S, Basconcillo R, Rabbani SA, Wu YY, Bradshaw RA, Herschman HR 2001 Expression of the urokinase plasminogen activator receptor is transiently required during “priming” of PC12 cells in nerve growth factor-directed cellular differentiation. J Neurosci Res 63:341–346 [DOI] [PubMed] [Google Scholar]
- Farias-Eisner R, Vician L, Silver A, Reddy S, Rabbani SA, Herschman HR 2000 The urokinase plasminogen activator receptor (UPAR) is preferentially induced by nerve growth factor in PC12 pheochromocytoma cells and is required for NGF-driven differentiation. J Neurosci 20:230–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom LA, Lochner J, Yeung W, Ciment G 1995 The metalloproteinase stromelysin-1 (transin) mediates PC12 cell growth cone invasiveness through basal laminae. Mol Cell Neurosci 6:56–68 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhu W, Wang YG, Liu XJ, Jiao L, Liu X, Zhang ZH, Lu CL, He C 2006 Interaction of SH2-Bβ with RET is involved in signaling of GDNF-induced neurite outgrowth. J Cell Sci 119:1666–1676 [DOI] [PubMed] [Google Scholar]
- Massie C, Mills IG 2006 The developing role of receptors and adaptors. Nat Rev Cancer 6:403–409 [DOI] [PubMed] [Google Scholar]
- Tu X, Batta P, Innocent N, Prisco M, Casaburi I, Belletti B, Baserga R 2002 Nuclear translocation of insulin receptor substrate-1 by oncogenes and Igf-I. Effect on ribosomal RNA synthesis. J Biol Chem 277:44357–44365 [DOI] [PubMed] [Google Scholar]
- Wu A, Tu X, Prisco M, Baserga R 2005 Regulation of upstream binding factor 1 activity by insulin-like growth factor I receptor signaling. J Biol Chem 280:2863–2872 [DOI] [PubMed] [Google Scholar]
- Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, Wilm M, Parton RG, Zerial M 2004 APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 116:445–456 [DOI] [PubMed] [Google Scholar]
- Blasi F, Carmeliet P 2002 uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol 3:932–943 [DOI] [PubMed] [Google Scholar]
- Romero F, Ramos-Morales F, Domínguez A, Rios RM, Schweighoffer F, Tocqué B, Pintor-Toro JA, Fischer S, Tortolero M 1998 Grb2 and its apoptotic isoform Grb3-3 associate with heterogeneous nuclear ribonucleoprotein C, and these interactions are modulated by poly(U) RNA. J Biol Chem 273:7776–7781 [DOI] [PubMed] [Google Scholar]
- Michaelson D, Silletti J, Murphy G, D'Eustachio P, Rush M, Philips MR 2001 Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol 152:111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Bjursell G, Kannius-Janson M 2006 Nuclear Jak2 and transcription factor NF1-C2: a novel mechanism of prolactin signaling in mammary epithelial cells. Mol Cell Biol 26:5663–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez D, Montserrat-Sentís B, Virgós-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J, Francí C, García de Herreros A 2003 Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol Cell Biol 23:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN 2001 Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol 21:6312–6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banninger G, Reich NC 2004 STAT2 nuclear trafficking. J Biol Chem 279:39199–39206 [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolás FJ, Hill CS 2002 Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol Cell 10:283–294 [DOI] [PubMed] [Google Scholar]
- Xu L, Kang Y, CölS, Massagué J 2002 Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol Cell 10:271–282 [DOI] [PubMed] [Google Scholar]
- Baranek C, Sock E, Wegner M 2005 The POU protein Oct-6 is a nucleocytoplasmic shuttling protein. Nucleic Acids Res 33:6277–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasca S, Canizares J, De Santa Barbara P, Mejean C, Poulat F, Berta P, Boizet-Bonhoure B 2002 A nuclear export signal within the high mobility group domain regulates the nucleocytoplasmic translocation of SOX9 during sexual determination. Proc Nat Acad Sci USA 99:11199–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehberg S, Lischka P, Glaser G, Stamminger T, Wegner M, Rosorius O 2002 Sox10 is an active nucleocytoplasmic shuttle protein, and shuttling is crucial for Sox10-mediated transactivation. Mol Cell Biol 22:5826–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Hubbard SR 2005 Structural characterization of a novel Cbl phosphotyrosine recognition motif in the APS family of adapter proteins. J Biol Chem 280:18943–18949 [DOI] [PubMed] [Google Scholar]
- Barrès R, Gonzalez T, Le Marchand-Brustel Y, Tanti JF 2005 The interaction between the adaptor protein APS and Enigma is involved in actin organisation. Exp Cell Res 308:334–344 [DOI] [PubMed] [Google Scholar]
- Wakioka T, Sasaki A, Mitsui K, Yokouchi M, Inoue A, Komiya S, Yoshimura A 1999 APS, an adaptor protein containing Pleckstrin homology (PH) and Src homology-2 (SH2) domains inhibits the JAK-STAT pathway in collaboration with c-Cbl. Leukemia 13:760–767 [DOI] [PubMed] [Google Scholar]
- Ahmed Z, Smith BJ, Pillay TS 2000 The APS adapter protein couples the insulin receptor to the phosphorylation of c-Cbl and facilitates ligand-stimulated ubiquitination of the insulin receptor. FEBS Lett 475:31–34 [DOI] [PubMed] [Google Scholar]