Abstract
Background
Only a small number of Pseudomonas putida strains possess the typical N-acyl homoserine lactone quorum sensing system (AHL QS) that consists of a modular LuxR family protein and its cognate LuxI homolog that produces the AHL signal. Moreover, AHL QS systems in P. putida strains are diverse in the type of AHLs they produce and the phenotypes that they regulate.
Results
We identified an unpaired LuxR solo (QS luxR homolog that occurs without the corresponding luxI homolog), which is highly conserved in both the AHL producing and non-AHL producing P. putida strains that we analyzed. In this study we report the cloning and functional characterization of this unpaired LuxR homolog designated PpoR. An AHL binding assay showed that PpoR protein binds to 3-oxo-C6-HSL. Studies using a ppoR promoter-lacZ reporter fusion revealed that it exhibits stringent growth phase dependent expression. Functional interaction of PpoR with the endogenous complete AHL QS systems of P. putida WCS358 (PpuI/R system) and PpoR was also investigated. Microarray analysis of P. putida WCS358 wild type and a PpoR over-expressing strain revealed several putative target genes that may be directly or indirectly regulated by PpoR.
Conclusion
Our results indicate that PpoR in P. putida strains may have a conserved role in detecting an AHL signal, either self or foreign, and regulating specific target genes.
Background
Most bacteria have a regulatory system, known as quorum sensing (QS), to modulate gene expression as a function of their cell density (for reviews see [1,2]). It usually works via the production of a signaling molecule that reaches a threshold concentration at high cell density allowing its detection by the bacterial population and resulting in the modulation of target gene expression. In gram negative, N-acyl homoserine lactone signaling molecules (AHLs) are thus far the most common signal molecules produced. A typical AHL QS system involves two major components: an AHL synthase gene (belonging to the LuxI protein family) and a modular transcriptional response-regulator (belonging to the LuxR protein family) which detects and responds to the AHL concentration [3].
AHL QS thus far is exclusively found in proteobacteria; 68 of 265 sequenced proteobacterial genomes possess at least one luxI/R family pair [4]. Interestingly, 90 genomes contained at least one luxR gene having the modular characteristics of the QS-family of regulators; however it was not associated with a cognate luxI-family gene. Of these, 45 genomes harbor at least one complete AHL QS system in addition to one or more luxR gene/s. These unpaired LuxR family proteins were firstly designated orphans [5] and recently they have been proposed to be renamed as LuxR 'solos' [6]; a few of these LuxR solos are beginning to be studied. ExpR of Sinorhizobium meliloti, BisR of Rhizobium leguminosarum bv. viciae and QscR of Pseudomonas aeruginosa, are LuxR solo proteins in AHL producing bacteria which have been well characterized and shown to be integrated with the resident complete AHL QS regulatory networks [7-10]. Only two solo LuxR homologs in non-AHL producing bacteria have thus far been investigated in some detail. One is called SdiA which is present in the Salmonella enterica and Escherichia coli and shown to be able to bind and detect AHLs produced by other bacteria. The other one is from plant pathogenic Xanthomonas spp. and in two Xanthomonas species it is involved in regulating virulence factors upon binding an unknown plant produced low molecular weight compound which is not an AHL [11-13]. This indicates that certain quorum sensing related LuxR family proteins are able to be involved in inter-kingdom signaling by detecting non-AHL compounds produced by eukaryotes.
Pseudomonas putida strains are mainly studied either for their ability to establish beneficial association with plants or due to their versatile catabolic potential. Previous studies have indicated that the majority of soil-borne or plant-associated P. putida strains do not produce AHLs; apparently only about one third of strains belonging to these species have a complete AHL QS system [14,15]. Furthermore, the type and role played by these AHL QS systems varies and is highly unpredictable [16]. P. putida strains appear to be rather unique in displaying such variation and lack of conservation in their AHL QS systems. In this study we report however that a LuxR solo is very well conserved in all P. putida strains we tested. This protein, which we designated PpoR, was shown to be able bind to AHLs, was not involved in rhizosphere colonization and was shown to be involved in the regulation of several loci. In addition its gene is stringently growth-phase regulated. The presence and sequence similarity of PpoR and its orthologs in all P. putida strains indicates that this protein might play a conserved role associated with the detection and response to bacterial endogenous and/or exogenous signaling compounds.
Results and Discussion
PpoR, an unpaired LuxR homolog protein is highly conserved in Pseudomonas putida
The model P. putida KT2440 has not been reported to possess an AHL QS system and its genome sequence does not encode for a LuxI homolog. As we were interested in studying solo QS LuxR homolog proteins in P. putida, the genome sequence of P. putida KT2440 (AE015451) was examined for the presence of such proteins that typically contain an N-terminal AHL binding domain (PFAM 03472) and a C-terminal helix-turn-helix DNA binding domain (PFAM 00196). A single ORF, PP_4647 of 705 bp was identified encoding a protein of 235 amino acids and named as PpoR (Pseudomonas putida orphan regulator). A BLAST search revealed high similarity to several other P. putida strains whose genome sequences, either complete or partial are available in the NCBI database. PpoR exhibits similarity to orthologs from P. putida F1 (ABQ80629.1; 97%), P. putida GB-1 (ABZ00528.1; 95%), P. putida W619 (ACA71296.1; 84%) as well as to its homolog from P. entomophila L48 (CAK17431; 75%). We were also interested to know if ppoR is present in two other P. putida strains; namely P. putida WCS358 and P. putida RD8MR3; these two P. putida strains also possess a complete AHL QS system, hence they are able to produce and respond to AHLs [16,17]. It was established that they possess a PpoR ortholog as we have cloned and sequenced ppoR from both strains (see Methods; Figure 1). Importantly, all these orthologs along with PpoR of P. putida KT2440 retain those five amino acids in their AHL-binding domain that are invariant in this family of proteins (Figure 1; [3]). These observations indicate that PpoR is highly conserved as it is present in all P. putida strains that we examined, suggesting that it might be part of the core genome of P. putida. On the other hand, approximately only one-third of P. putida strains possess a complete AHL QS; in addition, the type and role of these systems is not conserved [16]. Homologs of PpoR are also present in several groups of other bacteria which belong to γ-proteobacteria, β-proteobacteria as well as enterobacteria, all of which show a high similarity of around 50% at amino acid level, indicating conservation across species (data not shown).
Figure 1.
Alignment showing similarity of deduced sequence of PpoR to its orthologs. Multiple sequence alignment was performed using the ClustalW2 program (Thompson et al. 1994). The protein sequences used for the alignment are as follows; P. putida KT2440 (AAN70220.1), P. putida F1 (ABQ80629.1), P. putida RD8MR3 (this study; accession number FM992078), P. putida GB-1 (ABZ00528.1), P. putida WCS358 (this study; accession number FM992077) and P. putida W619 (ACA71296.1). The amino acids that are conserved in QS LuxR family proteins are indicated in bold [3]. In the alignment, all identical amino acids (*), similar amino acids (:) and completely different amino acids (.) at a particular position are indicated. Also indicated are the regions of the protein sequence of PpoR of P. putida KT2440 that constitutes the AHL binding domain (bold line from 17 to 162 amino acids; PFAM 03472) and the DNA binding domain (dashed line from 176 to 213 amino acids; PFAM 00196).
PpoR binds to AHL molecules
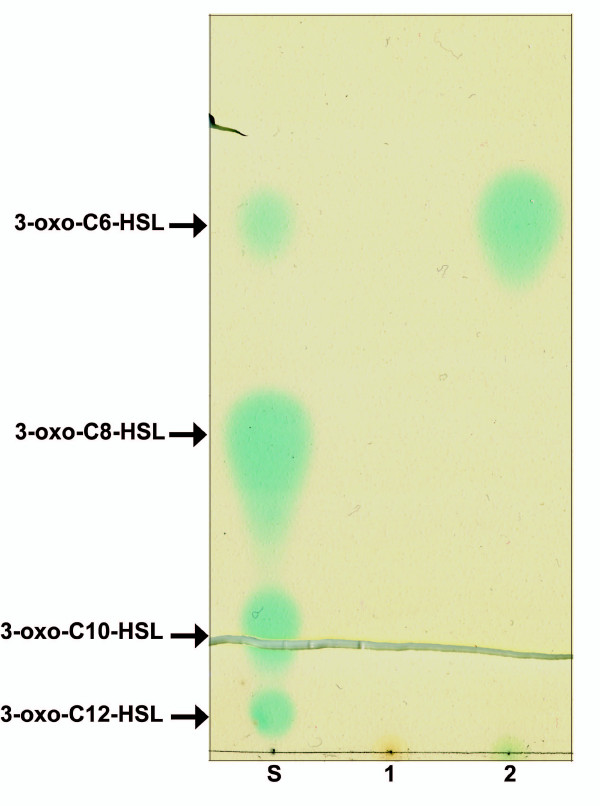
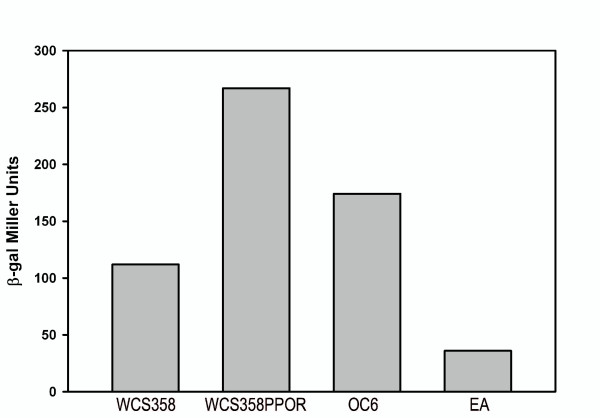
The presence of conserved amino acids in the AHL binding domain of PpoR of P. putida KT2440 indicated a possible binding to one or more AHLs. In order to identify if and which AHLs may bind PpoR, an AHL-binding assay was performed. E. coli strains that expressed PpoR protein or contained vector alone were grown in the presence of a set of externally supplemented AHLs (unsubstituted, oxo as well hydroxy AHLs) and any AHL that may bind to PpoR was visualized after purification via organic extraction, TLC and overlay with an AHL biosensor/indicator strain (as described in Methods). Purification of AHLs from E. coli over-expressing PpoR resulted in detection of 3-oxo-C6-HSL while E. coli cells which contained only the vector control, did not show any AHL (Figure 2). These results strongly indicate that PpoR most probably binds to 3-oxo-C6-HSL. Additionally, PpoR also exhibited probable binding to 3-oxo-C8-HSL and 3-oxo-C10-HSL, but to a lower extent at the concentrations of AHLs used in our experiment (data not shown). All the other AHLs tested in our assay could not be detected by TLC meaning over-expression of PpoR did not result in their purification. This could mean that they most probably do not bind to these AHLs or the binding is much lower than the sensitivity of this assay. It was concluded that PpoR of P. putida KT2440 and most probably other P. putida strains lacking a complete AHL QS system could be sensing and responding to AHL signals produced by neighboring bacteria. PpoR may also recognize endogenous AHL signals if the P. putida strain is able to produce AHLs. Interestingly, the few P. putida strains reported to possess a complete AHL QS system produce 3-oxo-C6-HSL [16-18], which as shown in this study could bind PpoR. In order to verify that P. putida WCS358 produced biologically active concentrations of 3-oxo-C6-HSL, we quantified these AHL levels in the wild type and in the ppoR mutant strains. As can be seen from Figure 3, strain WCS358 produced biologically active concentrations of AHLs and interestingly the ppoR mutant produced higher quantities. The reason for this is currently unknown, it cannot be excluded that lack of PpoR in the cells could result in higher quantities of free AHLs since as shown above, PpoR can bind and titrate away 3-oxo-C6-HSL.
Figure 2.
PpoR binds 3-oxo-C6-HSL. E. coli M15 (pRep4) containing either pQEPpoR or pQE30 were grown in LB in the presence of various AHLs (1 μM) added separately and protein expression induced with IPTG (1 μM). After 3.5 hours of growth post induction, AHLs were extracted from the cell pellets and visualized by TLC overlaid with A. tumefaciens NTL4 (pZLR4). The standards used are synthetic AHLs. In the figure, the lanes are marked as follows; S – AHL standards, 1 – AHL extracted from E. coli/pQE30 cell pellets grown with 3-oxo-C6-HSL supplementation and 2 – AHL extracted from E. coli/pQEPpoR cell pellets grown with 3-oxo-C6-HSL supplementation.
Figure 3.
3-oxo-C6-HSL measurement produced by P. putida WCS358 and by the ppoR mutant WCS358PPOR. AHLs were extracted from spent supernatants and levels were measured using a biosensor as described by Steindler et al., [15]. AHL levels were measured using a volume of extract corresponding to an amount of 5 × 108 cfu as described in the Methods section. 3-oxo-C6-HSL level is proportional to β-galactosidase activity (Miller Units); β-gal refers to β-galactosidase; OC6 refers to 0.05 μM of synthetic 3-oxo-C6-HSL used for the experiment and EA refers to ethyl acetate added as control in the experiment. β-galactosidase activity (Miller Units) represented as bars were obtained from one such experiment; similar values were obtained from additional experiments carried out with AHLs extracted independently from P. putida WCS358 and WCS358PPOR strains.
PpoR interacts with the endogenous AHL QS system of P. putida WCS358
To study the role of PpoR in P. putida WCS358 and P. putida RD8MR3 which also have a resident AHL QS system, knock-out mutants in ppoR were generated in both these strains (Table 1; Methods). The AHL production profile of the ppoR mutant was similar to the one of the WT with only a reproducible slight increase in the intensity of the signal for WCS358PPOR and lower intensity than wild type for RD8MR3PPOR (data not shown). Quantification of the amount of signal produced by the ppoR mutant (using two biosensors specifically detecting AHLs produced by WCS358 and one for the AHLs produced by strain RD8MR3), showed a similar trend of the ppoR mutants producing slightly more AHLs for WCS358 and slightly less AHLs for RD8MR3 (Figure 3 for data on quantification of 3-oxo-C6-HSL; for quantification of 3-oxo-C12-HSL data not shown).
Table 1.
Bacterial strains and plasmids used in this study
| Bacteria, plasmids or primers | Characteristics | Reference or source |
|---|---|---|
| Pseudomonas putida | ||
| P. putida RD8MR3 | Wild type; isolated from rhizosphere of rice roots | [24] |
| P. putida RD8MR3PPRI | [16] | |
| P. putida RD8MR3PPRR | pprR::Km of P. putida RD8MR3; Kmr | [16] |
| P. putida RD8MR3PPOR | ppoR::Km of P. putida RD8MR3, Kmr | This study |
| P. putida WCS358 | Wild type; plant growth promoting strain from the rhizosphere of potato roots | [21] |
| P. putida WCS358PPOR | ppoR::Km of P. putida WCS358, Kmr | This study |
| P. putida M17 | psrA178::Tn5 of P. putida WCS358, Kmr | [23] |
| P. putida MKO1 | rpoS880::Tn5 of P. putida WCS358, Kmr | [22] |
| P. putida IBE1 | gacA400::Tn5 of P. putida WCS358, Kmr | [17] |
| P. putida IBE2 | ppuR1793::Tn5 of P. putida WCS358, Kmr | [17] |
| P. putida IBE3 | rsaL1640::Tn5 of P. putida WCS358, Kmr | [17] |
| P. putida IBE5 | ppuI::Km of P. putida WCS358, Kmr | This study |
| E. coli | ||
| E. coli M15(pRep4) | Derivative of E. coli K-12, containing pREP4 plasmid ensuring the production of high levels of lac repressor protein; Kmr | Qiagen |
| E. coli Dh5α | F'/endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 (lacZYA-argF)U169 deoR [80dlac(lacZ)M15recA1] | [26] |
| A. tumefaciens NTL4 (pZLR4) | A. tumefaciens NT1 derivative carrying a traG::lacZ reporter fusion | [34] |
| Plasmid | ||
| pRK2013 | Tra+ Mob+ ColE1 replicon; Kmr | [31] |
| pMOSBlue i | Cloning vector, Ampr | Amersham-Pharmacia |
| pBBR mcs-5 | Broad-host-range vector, Gmr | [29] |
| pBBRPpoR | pBBR mcs-5 with 749-bp XbaI-KpnI fragment containing ppoR, Gmr | This study |
| pBluescript KS | Cloning vector, Ampr | Stratagene |
| pQE30 | Expression vector, Ampr | Qiagen |
| pQEPpoR | 721-bp containing ppoR of P. putida KT2440 cloned as SphI-HindIII fragment in pQE30 | This study |
| pMP220 | Promoter probe vector, IncP1; Tcr | [28] |
| pPUI220 | ppul promoter cloned in pMP220; Tcr | [17] |
| pPUR220 | ppuR promoter cloned in pMP220; Tcr | [17] |
| pRSA220 | rsaL promoter cloned in pMP220; Tcr | [17] |
| pPpoR1 | ppoR promoter of P. putida RD8MR3 cloned in pMP220; Tcr | This study |
| pPpoR2 | ppoR promoter of P. putida WCS358 cloned in pMP220; Tcr | This study |
| pMPpprIprom | Promoter of gene pprI cloned in pMP220 vector | [16] |
| pKNOCK-Km | Conjugative suicide vector; Kmr | [35] |
| pKNOCKppoR1 | Internal fragment of P. putida RD8MR3 ppoR cloned into KpnI-XbaI sites of pKNOCK-Km | This study |
| pKNOCKppoR2 | Internal fragment of P. putida WCS358 ppoR cloned into KpnI-XbaI sites of pKNOCK-Km | This study |
| pEXPPUIKm | pEXGm containing KpnI-SalI fragment of ppuI::Km | This study |
| pLAFRppoR | Cosmid clone containing P. putida RD8MR3 ppoR | [16] |
| pBS1 | pBluescript KS carrying the 598-bp pcr product of the P. putida RD8MR3 ppoR promoter region | This study |
| pBS2 | pBluescript KS carrying the 318-bp pcr product of the P. putida WCS358 ppoR promoter region | This study |
| pBS3 | pBluescript KS carrying the 721-bp pcr product of the P. putida KT2440 complete ppoR gene | This study |
| pBS4 | pBluescript KS carrying the 749-bp pcr product of the P. putida WCS358 complete ppoR gene | This study |
| pBS5 | pBluescript KS carrying the HindIII subclone of pLAFRppoR that contains ppoR | This study |
| pBS6 | pBluescript KS carrying the pKNOCK-Km insertion flanking sequences from P. putida WCS358PPOR genomic DNA | This study |
| pMOS1 | pMOSBlue vector carrying 394-bp internal portion of P. putida RD8MR3 ppoR gene | This study |
| pMOS2 | pMOSBlue vector carrying 385-bp internal portion of P. putida WCS358 ppoR gene | This study |
| pMOS3 | pMOSBlue vector carrying pcr product of 358_PpoRf and 4648 degR primers | This study |
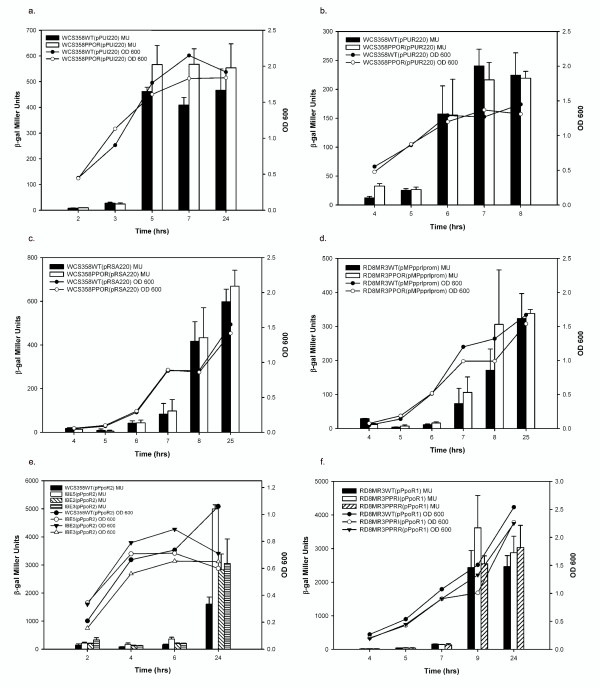
We also determined if PpoR was involved in transcriptional regulation of the QS systems ppuI/R of P. putida WCS358 and pprI/R of P. putida RD8MR3. To perform this experiment, lacZ-transcriptional promoter probe fusions of ppuI, ppuR and rsaL for P. putida WCS358 and pprI for P. putida RD8MR3 were monitored for expression throughout the growth phase in their respective wild type and ppoR mutant strains. For P. putida WCS358 QS-related gene promoters, it was observed that ppuR and rsaL promoters showed comparable expression levels in both wild type and ppoR mutant strains at different growth phases (Figures 4b &4c). On the other hand the ppuI promoter of P. putida WCS358 controlling the AHL synthase exhibited consistently higher expression levels in WCS358PPOR especially in the logarithmic growth phase which was statistically significant (Figure 4a). The pprI transcription levels in P. putida RD8MR3 were not significantly different from the wild type (Figures 4d)
Figure 4.
β-Galactosidase assays showing expression profile of ppoR and the QS system genes of P. putida WCS358 and RD8MR3. Bacterial cultures were started with an initial inoculum of 5 × 106 CFU per ml in 20 ml of minimal medium (M9-Cas) and β-Galactosidase activities were measured at different stages of growth. The growth curves of different mutants and the wild type strain are indicated in each graph. All experiments were performed in triplicate and the mean values of each time point along with standard deviations are shown in each graph. All the graphs were plotted using SigmaPlot version10.0. (a, b, and c) ppuI, ppuR and rsaL promoter activities of P. putida WCS358 in wild type and WCS358PPOR using plasmids pPUI220, pPUR220 and pRSA220. Paired t-test analysis of ppuI promoter activities revealed a significant difference between the mean values of wild type and WCS358PPOR at 7 hours of growth (p value 0.0184; t = 7.268 df = 2) at P < 0.05 significance level. (d) pprI promoter activity in P. putida RD8MR3 wild type and RD8MR3PPOR with the plasmid pMPpprIprom. (e) ppoR promoter activity in P. putida WCS358 wild type, ppuI knock-out (IBE5), ppuR (IBE2) and rsaL (IBE3) mutants with the plasmid pPpoR2. Anova analysis of sample means followed by Dunnett's multiple comparison test revealed that there is a significant difference between the means of wild type and IBE5 at P < 0.05 significance level at 4, 6 and 24 hours growth [F(3,8) = 6.278, F(3,8) = 22.97 and F(3,8) = 16.37 respectively] (f) ppoR promoter activity in P. putida RD8MR3 wild type, pprI (RD8MR3PPRI) and pprR (RD8MR3PPRR) mutants with the plasmid pPpoR1. β-gal, β-galactosidase; OD600, optical density at 600 nm; MU, Miller Units.
In order to understand whether ppoR expression is under the control of the QS systems of P. putida WCS358 and RD8MR3, ppoR promoter-lacZ reporter fusions of both strains were assayed in their respective QS system mutants and compared with wild type strains at different growth phases. Interestingly, in P. putida WCS358, ppoR expression shows substantial increase in the IBE5 ppuI AHL synthase mutant, indicating a QS system mediated repression of ppoR expression (Figure 4e). The ppoR promoter levels in this genetic background were not restored to WCS358 wild-type levels by adding exogenously the four AHLs (3-oxo-C6-, 3-oxo-C8-, 3-oxo-C10- and 3-oxo-C12-HSL) produced by WCS358 (data not shown). The reason for this is not known and we cannot exclude that QS is particularly sensitive to growth phase and AHL concentration, thus exogenous addition of AHLs might not necessarily re-establish the conditions present in the wild-type strain.
The expression levels of ppoR in P. putida WCS358 IBE2 & IBE3 (ppuR and rsaL mutant respectively), and P. putida RD8MR3PPRI and RD8MR3PPRR although higher were not statistically significant (Figures 4e &4f). These results suggest that ppoR interaction with the endogenous QS systems in these two P. putida strains may not be similar; in strain WCS358 negative regulation (albeit not very strong) of ppoR gene expression occurred in response to AHLs via a mechanism which could be independent of the cognate PpuR AHL sensor/regulator.
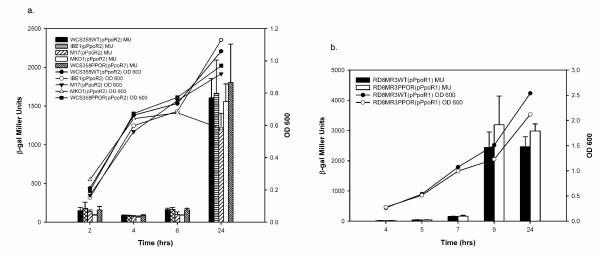
ppoR expression is growth phase regulated
In order to understand if PpoR expression patterns showed any correlation to its role in interacting with the endogenous QS system, ppoR expression levels were measured as β-galactosidase activities at different growth phases. Importantly, it was observed for both P. putida WCS358 and RD8MR3 that at low cell densities ppoR transcription showed minimal expression but was found to increase sharply when the culture enters the logarithmic phase of growth (Figure 5). This pattern of expression level was maintained even in WCS358PPOR and RD8MR3PPOR indicating a lack of regulation by PpoR of its own expression. To find out if ppoR expression is under the control of well known growth phase dependent global regulators, its expression level was monitored in P. putida WCS358 MKO1 (rpoS), M17 (psrA) and IBE1 (gacA). There was no significant difference in the expression pattern levels of ppoR promoter in the three mutants when compared to wild type suggesting that these three global growth-phase regulators were not involved in modulating ppoR expression levels (Figure 5). It was therefore concluded that ppoR gene expression is stringently growth phase regulated via a yet unidentified regulator.
Figure 5.
ppoR promoter activities in wild type and various mutant strains of P. putida WCS358 and RD8MR3. Bacterial cultures were started with an initial inoculum of 5 × 106 CFU per ml in 20 ml of minimal medium (M9-Cas) and β-galactosidase activities were measured at different stages of growth. The growth curves of different mutants and the wild type strain are indicated in each graph. All experiments were performed in triplicate and the mean values of each time point along with standard deviations are shown in each graph. All the graphs were plotted using SigmaPlot version10. (a) ppoR promoter activity in P. putida WCS358 wild type, gacA (IBE1), psrA (M17), rpoS (MKO1) and ppoR (WCS358PPOR) using plasmid pPpoR2. (b) ppoR promoter activity in P. putida RD8MR3 wild type and ppoR (RD8MR3PPOR) mutants using plasmid pPpoR1. β-gal, β-galactosidase; OD600, optical density at 600 nm; MU, Miller Units.
Rhizosphere colonization ability of P. putida WCS358PPOR & RD8MR3PPOR are not affected
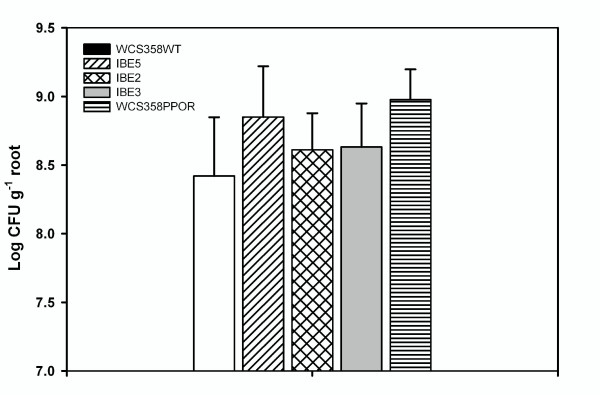
Traits involved in surface associated growth of P. putida may be regulated by their QS system and possibly also determine their fitness in the rhizosphere [19,20]. Rice root colonization was carried out following the protocol as previously reported [16] with P. putida WCS358 wild type, WCS358PPOR and WCS358 QS mutants. Our results revealed that wild type, IBE2 & IBE3 exhibited similar degree of colonization whereas IBE5 & WCS358PPOR were slightly better in colonization of rice roots (Figure 6). One way ANOVA analysis in conjunction with Dunnett's test (P < 0.01) was carried out to confirm that the means of the cell number were significantly different when compared to the wild type strain. Similar experiment with RD8MR3 wild type and RD8MR3PPOR showed that they colonized rice roots to the same extent (data not shown).
Figure 6.
Root colonization assay of P. putida WCS358 wild type and mutants. Colonization assays were performed as described previously (Steindler et al. 2008). The data presented are from one experiment. Anova analysis in combination with Dunnett's multiple comparison test revealed a significant difference between the mean values of wild type & IBE5 as well as between wild type & WCS358PPOR at P < 0.01 significance level [F(4,45) = 2.870].
Identification of putative target genes of PpoR by microarrayanalysis
In order to identify target genes directly or indirectly regulated by PpoR, global gene expression comparison was performed of P. putida WCS358 wild type with a strain over expressing ppoR (PpoR++). Microarray analysis was performed with a single biological sample for each strain with four technical replicates (as mentioned in Methods). Our results revealed that a total of 62 genes show differential expression of more than two fold (P < 0.05) in cultures that were grown in minimal medium (Table 2 and 3). Majority of genes that showed a down regulation of gene expression in the PpoR++ strain were those involved in amino acid catabolism. Genes that showed up regulation of expression in the PpoR++ were those that take part in protein synthesis and sulfur metabolism.
Table 2.
List of genes showing up regulation of gene expression in P. putida WCS358 PpoR++ strain
| Gene name as annotated in P. putida KT2440 | Function | Fold change | |
|---|---|---|---|
| 1. | PP0233 | Taurine ABC transporter, periplasmic taurine-binding protein | 5.016 |
| 2. | PP5172 | hypothetical protein | 4.503 |
| 3. | PP0237 | sulfonate ABC transporter, periplasmic sulfonate-binding protein SsuA | 3.801 |
| 4. | PP0236 | NADH-dependent FMN reductase | 3.751 |
| 5. | PP0170 | ABC transporter, periplasmic binding protein | 3.555 |
| 6. | PP0459 | 50S ribosomal protein L22 | 3.063 |
| 7. | PP0235 | antioxidant protein LsfA | 3.002 |
| 8. | PP0462 | 50S ribosomal protein L29 | 2.853 |
| 9. | PP0457 | 50S ribosomal protein L2 | 2.758 |
| 10. | PP0458 | 30S ribosomal protein S19 | 2.666 |
| 11. | PP5085 | malic enzyme | 2.665 |
| 12. | PP0461 | 50S ribosomal protein L16 | 2.631 |
| 13. | PP1465 | 50S ribosomal protein L19 | 2.626 |
| 14. | PP0463 | 30S ribosomal protein S17 | 2.602 |
| 15. | PP0455 | 50S ribosomal protein L4 | 2.592 |
| 16. | PP0464 | 50S ribosomal protein L14 | 2.563 |
| 17. | PP0460 | 30S ribosomal protein S3 | 2.455 |
| 18. | PP0465 | 50S ribosomal protein L24 | 2.431 |
| 19. | PP0453 | 30S ribosomal protein S10 | 2.426 |
| 20. | PP0721 | 50S ribosomal protein L25 | 2.334 |
| 21. | PP5168 | sulfate ABC transporter, ATP-binding protein | 2.297 |
| 22. | PP0466 | 50S ribosomal protein L5 | 2.236 |
| 23. | PP0475 | 50S ribosomal protein L36 | 2.213 |
| 24. | PP1600 | outer membrane protein OmpH | 2.205 |
| 25. | PP1464 | tRNA (guanine-N(1)-)-methyltransferase | 2.181 |
| 26. | PP0454 | 50S ribosomal protein L3 | 2.178 |
| 27. | PP0689 | 50S ribosomal protein L27 | 2.073 |
| 28. | PP0470 | 50S ribosomal protein L18 | 2.059 |
Table 3.
List of genes showing down regulation of gene expression in P. putida WCS358 PpoR++ strain
| Gene name as annotated in P. putida KT2440 | Function | Fold change | |
|---|---|---|---|
| 1. | PP3433 | 4-hydroxyphenylpyruvate dioxygenase | 18.116 |
| 2. | PP2335 | citrate synthase | 12.097 |
| 3. | PP1743 | acetate permease | 9.109 |
| 4. | PP4621 | homogentisate 1,2-dioxygenase | 7.574 |
| 5. | PP1742 | hypothetical protein | 7.057 |
| 6. | PP4064 | isovaleryl-CoA dehydrogenase | 6.120 |
| 7. | PP4065 | 3-methylcrotonyl-CoA carboxylase, beta subunit, putative | 6.042 |
| 8. | PP0882 | dipeptide ABC transporter, periplasmic dipeptide-binding protein | 5.896 |
| 9. | PP4402 | 2-oxoisovalerate dehydrogenase, beta subunit | 5.677 |
| 10. | PP4864 | branched-chain amino acid ABC transporter, ATP-binding protein | 5.553 |
| 11. | PP4619 | maleylacetoacetate isomerase, putative | 5.245 |
| 12. | PP0545 | aldehyde dehydrogenase family protein | 5.053 |
| 13. | PP2333 | transcriptional regulator, GntR family | 4.694 |
| 14. | PP4866 | branched-chain amino acid ABC transporter, permease protein | 4.469 |
| 15. | PP1140 | branched-chain amino acid ABC transporter, permease protein | 4.185 |
| 16. | PP1000 | ornithine carbamoyltransferase | 4.006 |
| 17. | PP0999 | carbamate kinase | 3.475 |
| 18. | PP0193 | hypothetical protein | 3.470 |
| 19. | PP1001 | arginine deiminase | 3.335 |
| 20. | PP1297 | general amino acid ABC transporter, periplasmic binding protein | 3.111 |
| 21. | PP0764 | hypothetical protein | 3.100 |
| 22. | PP4650 | ubiquinol oxidase subunit II, cyanide insensitive | 3.073 |
| 23. | PP0751 | malate:quinone oxidoreductase | 2.972 |
| 24. | PP0989 | glycine cleavage system protein H | 2.759 |
| 25. | PP0397 | hypothetical protein | 2.676 |
| 26. | PP4975 | long-chain acyl-CoA thioester hydrolase family protein | 2.601 |
| 27. | PP5258 | aldehyde dehydrogenase family protein | 2.507 |
| 28. | PP1690 | hypothetical protein | 2.469 |
| 29. | PP2738 | transcriptional regulator, putative | 2.463 |
| 30. | PP4814 | ATP-dependent protease La domain protein | 2.338 |
| 31. | PP3122 | CoA-transferase, subunit A, putative | 2.176 |
| 32. | PP4194 | citrate synthase | 2.162 |
| 33. | PP0684 | peptidyl-prolyl cis-trans isomerase, FKBP-type | 2.077 |
| 34. | PP5319 | hypothetical protein | 2.013 |
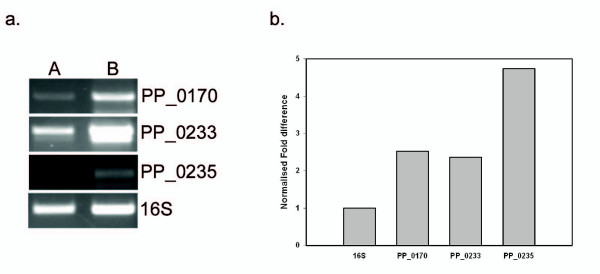
In order to validate the differential expression of genes observed in the microarray experiment, semi quantitative RT PCR analysis of three genes PP_0170, PP_0233 and PP_0235, was performed as they were among genes that showed maximum up-regulation in PpoR++ strains when compared to wild type. Briefly, PP_0170 codes for a putative ABC transporter periplasmic binding protein (3.55 fold up regulation in PpoR++ strain), PP_0233, designated as tauA, encodes a putative taurine ABC transporter periplasmic binding protein (5 fold up regulation in PpoR++ strain) and PP_0235, named lsfA, codes for a putative peroxidase (3 fold up regulation in PpoR++ strain). RT PCR analysis with two independent RNA isolations shows more than two fold increases in expression of these genes in PpoR++ strain when compared to wild type and is in agreement with the results obtained in microarray (Figure 7). As these genes take part in inorganic ion utilization and oxidative stress, it is possible that PpoR might play a functional role in these processes.
Figure 7.
RT-PCR analysis to validate expression of genes in P. putida WCS358. Total RNA isolations were carried out from bacterial cultures grown in minimal M9 medium using Ribopure RNA isolation kit (Ambion) and DNase treatment was carried out. cDNA synthesis was done using AMV Reverse Transcriptase (Promega) and second strand synthesis performed using Go Taq Flexi polymerase (Promega). RT-PCR analysis was performed with RNA obtained from two independent isolations and the figure shows results of one such experiment. (a) Agarose gel showing RT-PCR products for the genes PP_0170, PP_0233 and PP_0235. RT_PCR for 16S rRNA was carried out from the same RNA samples as control to ensure that equal amounts of RNA were taken. A. RT-PCR on RNA sample from P. putida WCS358 containing pBBR vector alone and B. RT-PCR on RNA sample from P. putida WCS358 containing pBBRPpoR. (b) Graph showing normalized fold difference of genes when compared to 16S rRNA expression levels. The gel image containing bands was analyzed by the ImageJ software and the bars indicate the fold increase in the intensity of the bands in PpoR++ strain (P. putida WCS358 containing pBBRPpoR) when compared to wild type (P. putida WCS358 containing pBBR vector alone).
Conclusion
The roles of solo QS LuxR proteins in inter-species as well inter-kingdom signaling are just beginning to be understood with a few recent studies on these proteins in non-AHL producing bacteria. The extent of the functional participation/interaction of these proteins in QS in AHL producing bacteria also differs depending on the strain. We have characterized PpoR, a solo LuxR homolog present in both AHL and non-AHL producing bacteria; its conservation indicates a significant role for this protein of P. putida. In fact most strains of P. putida do not harbor an AHL quorum sensing system, however they possess PpoR indicating that it is likely to be part of the core genome of this species. We have shown that PpoR binds AHLs and that it is highly conserved in P. putida; and this in our view represents the important novel finding of our study., In addition we believe that we are in a position to conclude that the results obtained using our strain represent what occurs in P. putida strains (including the ones which only have PpoR and do not contain a complete AHL QS system). Future studies will be directed towards understanding the regulation of target genes in response to exogenous AHLs in certain P. putida strains and also possibly endogenous AHLs in strains which harbor an AHL QS system.
Methods
Bacterial strains, plasmids and media
All strains, plasmids and primers used in this study are listed in Tables 1 and 4. P. putida [21-24] and E. coli strains were grown in Luria-Bertani (LB; [25]) medium at 30 and 37°C respectively. P. putida strains were also grown in M9 minimal medium [26] supplemented with 0.3% casamino acids (M9-Cas) at 30°C. Agrobacterium tumefaciens NTL4 (pZLR4) was grown in AB medium [27] at 28°C. Antibiotics when required were supplemented at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 100 μg/ml (Pseudomonas) or 50 μg/ml (E. coli); nalidixic acid, 25 μg/ml; tetracycline, 10 μg/ml (E. coli) or 40 μg/ml (Pseudomonas); and gentamicin, 10 μg/ml (E. coli) or 40 μg/ml (Pseudomonas). Transcriptional fusion constructs for ppoR promoter in pMP220 [28] were made as follows: a 598-bp fragment containing the ppoR promoter region was amplified from P. putida RD8MR3 genomic DNA with the primers 16orpF and 16orpR using Vent DNA polymerase (New England Biolabs) following supplier's instructions, cloned in pBluescript (Stratagene) yielding pBS1 and verified by DNA sequencing (Macrogen Inc., Korea). The ppoR promoter was removed as a KpnI-XbaI fragment from pBS1 and cloned in pMP220 yielding pPpoR1. Similarly, a 318-bp fragment was amplified from P. putida WCS358 genomic DNA using primers 358orpromF and 358orpromR and cloned in pBluescript yielding pBS2. The ppoR promoter was removed as KpnI-XbaI fragment from pBS2 and cloned in pMP220 yielding pPpoR2. To clone ppoR gene in pQE30, a 721-bp fragment containing the entire ppoR gene of P. putida KT2440 was amplified using primers KT_PpoRf and 4647R1 and cloned in pBluescript yielding pBS3. The ppoR gene was removed as SphI-HindIII fragment and cloned in pQE30 in the correct reading frame yielding pQEPpoR. To clone ppoR in pBBR [29], the 749-bp fragment containing the entire ppoR gene was amplified using P. putida WCS358 genomic DNA as the template using primers 358_PpoRf and 358_PpoRr and cloned in pBluescript yielding pBS4. ppoR gene was excised from pBS4 using XbaI-KpnI and cloned into pBBR mcs-5 yielding pBBRPpoR.
Table 4.
Primers used in this study
| Primer | Sequence | Reference or source |
|---|---|---|
| 16F | CGACGCAAACGCCACGGT | This study |
| 16R | TGCTCATCTTGGGCTGCG | This study |
| putidadegF | GCCTGCGCCACGG(AGCT)TGGAC(AGCT)CA | This study |
| putidadegR | GATCACGGAGCGGATGTG(AG)AA(AG)TT(AGCT)AC | This study |
| 16orpF | CATCAGCCTTGGTCACGCC | This study |
| 16orpR | GCGTCCTGCTTGTAGAACTC | This study |
| 358orpromF | GCACAGTGTCGCGCAAAGC | This study |
| 358orpromR | ATGTTCAGCCCCAAGTGTTC | This study |
| 358_PpoRf | CCATCCATAGGAGCGTTACG | This study |
| 358_PpoRr | GGTAGTCGCAGGGGTGGCTA | This study |
| KTPpoRf | AGGCATGCCTTCACTGGAAACCCGAG | This study |
| PP4647R1 | GGAAGCTTTCAGATCCAGCCACGCAA | This study |
| PP4647F | GGAATTCGAAACCCGAGTATCTTCGTG | This study |
| PP4647R | GGGTACCGGTGATCACGCTGCGGATATG | This study |
| 4648degR | CCCTGCAGGCGGATCAR(CT)TG(CT)TC(CT)TC | This study |
| 358PpoRintF | GCAGCACAATGAAAGCCAAG | This study |
| 358PpoRintR | GTGACAGCGACAGGATGGAG | This study |
| KT_16For | AGTTGGTGGGGTAATGGCTC | This study |
| KT_16Rev | TGTCAGTATCAGTCCAGGTG | This study |
| PP0170For | ATCGTCTACCTGCTGCTGAA | This study |
| PP0170Rev | AGCAAACAGCAAGGTCGGCG | This study |
| PP0233For | AAACCTTCCTCATCGCCACC | This study |
| PP0233Rev | TCCTTGCGGTAGTCGGCGTA | This study |
| PP0235For | GCTGAAGGACGAGTTTGCCA | This study |
| PP0235Rev | ATCACCACTTCGTCGCCGTC | This study |
Recombinant DNA techniques
DNA manipulations, including digestion with restriction enzymes, agarose gel electrophoresis, purification of DNA fragments, ligation with T4 ligase, end filling with the Klenow enzyme, hybridization, radioactive labeling by random priming, and transformation of E. coli, were performed as described previously [26]. Southern hybridizations were performed by using N+Hybond membranes (Amersham Biosciences); plasmids were purified by using Jet star columns (Genomed GmbH, Löhne, Germany); and total DNA from Pseudomonas was isolated by Sarkosyl-pronase lysis as described previously [30]. Triparental matings between E. coli and P. putida were carried out with the helper strain E. coli DH5α (pRK2013) [31].
AHL binding assay
50 ml cultures of E. coli M15(pRep4) strains containing either pQE30 or pQEPpoR were grown at 37°C to an initial optical density at 600 nm of 0.1 (OD 600). 5 ml aliquots of these cultures were supplemented with different types of AHLs at 1 μM concentration and grown at 30°C until OD 600 of 0.6. Protein expression was induced by IPTG (1 μM final concentration) and the cultures were incubated further with shaking for 3.5 hours. OD 600 was measured and equal numbers of cells were processed for all samples as follows; the cell pellets were washed three times with PBS before finally resuspending in equal volume of PBS. These cell suspensions were extracted once with the same culture volume of ethyl acetate-0.1% acetic acid. The extracts were then dried, resuspended in ethyl acetate and analyzed by thin layer chromatography (TLC) performed with C18 reverse-phase chromatography plates (Shaw et al., 1997). The AHL molecules were detected by overlaying the TLC plate with a thin layer of AB top agar seeded with A. tumefaciens NTL4 (pZLR4) in presence of 100 μg/ml X-gal.
AHL extraction, visualization and quantification
AHLs were purified from bacterial strains and analyzed either on a C18 reverse-phase chromatography TLC plates as previously described (Shaw et al., 1997) or quantified as previously described Rampioni et al [32] for 3-oxo-C12-HSL or by Steindler et al., [16] for 3-oxo-C6-HSL. For visualization on TLC, the extracts were placed on a TLC plate and AHLs were separated as previously described [33] and the plate was then overlaid with a thin layer AB top agar seeded with A. tumefaciens NTL4 (pZLR4) [34] in presence of 100 μg/ml X-gal, as described previously [33].
Cloning of the ppoR gene of P. putida RD8MR3 and WCS358, generation of ppoR mutants in both strains and of a ppuI mutant in WCS358
The P. putida RD8MR3 ppoR gene was cloned as follows; P. putida KT2440 partial ppoR gene was amplified using primers PP_4647F and PP_4647R and used as probe to screen a cosmid library of P. putida RD8MR3 [16] by colony hybridization. Cosmid pLAFRppoR was identified, ppoR gene localized to a 4.5-kb HindIII fragment and cloned in pBluescript to yield pBS5 which was sequenced using vector specific primers and by primer walking to obtain 1735-bp containing RD8MR3 ppoR. To generate a ppoR mutant in strain RD8MR3, we constructed pKNOCKppoR1 as follows; a 394-bp internal fragment of P. putida RD8MR3 ppoR gene was amplified by PCR using primers 16F and 16R and cloned in pMOSblue yielding pMOS1. ppoR internal fragment was excised from pMOS1 using XbaI-KpnI and cloned into pKNOCK-Km [35] to yield pKNOCKppoR1. pKNOCKppoR1 was used as suicide vector to create knockout mutants of ppoR by homologous recombination in P. putida RD8MR3 designated RD8MR3PPOR. The fidelity of the marker exchange events was confirmed by Southern analysis of mutants.
In order to generate a ppoR mutant in strain WCS358, we constructed pKNOCKppoR2 as follows; a 385-bp internal fragment of P. putida WCS358 ppoR gene was amplified by PCR using degenerate primers putidadegF and putidadegR and cloned in pMOSblue yielding pMOS2. ppoR internal fragment was excised from pMOS2 using XbaI-KpnI and cloned into pKNOCK-Km generating pKNOCKppoR2. pKNOCKppoR2 was then used as a suicide vector to create knockout mutants of ppoR by homologous recombination in WCS358 designated WCS358PPOR. The fidelity of the marker exchange events was confirmed by Southern analysis of mutants. In order to clone the ppoR gene from P. putida WCS358, the genomic DNA of WCS358PPOR (generated as mentioned above) was digested with an enzyme flanking vector insertion on one side and cloned into pBluescript to yield pBS6. Sequencing of this clone using vector specific primers yielded an 1148-bp sequence covering the promoter and the first 570-bp of ppoR. The last 135-bp of the ppoR gene was obtained by amplification of this region from P. putida WCS358 wild type using primers 358_PpoRf and 4648degR (a degenerate primer based on available P. putida sequences of the downstream gene PP_4648), cloning in pMOS to yield pGEM3 and sequencing of pMOS3 with vector specific primers.
In order to generate a ppuI AHL synthase mutant in WCS358 we introduced a Km resistance cassette removed as a HincII fragment into the BsaAI site of ppuI harbored in pBQS1 generating pQS1:Km. ppuI::Km was then cloned into pEXGm as a KpnI-SalI fragment generating pEXPPUIKm. This latter plasmid was used in generating a ppuI knock mutant, designated P. putida IBE5, via homologous recombination and selection as previously described [36].
Reporter gene fusion assay and root colonization
β-galactosidase activities were determined during growth in M9-Cas essentially as described by Miller [25] with the modifications of Stachel et al[37]. All experiments were performed in triplicate, and the mean values are given. Statistical significance of the values were calculated by paired t-test (to compare two sample mean values; P < 0.05) or one way anova in combination with Dunnett's test (to compare multiple sample mean values; P < 0.05). β-galactosidase activities were determined at various times after a 20-ml M9-Cas culture was started with an initial inoculum of 5 × 106 CFU. Root colonization assays were performed exactly as described by Steindler et al [16].
Total RNA isolation
An overnight culture of P. putida WCS358 strains carrying pBBR mcs-5 or pBBRPpoR grown in M9-Cas was used to obtain an initial OD 600 of 0.1. The cultures were incubated at 30°C on a rotary shaker at 180 rpm until they reached an OD 600 of approximately 1.2. RNA isolation was carried out from 2 × 109 cells using Ribopure™-bacteria RNA isolation kit (Ambion Inc., Austin, USA) as per manufactures's instructions. DNase treatment of RNA was done at 37°C for 1 hour (Ambion), if necessary twice and RNA purified. The purity of RNA was assessed by performing a PCR on a fixed quantity of total RNA (250 ng) with GoTaq polymerase (Promega) using genomic DNA as control with 358PpoRintF and 358PpoRintR primers specific for P. putida WCS358 ppoR. The RNA quality was assessed by spectrophotometric measurement at 260 nm and 280 nm and its intact nature verified by visualizing RNA samples on an agarose gel.
Microarray analysis
A customized high-density oligonucleotide whole genome expression array (NimbleGen Systems Inc., Madison, WI) was designed for P. putida KT2440 using the genome sequence and open reading frame (ORF) predictions available from GenBank accession number NC_002947. The 6,181,863-bp chromosome of KT2440 contains 5,350 predicted ORFs and 96 RNAs. 60-mer probes paired with perfect-match (PM) oligonucleotides and their corresponding mismatch oligonucleotides were selected for 5350/5350 sequences with the median number of probes/sequence being 18. Each probe was replicated 4 times on the chip representing a technical replicate. The cDNA synthesis, hybridization, and scanning were performed by NimbleGen Systems Inc. Microarray data analysis was performed using the robust multiarray average method [38] based on the log2 values of the absolute signal intensities for PM probes only. Student's t test for each PM probe and each technical replicate, followed by the Bonferroni correction, was used to identify genes showing differential expression patterns (P < 0.05). The data presented are the results from one experiment.
Semi quantitative RT PCR and analysis
Reverse transcription was performed in a 20-μl reaction mixture containing 2 μg of total RNA, 100 ng of random primers/μg of RNA and 5 U of AMV reverse Transcriptase (Promega, Madison, WI) following manufacturer's instructions. After denaturing RNA and random primers at 65°C for 3 min, the remaining reagents were added and the mixture incubated at 25°C for 10 min, 42°C for 90 min and held at 70°C for 10 min to inactivate the enzymes. The KT_16For and KT_16Rev primers were used to measure the transcription of 16S rRNA. Second strand synthesis was performed using Go Taq Flexi polymerase (Promega) using 1 μl of cDNA reaction as template; for 16S rRNA, 1 μl of 1:100 diluted cDNA reaction was used. The number of PCR cycles to be performed for each gene was standardized so that the product amplification is in the linear range and proportional to the amount of input sample. 10 μl of the PCR reaction was analyzed by agarose gel electrophoresis. The intensity of the bands obtained were measured and normalized to that of 16S rRNA using the ImageJ software [39] to obtain the fold difference. Each gene was validated twice by RT PCR analysis of RNA samples from two independent isolations.
Nucleotide sequence accession numbers
All DNA sequences were performed at Macrogen http://www.macrogen.com and the nucleotide sequences were deposited in GenBank/EMBL/DDBJ; ppoR gene of P. putida RD8MR3 is given under accession number FM992078 whereas the ppoR gene of P. putida WCS358 is given under accession number FM992077.
Authors' contributions
SS carried out all the experimental studies and participated in experimental design and drafting the manuscript. VV designed, coordinated the study and drafted the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Sujatha Subramoni, Email: subramoni@icgeb.org.
Vittorio Venturi, Email: venturi@icgeb.org.
Acknowledgements
We thank Iris Bertani for constructing the WCS358 ppuI mutants and Zulma R. Suarez-Moreno for assistance in editing the manuscript and figures. SS is beneficiary of an ICGEB fellowship. VV's laboratory is supported by ICGEB, Fondazione Cassamarca (TV, Italy) and the Italian Cystic Fibrosis Research Foundation (VR, Italy).
References
- Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Winans SC, Greenberg EP. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- Case RJ, Labbate M, Kjelleberg S. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. Isme J. 2008;2:345–349. doi: 10.1038/ismej.2008.13. [DOI] [PubMed] [Google Scholar]
- Fuqua C. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J Bacteriol. 2006;188:3169–3171. doi: 10.1128/JB.188.9.3169-3171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramoni S, Venturi V. LuxR-family 'solos': bachelor sensors/regulators of signalling molecules. Microbiology. 2009. [DOI] [PubMed]
- Danino VE, Wilkinson A, Edwards A, Downie JA. Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol Microbiol. 2003;50:511–525. doi: 10.1046/j.1365-2958.2003.03699.x. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lequette Y, Greenberg EP. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol Microbiol. 2006;59:602–609. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol. 2006;188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh M, Krol E, Becker A. Competitive and cooperative effects in quorum-sensing-regulated galactoglucan biosynthesis in Sinorhizobium meliloti. J Bacteriol. 2008;190:5308–5317. doi: 10.1128/JB.00063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferluga S, Bigirimana J, Hofte M, Venturi V. A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol Plant Pathol. 2007;8:529–538. doi: 10.1111/j.1364-3703.2007.00415.x. [DOI] [PubMed] [Google Scholar]
- Ferluga S, Venturi V. OryR is a LuxR-family protein involved in inter-kingdom signaling between pathogenic Xanthomonas oryzae pv. oryzae and rice. J Bacteriol. 2008. [DOI] [PMC free article] [PubMed]
- Zhang L, Jia Y, Wang L, Fang R. A proline iminopeptidase gene upregulated in planta by a LuxR homologue is essential for pathogenicity of Xanthomonas campestris pv. campestris. Mol Microbiol. 2007;65:121–136. doi: 10.1111/j.1365-2958.2007.05775.x. [DOI] [PubMed] [Google Scholar]
- d'Angelo-Picard C, Faure D, Penot I, Dessaux Y. Diversity of N-acyl homoserine lactone-producing and -degrading bacteria in soil and tobacco rhizosphere. Environ Microbiol. 2005;7:1796–1808. doi: 10.1111/j.1462-2920.2005.00886.x. [DOI] [PubMed] [Google Scholar]
- Elasri M, Delorme S, Lemanceau P, Stewart G, Laue B, Glickmann E, Oger PM, Dessaux Y. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl Environ Microbiol. 2001;67:1198–1209. doi: 10.1128/AEM.67.3.1198-1209.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler L, Bertani I, De Sordi L, Bigirimana J, Venturi V. The presence, type and role of N-acyl homoserine lactone quorum sensing in fluorescent Pseudomonas originally isolated from rice rhizospheres are unpredictable. FEMS Microbiol Lett. 2008;288:102–111. doi: 10.1111/j.1574-6968.2008.01344.x. [DOI] [PubMed] [Google Scholar]
- Bertani I, Venturi V. Regulation of the N-Acyl Homoserine Lactone-Dependent Quorum-Sensing System in Rhizosphere Pseudomonas putida WCS358 and Cross-Talk with the Stationary-Phase RpoS Sigma Factor and the Global Regulator GacA. Appl Environ Microbiol. 2004;70:5493–5502. doi: 10.1128/AEM.70.9.5493-5502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidle A, Allesen-Holm M, Riedel K, Berg G, Givskov M, Molin S, Eberl L. Identification and characterization of an N-acylhomoserine lactone-dependent quorum-sensing system in Pseudomonas putida strain IsoF. Appl Environ Microbiol. 2002;68:6371–6382. doi: 10.1128/AEM.68.12.6371-6382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo-Ferro C, Reil G, Gorg A, Eberl L, Riedel K. Biofilm formation of Pseudomonas putida IsoF: the role of quorum sensing as assessed by proteomics. Syst Appl Microbiol. 2005;28:87–114. doi: 10.1016/j.syapm.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dubern JF, Lugtenberg BJ, Bloemberg GV. The ppuI-rsaL-ppuR quorum-sensing system regulates biofilm formation of Pseudomonas putida PCL1445 by controlling biosynthesis of the cyclic lipopeptides putisolvins I and II. J Bacteriol. 2006;188:2898–2906. doi: 10.1128/JB.188.8.2898-2906.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geels FP, Schippers B. Reduction in yield depression in high frequency potato cropping soil after seed tuber treatments with antagonist fluorescent Pseudomonas spp. Phytopathol Z. 1983;108:207–221. [Google Scholar]
- Kojic M, Degrassi G, Venturi V. Cloning and characterisation of the rpoS gene from plant growth-promoting Pseudomonas putida WCS358: RpoS is not involved in siderophore and homoserine lactone production. Biochim Biophys Acta. 1999;1489:413–420. doi: 10.1016/s0167-4781(99)00210-9. [DOI] [PubMed] [Google Scholar]
- Kojic M, Venturi V. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J Bacteriol. 2001;183:3712–3720. doi: 10.1128/JB.183.12.3712-3720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassak K, Van Holm L, Duchateau L, Vanderleyden J, De Mot R. Isolation and characterization of fluorescent Pseudomonas associated with the roots of rice and banana grown in Sri Lanka. Plant and Soil. 1992;145:51–63. [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, N.Y; 1972. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor, N.Y; 1989. [Google Scholar]
- Chilton MD, Currier TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink HP, Okker RJH, Wijffelmann CA, Pees E, Lugtemberg BJJ. Promoter in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Better M, Lewis B, Corbin D, Ditta G, Helinski DR. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell. 1983;35:479–485. doi: 10.1016/0092-8674(83)90181-2. [DOI] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampioni G, Polticelli F, Bertani I, Righetti K, Venturi V, Zennaro E, Leoni L. The Pseudomonas Quorum-Sensing Regulator RsaL Belongs to the Tetrahelical Superclass of H-T-H Proteins. J Bacteriol. 2007;189:1922–1930. doi: 10.1128/JB.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PD, Ping G, Daly SL, Cha C, Cronan JE Jr, Rinehart KL, Farrand SK. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrand SK, Qin Y, Oger P. Quorum-sensing system of Agrobacterium plasmids: analysis and utility. Methods Enzymol. 2002;358:452–484. doi: 10.1016/s0076-6879(02)58108-8. [DOI] [PubMed] [Google Scholar]
- Alexeyev MF. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques. 1999;26:824–826. doi: 10.2144/99265bm05. 828. [DOI] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Stachel SE, An G, Flores C, Nester EW. A Tn3 lacZ transposon for the random generation of b-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. Embo J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]