Abstract
Plants and invertebrates can protect themselves from viral infection through RNA silencing. This antiviral immunity involves production of virus-derived small interfering RNAs (viRNAs) and results in specific silencing of viruses by viRNA-guided effector complexes. The proteins required for viRNA production as well as several key downstream components of the antiviral immunity pathway have been identified in plants, flies, and worms. Meanwhile, viral mechanisms to suppress this small RNA-directed immunity by viruses are being elucidated, thereby illuminating an ongoing molecular arms race that likely impacts the evolution of both viral and host genomes.
Introduction
Two major lines of evidence indicate that several RNA silencing components form the core of an antiviral defense in plants and invertebrates. First, virus-derived siRNAs (viRNAs) targeting viral RNAs accumulate during infections. Second, to counteract this defense, viruses produce essential virulence factors called viral suppressors of RNA silencing (VSRs). Here, we review recent progress in understanding the mechanisms for production and function of viRNAs in plants and invertebrates. We emphasize how deciphering the genetics of antiviral RNA silencing in these organisms has been instrumental in elucidating the mode of action of VSRs. We also discuss emerging evidence that viruses often impinge on or subvert small RNA-directed functions, notably in mammals, and we underline the consequences of the intimate interactions between viruses and RNA silencing pathways for the evolution of both viral and host genomes.
Viruses and RNA Silencing
Viruses are obligate intracellular pathogens that infect all forms of life. Their genomes, packaged into virions, comprise DNA or RNA that is either single-stranded (ss) or double-stranded (ds). Both dsRNA and ssRNA viruses— the latter being further divided into positive-sense (+) and negative-sense (−) ssRNA viruses—encode their own RNA-dependent RNA polymerase (RdRP, or RDR for cellular RdRPs) for error-prone replication. RNA genomes can be replicated via DNA intermediates through the action of viral reverse transcriptases, as exemplifed by retroviruses, or via RNA intermediates through the activity of host DNA-dependent RNA polymerases, exemplifed by viroids. Viroids are composed of ~300 nucleotide circular ssRNAs and are pathogenic to many plant species.
Organisms have diverse mechanisms for combating viral infections. One mechanism—discovered first in plants and subsequently in invertebrates—is through RNA silencing. This mechanism of gene silencing depends upon small RNAs that are 21–30 nucleotides in length and are divided into three main classes: small interfering RNAs (siRNAs), microRNAs (miRNAs), and Piwi-associated interfering RNAs (piRNAs). siRNAs and miRNAs are processed as duplexes from dsRNA precursors by an RNaseIII enzyme called Dicer (Hammond, 2005). Perfectly base-paired dsRNAs are the precursors of siRNA populations. In contrast, primary miRNA transcripts (pri-miRNA) contain imperfect intramolecular stem loops and are first processed within the nucleus. The resulting precursor-miRNAs (pre-miRNA) are then converted into a single mature miRNA species in the cytoplasm (Bartel, 2004). In plants, however, processing of miRNAs occurs entirely in the nucleus (Vaucheret, 2006). miRNAs regulate important biological processes, and, hence, plants and animals with compromised miRNA functions display severe developmental defects (Bartel, 2004; Vaucheret, 2006). In the fruit fly Drosophila, miRNAs and siRNAs are products of two distinct Dicers, Dicer-1 (Dcr1) and Dicer-2 (Dcr2), respectively (Hammond, 2005). On the other hand, worms and vertebrates only have one Dicer that produces both miRNAs and siRNAs. The model plant Arabidopsis thaliana encodes four Dicer-like proteins (DCL1 to DCL4): DCL1 primarily synthesizes miRNAs (Bartel, 2004), whereas DCL2, DCL3, and DCL4 process long dsRNA molecules of various cellular origins into siRNA populations that are 22, 24, and 21 nucleotides in length, respectively (Brodersen and Voinnet, 2006). In plants, fungi, and worms, siRNA production is amplified through de novo dsRNA synthesis by cellular RNA-dependent RNA polymerases, leading to secondary siRNA accumulation (Wassenegger and Krczal, 2006). In contrast to siRNAs and miRNAs, piRNAs—which are ~30 nucleotides in length and are found in the germline of flies and vertebrates—are Dicer independent (Zamore, 2007).
Effector complexes called RNA-induced silencing complexes (RISCs) are assembled upon loading of one selected small RNA strand into one member of the Argonaute (Ago) protein family (Tolia and Joshua-Tor, 2007). Drosophila Ago1 and Ago2 recruit miRNAs and siRNAs, respectively, whereas piRNAs interact with one of three related proteins, Piwi, Aubergine, and Ago3 (Zamore, 2007). Ago proteins are often named Slicer proteins because they cleave target ssRNAs at the duplex formed with the guide-strand small RNA (Tolia and Joshua-Tor, 2007). Nonetheless, miRNAs can guide gene silencing through translational arrest without slicing (Bartel, 2004), and siRNAs can mediate transcriptional nuclear gene silencing through DNA and/or histone methylation. For example, in Arabidopsis, DCL3-dependent 24 nucleotide siRNAs recruit AGO4 to transcriptionally silence transposons and DNA repeats through chromatin modifications (Matzke and Birchler, 2005). Small RNAs also accumulate upon virus infection in plants and invertebrates, but their origin and the involvement of Dicer and Argonaute proteins in their processing and action were not well understood until recently.
Small RNA-Directed Antiviral Immunity
Origin, Processing, and Stability of Virus-Derived siRNAs
viRNAs from (+)ssRNA viruses were thought to be populations of siRNAs produced from long dsRNA replication intermediates (Figure 1). Accordingly, cloning and sequencing showed approximately equal ratios of (+) and (−) strand viRNAs derived from several (+)ssRNA plant viruses (Ho et al., 2006; Yoo et al., 2004). However, far more abundant (+)viRNAs than (−)viRNAs were detected in plants infected with tombusviruses and carmoviruses, and, notably, many (+)viRNAs mapped to discrete intramolecular hairpins within viral genomic RNA (Ho et al., 2006; Molnar et al., 2005; Figure 1). This finding was surprising, as those secondary structures should be strongly selected against by viruses, unless they are required for viral genome expression, replication, or encapsidation. For example, the 35S polycistronic transcript of the dsDNA plant virus Cauliflower mosaic virus contains an extensive secondary structure, the “35S leader” (Figure 1), which, in spite of being a major viRNA source, is preserved in all Cauliflower mosaic virus strains, possibly because it is crucial for ribosome shunting during translation (Moissiard and Voinnet, 2006). Similarly, viroid genomic-strand viRNAs accumulate much higher than antigenomic (replicating)-strand viRNAs, suggesting that they largely originate from hairpins formed by extensive folding of the covalently closed viroid RNA genome, necessary for virulence and spread (Itaya et al., 2007). Therefore, plant antiviral Dicers are probably stimulated by both long dsRNA replication intermediates and imperfect RNA hairpins required for the pathogen’s biology. Other plant viRNA sources include dsRNA segments formed by overlapping sense-antisense transcripts produced by circular geminivirus ssDNA genomes (Figure 1). The origin of viRNAs from invertebrate viruses is yet to be characterized.
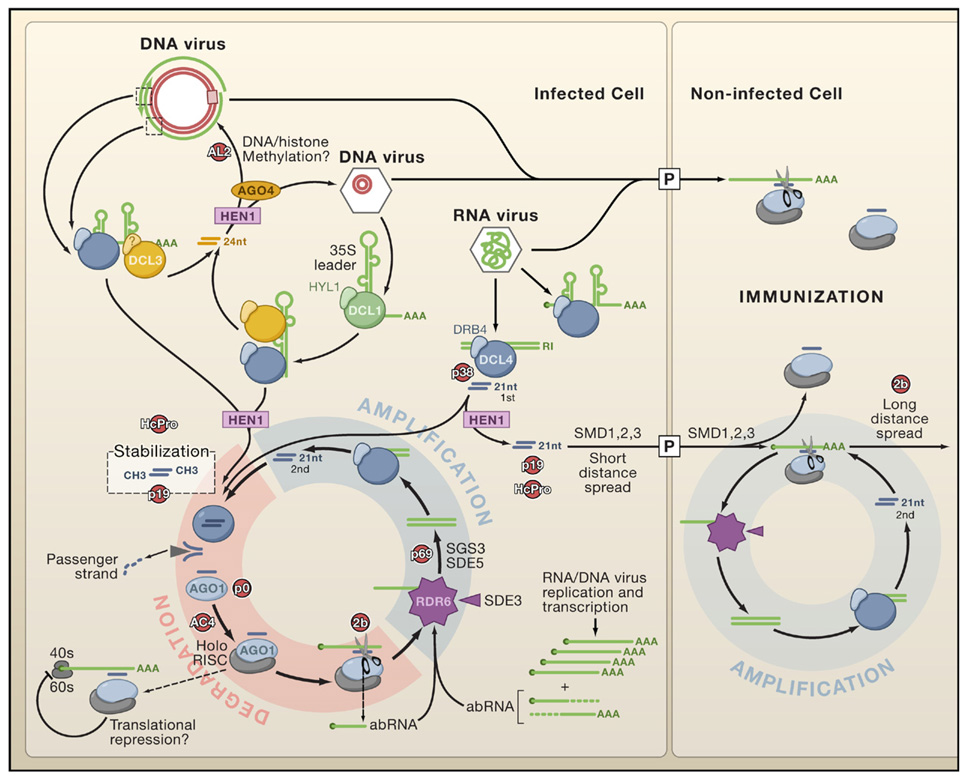
Figure 1. Antiviral Silencing in Arabidopsis.
Dicer-like proteins (DCLs) are represented in association with known and unknown cognate double-stranded (ds)RNA-binding proteins (DRBs). Note the indirect implication of DCL1 in viRNA biogenesis from DNA viruses (e.g., Cauliflower mosaic virus) and the putative contribution of DCL3-dependent viRNAs to viral DNA/histone methylation. DCL4 is the primary Dicer to detect RNA viruses and is replaced by DCL2 if suppressed (for example by the VSR P38; see also Figure 3B). AGO1 is presented as a major antiviral slicer, but other AGO paralogs are likely to be involved, potentially also mediating translational repression. All viRNAs are stabilized through HEN1-dependent 2′-O-methylation. The figure shows how primary viRNAs (1st) are amplified into secondary viRNAs (2nd) in the RDR6-dependent pathway. A similar scheme is anticipated with the salicylic acid-induced RDR1 (not shown; Diaz-Pendon et al., 2007). Aberrant (ab) viral mRNAs lacking a cap or polyA tail (AAA) can enter RNA-dependent RNA polymerase pathways independently of 1st viRNA synthesis. A DCL4-dependent silencing signal (arbitrarily depicted as free 21 nucleotide viRNAs) moves through the plasmodesmata (P) to immunize neighboring cells. Movement may be enhanced through further rounds of amplification involving viral transcripts that enter immunized cells. VSRs and potential endogenous silencing suppressors (red) represent genetic rather than direct physical interactions with host silencing components.
In Drosophila, mutations in the two Dicer genes uncouple miRNA from siRNA biogenesis (Hammond, 2005). Because dcr2 knockout adults develop normally, they were used to establish Dcr2 as a major immunity determinant against four distinct (+)ssRNA insect viruses: flock-house virus, Drosophila C virus, cricket paralysis virus, and Sindbis virus (Galiana-Arnoux et al., 2006; van Rij et al., 2006; Wang et al., 2006). Flock-house virus titers are also increased in flies deprived of R2D2, a dsRNA-binding protein required downstream of dicing (Figure 2A), but, unlike in dcr-2 mutants, viRNAs accumulate normally in r2d2 mutants (Wang et al., 2006), implying that Dcr2 produces these molecules. In contrast, accumulation of Drosophila X virus (dsRNA genome) was unaffected in dcr2 mutant flies (Zambon et al., 2006), suggesting that Dcr-1 or perhaps the nuclease involved in piRNA biogenesis may be involved. Nonetheless, viRNAs remain to be detected in Drosophila X virus-infected flies.
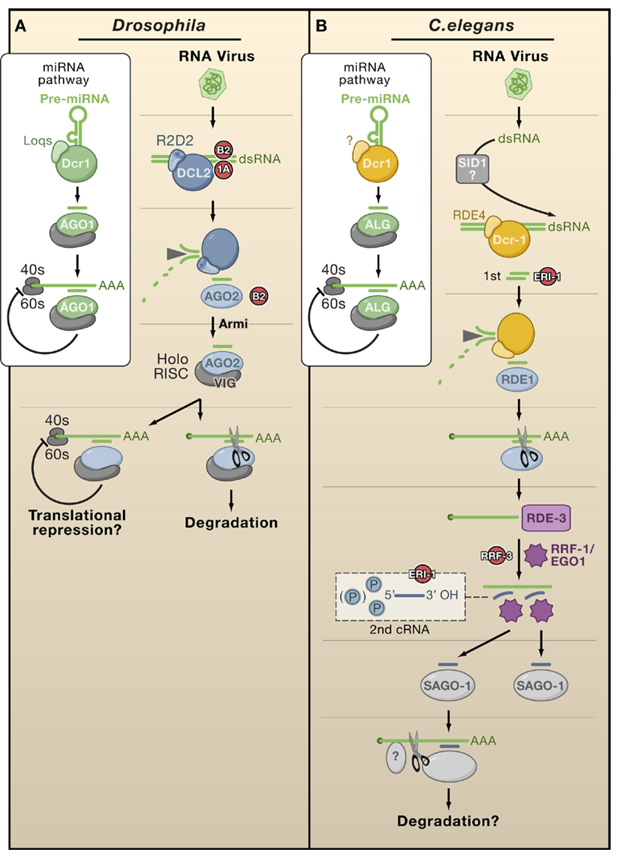
Figure 2. Antiviral Silencing in Flies and Worms.
(A) The Drosophila pathway is conceptually similar to a linear antiviral silencing pathway in plants. Although R2D2 heterodimerizes with Dcr2, it is required for loading but not dicing of viRNA; the Armitage (Armi) protein allows assembly of the RISC holoenzyme. The box illustrates the involvement of Dcr1 and AGO1 in the miRNA pathway leading to translational repression. This pathway can be disrupted at multiple points by VSRs (red).
(B) Antiviral silencing in C. elegans has been inferred through studies of artificial infection systems. ALG, RDE-1, and SAGO are worm AGOs that recruit miRNAs, 1st siRNAs, and 2nd siRNAs, respectively. RRF-1 is thought to produce 2nd siRNAs or to copy RNAs (cRNAs) directly from RDE3-stabilized templates. SID-1 may possibly take up viral dsRNAs into cells. These pathways can be disrupted at multiple points by endogenous suppressors (red).
Although natural C. elegans viruses have not been isolated, replication of flock-house virus and vesicular stomatitis virus—a (−)ssRNA virus that naturally infects biting flies and livestock—induces antiviral silencing in adult worms and embryonic cells, respectively, and detection of vesicular stomatitis virus-specific viRNAs was reported (Lu et al., 2005; Schott et al., 2005; Wilkins et al., 2005). Because vesicular stomatitis virus replication is enhanced in cells partially deprived of Dcr-1 (Schott et al., 2005), the sole Dicer of C. elegans likely has antiviral roles in worms (Figure 2B). However, Dcr-1 processing of viral RNA awaits biochemical demonstration.
Individual mutations in the four Arabidopsis DCLs had not been convincingly linked to changes in virus susceptibility, suggesting functional redundancy in antiviral immunity (Brodersen and Voinnet, 2006). Accordingly, recent studies demonstrate that inactivating both DCL4 and DCL2 is necessary and sufficient to elicit the highest susceptibility to several (+)ssRNA viruses and to dramatically decrease viRNA accumulation (Bouché et al., 2006; Deleris et al., 2006; Fusaro et al., 2006; Diaz-Pendon et al., 2007). DCL4 is the primary antiviral Dicer against these (+)ssRNA viruses and produces 21 nucleotide-long viRNAs. viRNA synthesis by DCL2 is hardly detectable when DCL4 is functional, but DCL2 rescues antiviral silencing if DCL4 is genetically inactivated or suppressed, producing 22 nucleotide-long viRNAs (Figure 1). DCL3-dependent synthesis of 24 nucleotide-long viRNAs has been only detected in wild-type plants infected with Tobacco rattle virus and Cucumber mosaic virus and in single or double dcl2/dcl4 mutants infected with Turnip crinkle virus. Although loss of DCL3 activity in dcl2/dcl4 mutants further increased virus titers, 24 nucleotide viRNAs were inactive in targeting homologous mRNA for degradation, presumably because DCL3 products normally guide chromatin modifications (Figure 3C). The contribution of the miRNA-specific DCL1 to immunity against (+)ssRNA viruses is negligible because dcl2/dcl3/dcl4 and dcl1/dcl2/dcl3/dcl4 mutants showed unaltered susceptibility to Cucumber mosaic virus or Turnip crinkle virus, and DCL1-dependent viRNAs were hardly detectable even in the dcl2/dcl3/dcl4 mutant. Therefore, hierarchy and redundancy link Arabidopsis DCLs to dsRNA produced by cytoplasmically replicating (+)ssRNA viruses. In contrast, the four DCLs cooperate in defense against two DNA viruses replicating in the nucleus: Cauliflower mosaic virus and a geminivirus (ssDNA virus). DCL4 and DCL3 were the prevalent Dicers, whereas DCL2 activity was mostly evident following DCL4 inactivation (Blevins et al., 2006; Moissiard and Voinnet, 2006). Increased susceptibility to Cauliflower mosaic virus and reduced viRNA accumulation were only observed if DCL4, DCL2, and DCL3 were all disabled (Moissiard and Voinnet, 2006). Thus, unlike in (+)ssRNA virus infections, DCL3 has clear antiviral roles in natural DNA virus infections. Moreover, DCL1 was required for optimal accumulation of DCL4- and DCL3-dependent viRNAs from the Cauliflower mosaic virus 35S leader (Figure 1). DCL1 likely excises such hairpins from primary transcripts and thereby facilitates their subsequent access by the other DCLs, a process resembling the nuclear and DCL1-dependent pri-miRNA to pre-miRNA conversion step. This process is not likely to affect hairpin structures within RNA viruses, which are cytoplasmic.
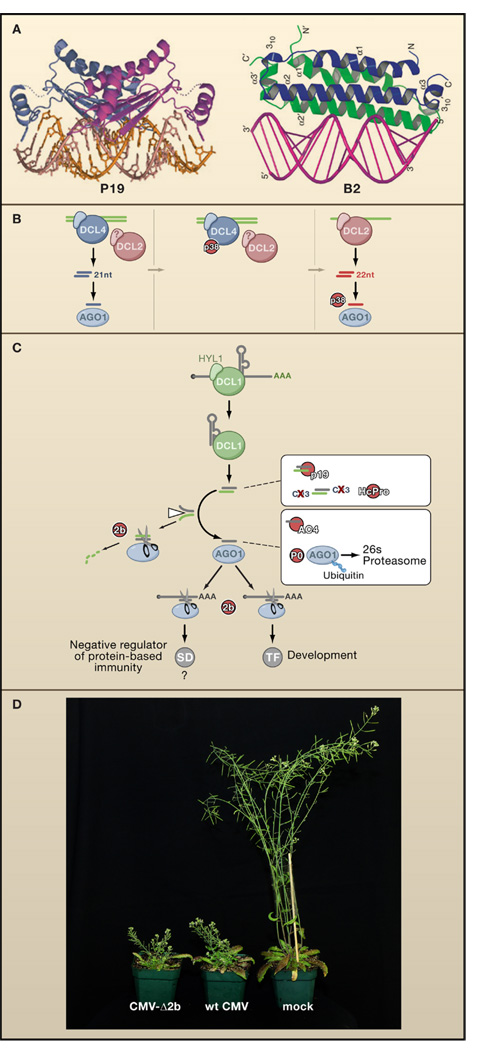
Figure 3. VSRs of Plant and Fly Viruses.
(A) The VSRs P19, encoded by Cymbidium ringspot virus, and B2, encoded by flock-house virus, both bind dsRNA but with very different structural requirements. P19 acts as a head-to-tail homodimer that binds to and specifically measures 21 bp duplexes that are the products of DCL4. In contrast, B2 forms a four-helix bundle that binds to one face of an A-form RNA duplex, independent of its length. P19 in complex with siRNA, reprinted by permission from MacMillan Publishers Ltd: Nature (98), copyright 2003, and B2 in complex with dsRNA, adapted by permission from MacMillan Publisher Ltd: Nat. Struct. Mol. Biol. (109), copyright 2005.
(B) Inhibition of DCL4 by the P38 VSR of Turnip crinkle virus reveals the redundant antiviral function of DCL2 which generates 22 nucleotide instead of 21 nucleotide viRNAs in Turnip crinkle virus-infected Arabidopsis. The antiviral activity of DCL2-dependent viRNAs is in turn further compromised by P38, possibly through inhibition of AGO1.
(C) Transgenically expressed VSRs interfere with the Arabidopsis DCL1-dependent miRNA pathway. In contrast to the genetic interactions in Figure 1, these interactions are likely to be physical. VSRs may interfere both with developmental programs mediated by transcription factors (TF) and innate immune pathways negatively regulated by suppressors of defense (SD).
(D) Both wild-type Cucumber mosaic virus (middle) and a version lacking the VSR 2b (CMV-Δ2b, left) induce similarly severe stunting symptoms in dcl2/dcl4 double mutant plants, demonstrating that the VSR is dispensable for infection and disease induction in a host defective in small RNA-directed immunity (Diaz-Pendon et al., 2007). Image courtesy of R. Lu.
The hierarchy of DCL action may be influenced by the subcellular localization of viral dsRNA and DCLs and by intrinsic DCL affinities for various dsRNA forms. All four Arabidopsis DCLs, perhaps with the exception of DCL2, appeared to localize to the nucleus based on reportergene fusion experiments (Vaucheret, 2006). Yet, under appropriate genetic circumstances, each DCL can process viral dsRNA of either cytosolic or nuclear origin. Thus, either the DCL localization data are inaccurate or some cytoplasmic viral dsRNAs are imported into the nucleus for processing. Alternatively, DCLs might relocalize during infection. How, when, and where are Dicers expressed during infection? How does their cellular distribution compare with the sites of viral RNA replication, translation, or encapsidation? These questions deserve careful attention both in plants and animals.
The different DCL affinities for various dsRNA substrates could depend on dsRNA-binding proteins (DRB) known to physically and specifically interact with Dicers in several organisms. In Arabidopsis, DRB1 (also known as HYL1) facilitates DCL1-dependent miRNA production from their fold-back precursors (Bartel, 2004; Vaucheret, 2006), and DRB4 enhances DCL4 processing of long dsRNAs involved in endogenous gene silencing (Vaucheret, 2006; Figure 1). Drosophila Dcr1 interacts with Loquacious (Loqs or R3D1) to process miRNAs, and Dcr2 associates with R2D2 during RNA interference (RNAi; Hammond, 2005). The closest C. elegans R2D2 homolog, RDE-4, interacts with Dcr-1 (Hammond, 2005). Mutations in rde-4 enhance vesicular stomatitis virus replication in worm cells (Schott et al., 2005), and r2d2 mutant flies accumulate greater titers of flock-house virus, cricket paralysis virus, and Drosophila X virus (Wang et al., 2006; Zambon et al., 2006), indicating a key role for those dsRNA-binding proteins in antiviral immunity (Figures 2A and Figure 2B). Assigning such a role to the fve Arabidopsis dsRNA-binding proteins has been comparatively difficult because of possible functional redundancy.
Specific proteins influence small RNA stability upon dicing. Arabidopsis HEN1 methylates the 2′ hydroxy groups at the 3′-end termini of cellular small RNAs, protecting them from degradation (Chen, 2007). Resistance to beta-elimination indicates that plant viRNAs are also methylated (Blevins et al., 2006; Figure 1). Moreover, hen1 mutants accumulate less viRNAs from RNA and DNA viruses and exhibit reduced virus-induced gene silencing (Blevins et al., 2006). 3′-end methylation of animal piRNAs has just been reported (Chen, 2007), and, thus, animal viRNAs might carry similar protective modifications. In C. elegans, several mutations enhance RNAi, one of which is Eri-1, encoding an exonuclease thought to degrade worm small RNAs (Grishok, 2005; Figure 2B). Embryonic cells and adult worms lacking Eri-1 function are only slightly more resistant to vesicular stomatitis virus, an effect strongly enhanced by additional mutations in lin-15B, a suppressor of somatic RNAi (Schott et al., 2005).
Antiviral RNA-Induced Silencing Complexes
The mere existence of plant and invertebrate antiviral RISCs has been debated because Dicer-mediated processing of viral dsRNA is, in principle, sufficient to impede virus replication. Nonetheless, viRNAs inhibit expression of homologous cellular transcripts in trans. This has been shown with plant and insect recombinant viruses whose genomes incorporate fragments of host or reporter transcripts. Upon infection, the viral symptoms mimic knockdown mutations in the corresponding mRNA, a process called virus-induced gene silencing (Ruiz et al., 1998; Uhlirova et al., 2003). Virus-induced gene silencing elicited by Tobacco rattle virus was used to explore the requirements of a putative RISC in Arabidopsis antiviral defense (Deleris et al., 2006). Virus-induced gene silencing was unaltered in dcl2/dcl3 and dcl3/dcl4 double mutants, but was abolished in dcl2/dcl4 plants, which exhibited strong symptoms and high viral titers. However, there were equivalent levels of 21, 22, and 24 nucleotide viRNAs in dcl2/dcl3, dcl3/dcl4, and dcl2/dcl4 plants, respectively, suggesting that dicing per se is not sufficient for antiviral immunity (Deleris et al., 2006). Direct evidence for a viRNA-loaded RISC came from studies of Cymbidium ringspot virus (a (+)ssRNA tombusvirus; Pantaleo et al., 2007). Because Cymbidium ringspot virus viRNAs derive mostly from discrete hot spots within the (+)RNA strand, a viRNA-guided RISC was expected to target symmetrical positions in the (−)RNA strand. These cleavage fragments were indeed detected and carried nontemplated U residues at predicted cut sites, a signature of RISC-directed cleavage. A likely antiviral slicer in plants is AGO1, a major component of the miRNA-guided RISC (Figure 1 and Figure 3C). First, AGO1 has the catalytic residues diagnostic of cleavage-competent AGOs (Baumberger and Baulcombe, 2005). Second, ago1 hypomorphic mutants are more susceptible to viruses (Morel et al., 2002). Third, AGO1 immunoprecipitates with viRNAs in infected tissues (Zhang et al., 2006), and fourth, Cymbidium ringspot virus-derived viRNAs and cellular miRNAs cofractionate in two protein complexes that likely correspond to free AGO1 (~150 kDa) and partially or fully assembled (holo) RISC (~650 kDa), respectively (Pantaleo et al., 2007). A similarly sized complex was isolated in separate experiments involving a virus related to Cymbidium ringspot virus. This complex contained viRNAs and exhibited in vitro nuclease activity that was virus sequence preferential and ssRNA specific (Omarov et al., 2007).
Drosophila Ago2 was the first component identified in the antiviral RNA silencing pathway of the animal kingdom (Li et al., 2002). Dcr2 expression fails to restrict flock-house virus replication in fly embryos deprived of the siRNA-guided slicer Ago2 (Wang et al., 2006), whereas ago2 mutants are hypersusceptible to Drosophila C virus and Drosophila X virus (van Rij et al., 2006; Zambon et al., 2006). Likewise, knockdown of Anopheles gambiae Ago2 increases viral titers in cell culture and adults (Li et al., 2004; Keene et al., 2004). Therefore, as in plants, dicing alone is insufficient for insect antiviral immunity, and the additional function of Ago2 is required (Figure 2A). Other mutations downstream of dicing in the dsRNA-siRNA pathway enhance Drosophila and C. elegans susceptibility to viruses. Flock-house virus and vesicular stomatitis virus replicate more in worms deprived of the Ago2 paralog, RDE1, and of RDE4, which links Dcr-1 to RDE1 (Lu et al., 2005; Schott et al., 2005; Wilkins et al., 2005). Similarly, components of the Ago2-associated RISC restrict Drosophila X virus accumulation in fly, including the Vasa intronic gene dsRNA helicase (VIG), armitage, and r2d2, required for RISC assembly and siRNA loading (Zambon et al., 2006; Figure 2). Nonetheless, although these factors likely form an antiviral holoRISC, sliced viral RNA fragments await characterization in Drosophila and C. elegans.
viRNA-loaded RISCs could also inhibit viral mRNA translation, as do plant and animal miRNA-loaded RISCs (Bartel, 2004). Furthermore, RISCs directing DNA/histone methylation could recruit viRNAs to impact DNA viruses. For example, de novo asymmetric cytosine methylation—a hallmark of DCL3/Ago4-directed hetero-chromatic silencing—occurs on both DNA strands of Tomato leaf curl virus and restricts its replication (Bian et al., 2006; Figure 1). A general issue of antiviral RISCs is functional diversification: there are 26, 10, and 5 AGO-like proteins in C. elegans, Arabidopsis, and Drosophila, respectively, and many have the catalytic residues of authentic Slicers (Tolia and Joshua-Tor, 2007). Perhaps there is no single antiviral RISC in those organisms but, rather, a multilayered network of defensive complexes (Lu et al., 2005) with redundancy and specialization. Hence, the antiviral function of worm and fly miRNA-directed Agos has not been examined yet. Moreover, Piwi and Aubergine, two Drosophila Slicers involved in the germline piRNA pathway that silences retrotransposons, inhibit Drosophila X virus replication (Zambon et al., 2006). Another unresolved issue concerns target accessibility: the quasi-rod-shaped RNA genome of viroids is an effective DCL substrate but is largely inaccessible to slicing, presumably because RISC cannot resolve extensive intramolecular RNA folds (Itaya et al., 2007). By extension, dicing hot spots found within virion RNA strands may not be efficiently sliced, agreeing with the mapping of several RISC-directed cleavage sites on the Cymbidium ringspot virus genomic (+)ssRNA (Pantaleo et al., 2007). Interestingly, the same study showed no cleavage of the replicating (−)ssRNA even through cleavage was detected if a homologous RNA was expressed transgenically, suggesting that replication within membrane-bound vesicles normally protects the (−)ssRNA from (+)viRNA-loaded RISC.
Amplification and Spread of Antiviral Silencing
RNA silencing can be amplified by cellular RNA-dependent RNA polymerases identified in plants, C. elegans, and fungi, but not in insects or vertebrates which do not possess these enzymes (Wassenegger and Krczal, 2006). In plants, amplification entails at least two mechanisms. In the first mechanism, primary siRNAs derived from viral- or transgene-dsRNA recruit RNA-dependent RNA polymerases to homologous ssRNA, from which complementary RNA synthesis and subsequent secondary siRNA production ensues. These secondary RNAs are not related to the primary targeted site. This process—called transitivity—occurs in virus-induced gene silencing of transgenes, during which transgene-specific siRNAs from regions absent in the recombinant viral genome accumulate (Voinnet, 2005). Targeting transgenes through transitivity in Arabidopsis requires RDR6, the RNA-helicase SDE3, and SDE5, which resembles human mRNA export factor TAP (Voinnet, 2005; Hernandez-Pinzon et al., 2007). Together with AGO1 and the coiled-coil protein SGS3, RDR6, SDE3, and possibly SDE5 mediate a second RNA-dependent RNA polymerase mechanism, in which primary siRNA production is dispensable (Voinnet, 2005; Vaucheret, 2006). This mechanism likely perceives aberrant RNAs spuriously produced by transgenes, transposons, or viruses and converts them de novo into dsRNA. Aberrations include the lack of 5′ cap or polyA tail, frequent in aborted viral transcription products (Figure 1; Brodersen and Voinnet, 2006).
The two RNA-dependent RNA polymerase mechanisms are not mutually exclusive, and both are likely required to keep pace with viral replication. Arabidopsis mutants lacking components of the AGO1-RDR6-SGS3-SDE5 pathway support higher Cucumber mosaic virus titers comparable to those in dcl2/dcl4 mutant plants (Bouché et al., 2006; Deleris et al., 2006; Vaucheret, 2006). These results—together with those from recent studies of transgene silencing (Moissiard et al., 2007)—suggest that the same DCL2-DCL4 consortium is required for synthesis of primary and secondary small RNAs, although secondary viRNA accumulation has been experimentally verified only recently (Diaz-Pendon et al., 2007). Tobacco plants with reduced RDR6 activity also exhibit temperature-dependent hypersusceptibility to several unrelated (+)ssRNA viruses (Qu et al., 2005; Schwach et al., 2005). No antiviral role has been ascribed so far to RDR2, involved in the DCL3-AGO4 heterochromatic silencing pathway (Matzke and Birchler, 2005). However, the existence of six Arabidopsis RNA-dependent RNA polymerase paralogs (of which only RDR1, RDR2, and RDR6 have established functions) suggests both redundancy and specialization in antiviral defense, as seen with DCLs and, possibly, with DRBs and AGOs. Consistent with this idea, reducing the activity of RDR1 in tobacco increases susceptibility to tobamo-, tobra-and potex-viruses, but not to Cucumber mosaic virus (Yang et al., 2004; Yu et al., 2003).
C. elegans RNA-dependent RNA polymerase mechanisms differ from those of plants. In worms, siRNAs accumulating in response to exogenous dsRNA are predominantly secondary siRNAs produced through unprimed RNA synthesis (Grishok, 2005). Unlike Dcr-1-dependent primary siRNAs, secondary siRNAs are exclusively antisense, have 5′ di- or triphosphates, and are thought to occur as individual RNA-dependent RNA polymerase products, or “copy” (c)RNA, directly synthesized from target mRNA upon primary siRNA-guided cleavage (Pak and Fire, 2007; Sijen et al., 2007; Figure 2B). Three of the four C. elegans RNA-dependent RNA polymerases have been studied: RRF-1 is mandatory for somatic RNAi, and EGO-1 is its germline-specific counterpart; RRF-3 negatively regulates RRF-1 and EGO-1, possibly by competing for RNAi effectors (Grishok, 2005). RRF-1 function might require the RNAi component RDE-3, a putative beta-nucleotidyltransferase that could stabilize RRF-1 templates. Because vesicular stomatitis virus replication is enhanced in rrf-1 and rde-3 mutant cells and decreased in rrf-3, RNAi amplification likely contributes to antiviral immunity (Schott et al., 2005; Wilkins et al., 2005; Figure 2B). It will be important to determine which proportion of viRNAs have the distinctive features of (c)RNAs because the (c)RNAs involved in experimental RNAi are only in their minority associated with the AGO homolog RDE1 (Yigit et al., 2006), which is, nonetheless, crucial for antiviral defense.
In plants, RNA-dependent RNA polymerases influence the extent to which silencing moves from cell-to-cell through plasmodesmata and over long distances through the phloem (Voinnet, 2005). Cell-to-cell RNAi spread involves two interrelated processes differing in their requirement for transitivity. In short-range signaling (10–15 cells) the specific activity of DCL4, but not of RDR6 or SDE3, is mandatory for synthesis or detection of a signal, whose movement necessitates at least three SILENCING MOVEMENT DEFICIENT genes (SMD1 to −3; Voinnet, 2005). Long-range cell-to-cell signaling requires that RDR6, SDE3, and possibly SDE5 convert homologous transcripts into new dsRNA in cells receiving the short-range signal (Voinnet, 2005). DCL4 processes the new dsRNA into secondary 21 nucleotide siRNAs, and movement proceeds over another 10–15 cells. Short- and long-range signaling events are related to primary and secondary viRNA synthesis, respectively, and likely impact virus movement (Figure 1). Hence, VSR-deficient Cymbidium ringspot virus and Turnip crinkle virus accumulate in vascular bundles, yet fail to unload into neighboring cells, which, although virus-free, exhibit sequence-specific resistance to secondary infection (Deleris et al., 2006; Havelda et al., 2003). As this phenomenon is alleviated in Arabidopsis loss-of-function dcl4 mutant, cell-to-cell spread and amplification of DCL4-dependent signals likely immunize tissues ahead of the infection (Deleris et al., 2006; Figure 1). Long-distance silencing movement through the vasculature, demonstrated with Potato virus × (Voinnet, 2005), also likely has antiviral roles because it is precluded by the Cucumber mosaic virus-encoded VSR protein 2b (Guo and Ding, 2002). Upon vascular transport, silencing amplification helps immunizing recipient tissues, as RDR6 activities enabling detection/amplification of longdistance transgene silencing exclude viruses from meri-stems—the stem cell niches at apical growing points (Qu et al., 2005; Schwach et al., 2005).
Systemic RNAi in C. elegans involves many tissue-specific components (Voinnet, 2005). However, a near-ubiquitous player is SID-1, a transmembrane channel enabling long dsRNA uptake in target organs (Figure 2B). Although no biological function has been ascribed to SID-1, systemic antiviral defense is now a testable possibility due to the establishment of flock-house virus replication in adult worms (Lu et al., 2005). The impact of RNA-dependent RNA polymerases in systemic antiviral silencing might be harder to evaluate because their function is, above all, mandatory for cell-autonomous silencing. Nonetheless, systemic transitive RNAi observed in C. elegans (Voinnet, 2005) could entail antiviral effects of RRF-1 and RDE-3 in whole organisms. The cell-autonomy of RNAi triggered by an inverted-repeat transgene and the absence of recognizable genome-encoded RNA-dependent RNA polymerases in Drosophila have prompted the idea that arthropods lack systemic silencing. However, injected dsRNA allows robust systemic gene knockdown in adult Drosophila, Tribolium castaneum and A. gambiae (Voinnet, 2005). Furthermore, injected dsRNA, but not siRNAs, confers systemic, sequence-specific antiviral immunity to adult shrimps (Robalino et al., 2005). Therefore, dsRNA dissemination mechanisms, together with recently discovered dsRNA uptake mechanisms based on receptor-mediated endocytosis (Saleh et al., 2006), may well contribute to arthropod antiviral defense.
Viral Suppressors of RNA Silencing
dsRNA Binding by VSRs: Separating the Wheat from the Tares
More than 35 individual VSR families have been identifed from virtually all plant virus types, unraveling a necessary and ubiquitous counterstrategy (Li and Ding, 2006). VSRs were also isolated from insect and fungus viruses including flock-house virus (B2), cricket paralysis virus/Drosophila C virus (1A), and Cryphonectria para-sitica hypovirus (P29; Li et al., 2002; Segers et al., 2006; Wang et al., 2006; van Rij et al., 2006). VSRs are strikingly diverse within and across kingdoms because they are often encoded by novel, out-of-frame overlapping genes contained within more ancient genes. Their acquisition is through fast evolutionary convergence, confining VSRs within tight lineages in virus phylogenies. Consequently, similar silencing suppression strategies may evolve independently several times such that unrelated VSRs might share analogous biochemical properties. For example, both Cymbidium ringspot virus-encoded P19 and fock-house virus-encoded B2 proteins display dsRNA-binding activities required for suppression, but X-ray crystallography studies reveal very distinct protein folds in these proteins (Figure 3A). B2 dimerizes into a four-helix bundle that binds A-form RNA duplexes independently of length (Chao et al., 2005). In contrast, P19 head-to-tail homodimers form a “siRNA caliper” that specifically sequesters DCL4-dependent 21 bp RNA duplexes (Vargason et al., 2003; Ye et al., 2003). B2 inhibits long dsRNA processing by Dcr2 in vitro, as does the Drosophila C virus-encoded 1A protein (Lu et al., 2005; Fenner et al., 2007; van Rij et al., 2006). Moreover, point mutations disrupting dsRNA binding by B2 and 1A abolish their VSR activity. viRNA sequestration by P19 prevents their incorporation into RISC, and plants infected with P19-defcient Cymbidium ringspot virus (CymRSV-ΔP19) contain vastly reduced virus titers (Lakatos et al., 2004). Therefore, dsRNA binding by B2, 1A, and P19 is necessary to silencing suppression. These examples have promoted dsRNA binding as a popular feature in characterizing VSRs, but recent results obtained with many additional recombinant viral proteins with suspected or established VSR functions should be cautiously interpreted (Lakatos et al., 2006). First, RNA binding is often nonspecific: the octameric ring formed by monomers of P21, a plant closterovirus VSR, shows equal affinity for long, short, single-, or double-stranded RNAs, providing no obvious insight into its mode of action (Ye and Patel, 2005). In fact, dsRNA binding might often refect additional VSR functions, unrelated to suppression, that require close association to viral nucleic acids. Moreover, in vitro binding assays for many VSRs usually lack crucial negative controls involving stable loss-of-function VSR alleles. Consequently, whether dsRNA binding is a bona fide feature of silencing suppression remains, in most cases, unresolved.
Emerging Themes in VSR Functions
VSRs are often studied as isolated proteins that are transgenically or transiently expressed with a second reporter transgene (Li and Ding, 2006), an approach showing increasing limitations. Any given virus might produce multiple VSRs—as do Citrus tristesa virus and geminiviruses (Lu et al., 2004; Vanitharani et al., 2004)—with probable temporally or spatially restricted functions that could be overlooked in transgenic/transient expression. The experimental silencing systems used in VSR studies can also have misleading outputs: VSRs acting upstream of viRNA synthesis will be inactive against siRNA-induced RNAi; those inhibiting secondary viRNA synthesis will not be recognized in the context of silencing triggered by inverted-repeat-transgenes that produce fold-back RNAs, which does not require RDR activities. Additionally, the level and timing of VSR and silencing trigger expression are usually set arbitrarily such that the results from independent systems might not be comparable and may provide exaggerated or inaccurate views of VSR function during authentic infections. These and other caveats emphasize the value of recent genetic rescue experiments involving viruses with disabled or modified VSRs. For example, out of many dcl combination mutants, only in dcl2/dcl4 double mutants were the VSR-deficient mutants of Turnip crinkle virus (TCV-ΔP38) and Cucumber mosaic virus (CMV-Δ2b) as virulent as their wild-type counterparts in wild-type plants (Deleris et al., 2006; Diaz-Pendon et al., 2007). This clearly implicates pathways initiated by DCL2 and DCL4 as genetic targets of both P38 and 2b (Figures 3B and 3D). Similarly, rescue of B2-defcient flock-house virus in Ago2-depleted cells and in dcr2 and ago2 Drosophila mutant embryos unequivocally implicated the Dcr2-Ago2 pathway in anti-flock-house virus silencing, establishing a molecular framework to characterize B2 function (Li et al., 2002; Wang et al., 2006).
Studying TCV-ΔP38 provided insights into the P38 mode of action and simultaneously unraveled the functional redundancy between DCL2 and DCL4 in viRNA synthesis. DCL2-dependent, 22 nucleotide viRNAs normally accumulate in Turnip crinkle virus-infected wild-type Arabidopsis. However, 21 nucleotide, instead of 22 nucleotide, viRNAs accumulate in response to TCV-ΔP38, indicating that, although DCL4 is the primary Dicer to access Turnip crinkle virus dsRNA, its action is suppressed by P38. Consequently, DCL2 substitutes DCL4 to produce viRNAs 22 nucleotide in length whose activity is, in turn, also compromised by P38 (Deleris et al., 2006; Figure 3B). This dual P38 action readily explains why inactivating DCL2 or DCL4 separately has little impact on wild-type Turnip crinkle virus infection. Therefore, a full appreciation of antiviral silencing requires the use of VSR-deficient viruses because only under these conditions are the redundant effects of VSRs and those of genetic mutations in host silencing factors uncoupled. Further emphasizing this notion, only with CymRSV-ΔP19 was the demonstration of an active viRNA-loaded RISC possible, because most viRNAs are normally sequestered by P19 in wildtype Cymbidium ringspot virus infection (Lakatos et al., 2004; Pantaleo et al., 2007).
Viral Antisilencing Strategies
In Drosophila, the flock-house virus B2 and Drosophila C virus/cricket paralysis virus 1A proteins directly inhibit viral dsRNA processing (Lu et al., 2005; Fenner et al., 2007; van Rij et al., 2006). Nonetheless, VSRs may suppress many additional steps, sometimes simultaneously (Figure 1 and Figure 3C). Tobamovirus infection and the potyviral VSR HcPro mimic mutations in hen1: viRNAs become oligo-urydilated and partially degraded because they lack 2′-O methylation (Ebhardt et al., 2005; Yu et al., 2006), as do P19-bound viRNA duplexes (Yu et al., 2006). The geminivirus VSR AC4 seems to prevent holoRISC assembly by capturing single-stranded small RNAs normally bound by AGOs (Chellappan et al., 2005), whereas the Cucumber mosaic virus 2b protein physically interacts with siRNA-loaded AGO1 and inhibits slicing (Zhang et al., 2006). Because AGO1 immunoprecipitates with Cucumber mosaic virus-derived viRNAs, this finding supports the idea that AGO1 is an antiviral slicer, as does the observation that the P0 VSR of poleroviruses contains an F-box-like domain that likely promotes ubiquitin-dependent proteolysis of AGO1 (Pazhouhandeh et al., 2006). Suppression of sense transgene silencing (RDR6-dependent) but not of silencing induced by inverted repeats (RDR6-independent) by the tymoviral P69 protein suggests that it inhibits viRNA amplification (Chen et al., 2004), although direct suppression of an RNA-dependent RNA polymerase activity is yet to be documented. Several VSRs that sequester, degrade, or inactivate primary viRNAs also inhibit transitivity and de novo dsRNA production by RNA-dependent RNA polymerases (Moissiard et al., 2007), providing possible insights into the suppression of silencing movement ahead of the infection (Figure 1).
AC2 from begomoviruses is a viral transcriptional activator whose nuclear localization, DNA-, and zinc-binding domains are all necessary to its VSR function. Transgenic AC2 indeed acts on host DNA by inducing ~30 genes, of which one possibly encodes a cellular silencing suppressor (Trinks et al., 2005). Similarly, HcPro interacts with a calmodulin-related protein (rgs-CaM) that suppresses silencing when overexpressed (Anandalakshmi et al., 2000). Silencing suppression by AL2, the AC2 paralog of curtoviruses (another genus of the geminiviridae), relies, in contrast, on direct protein-protein interaction with adenosine kinase (Wang et al., 2005). Because adenosine kinase sustains the cellular methyl levels, its inhibition could prevent methylation of viral episomes by DCL3-dependent viRNAs. RNA- rather than protein-based suppression has been reported with Red clover mosaic virus, where RNA elements required for (−)-strand synthesis likely recruit and titrate DCL activities (Takeda et al., 2005). Suppression might not even entail specific VSR functions. Hence, the silencing defense response of Arabidopsis against transfer-DNAs (T-DNA) of virulent Agrobacterium tume-faciens is overcome by expression of T-DNA-encoded oncogenes promoting host cell proliferation (Dunoyer et al., 2006). The dramatic reduction of DCL activities during cell proliferation may also benefit plant geminiviruses that typically reactivate host DNA replication to amplify their genome (Matthews, 1991). Viroid genomes encode no protein yet resist silencing because they are structurally protected against RISC activity (Itaya et al., 2007). Viral genome association with capsid or movement proteins also likely contributes to reduced access by silencing ribonucleases, as does the compartmentalization of their replication into vesicles or organelles. Finally, the high rates of replication and movement of viruses might often outcompete the silencing machinery, both at cellular and tissue levels.
Suppression of Cellular Silencing Pathways
Many antiviral silencing factors are components of cellular pathways regulating host gene expression, and, thus, VSRs are expected to interfere with those pathways. VSRs that sequester small RNAs (e.g., P19) or inhibit slicing by AGO1 (e.g., 2b) commonly stabilize accumulation of host miRNAs in an inactive duplex form because the normally labile passenger strand, called miRNA*, is no longer unwound and degraded (Figure 3C). Consequently, cellular miRNA targets, including developmentally important transcription factor mRNAs, accumulate in cells where they should be cleared by AGO1-dependent miRNA activities (Figure 3C). Therefore, VSR transgenic plants frequently exhibit recurrent developmental anomalies resembling those of hyl1, ago1, or dcl1 mutants (Chapman et al., 2004; Dunoyer et al., 2004; Zhang et al., 2006; Figure 3D). However, the fact that incidental inhibition of miRNA-directed functions brings out some of the symptoms elicited by viruses is probably true only to some extent because these studies involve constitutive or inducible VSR expression in a much broader tissue range than in natural infections. Second, miRNAs and other cellular small RNAs have recently emerged as key regulators of basal and race-specific disease resistance in Arabidopsis (Katiyar-Agarwal et al., 2006; Navarro et al., 2006). These protein-based processes protect plants against many pathogens, including viruses. Therefore, inhibition of endogenous small RNA pathways by VSRs might refect a deliberate viral strategy to inhibit such immune systems (Figure 3C). Studies of the virulence-attenuating hypovirus of Cryphonectria parasitica (chestnut blight fungus) also illustrate how VSR might affect important host traits. The decreased fungal pathogenicity is partly due to reduced asexual sporulation (conidiation), recently ascribed to the hypovirus-encoded P29 papain-like protease (Segers et al., 2006). Because P29 resembles the potyviral HcPro and has VSR activities in C. parasitica, perturbation of fungal RNA silencing pathways required for conidiation could contribute to hypovirus-mediated attenuation of fungal fitness.
RNA Silencing in Vertebrate Viral Infections
Toll-like receptor (TLR)-mediated innate immunity was first established in Drosophila, and subsequent studies showed that TLR-dependent NF-κB signaling is highly conserved between flies and mammals (Imler et al., 2004), as is the case for RNAi. The demonstration of an antiviral role for RNAi in insects intuitively suggested a similar function for RNAi in mammals, and, indeed, some mammalian viral proteins suppress antiviral silencing in fly cells, and experimental RNAi in mammalian cells (Li and Ding, 2006). However, despite their requirement for viral infection, suppression of antiviral silencing has never been demonstrated in mammalian cells for any of these factors. Furthermore, cloning and sequencing of small RNAs from (+ss)RNA virus-infected mammalian cells failed to identify viRNAs (Pfeffer et al., 2004), which are otherwise readily detected in infected plants and invertebrates. On the one hand, mammalian virus infections commonly produce dsRNA, known to trigger broad-spectrum immune responses via extra- (TLR3) and intracellular (PKR/RIG-I/MDA-5) sensors. These responses are modulated by host proteins, such as PACT and Tar-binding protein (TRBP; Gupta et al., 2003; Garcia-Sastre and Biron, 2006). On the other hand, human Dicer processes dsRNA into siRNAs in vitro and in some human cultured cells (Hammond, 2005), and both PACT and TRBP play important roles in mammalian RNA silencing (Lee et al., 2006). Therefore, the mammalian RNAi machinery may have roles in dsRNA-mediated immunity in a manner awaiting further characterization.
It is clear, however, that mammalian viruses entertain intimate interactions with the host miRNA pathway. Several studies suggest that virus infection in mammalian cells might be indirectly counteracted by cellular miRNAs, in contrast to plants and invertebrates, in which viRNAs derive from viral genomes. The miRNA target is the virus itself in the case of the primate foamy retrovirus and vesicular stomatitis virus (Lecellier et al., 2005; Otsuka et al., 2007), whereas a host factor critical to viral gene expression is suppressed in HIV-infected cells (Triboulet et al., 2007). Accordingly, knockdown of Dicer or Drosha enhance HIV replication, whereas accumulation of the cellular antiviral miRNAs is suppressed during infection. Other examples linking viruses to mammalian miRNAs include PACT and TRBP—activator and inhibitor of antiviral innate immunity respectively (Gupta et al., 2003)—both of which are required for human miRNA biogenesis/activity (Tolia and Joshua-Tor, 2007; Lee et al., 2006), whereas the adenoviral PKR-antagonist VA1 RNA inhibits miRNA processing and nuclear export (Cullen, 2006). Several groups of DNA viruses replicating in the nucleus intercept the mammalian miRNA pathway, producing viral miRNAs that may act both in cis, to regulate viral genome expression, and in trans, to alter host gene expression (Cullen, 2006). Cis effects are illustrated by SV40-encoded miRNAs that mediate slicing of the perfectly complementary SV40 early transcripts (Sullivan et al., 2005). This decreases viral T antigen expression, attenuating susceptibility to cytotoxic T cells without reducing virus yield. Trans effects are exemplified by the HSV1-encoded anti-apoptotic miR-LAT, contributing to virus persistence in sensory neurons (Gupta et al., 2006). Several additional studies support the view that vertebrate viruses might extensively usurp the host miRNA pathway to complete their infection cycle (Jopling et al., 2005; Stern-Ginossar et al., 2007). For instance, hepatitis C virus (HCV) subverts the liver-specific human miR-122 to facilitate its replication (Jopling et al., 2005), although the mechanism involved is unknown.
The Molecular Arms Race
RNA Silencing and Evolution of Virus and Host Genomes
The reprogramming of host gene expression by mammalian virus-encoded miRNAs has parallels in plant-virus interactions. Several viRNAs derived from the Cauliflower mosaic virus 35S leader exhibit near perfect complementarity to Arabidopsis transcripts that are effectively targeted for sequence-specific downregulation during infection (Moissiard and Voinnet, 2006). In fact, dozens, if not hundreds of host transcripts might be targeted by viRNAs from Cauliflower mosaic virus, and at least two plant RNA viruses (Moissiard and Voinnet, 2006; O.V. and V.R. Ferrer, unpublished data). Host mRNA silencing by viRNAs might be fortuitous, with little consequence on virus fitness. However, some viRNAs could be selected by viruses, for instance, if they target host defense factors. At present, the sheer density and diversity of plant viRNAs precludes a clear estimation of the role of positive selection versus chance in this process, but this certainly prompts a re-evaluation of the current models for antiviral silencing. As much as some viRNAs might promote defense, others might benefit viruses (Figure 4A). Cell-to-cell silencing spread, thought to immunize naive tissues, might similarly be hijacked by viruses to create an optimal environment in tissues about to be invaded. These notions deserve careful attention, as they could partly explain the profound modifications in cell metabolism commonly elicited by plant viruses, some of which occur several cells ahead of infection fronts. A further layer of complexity in this double-faced scheme involves the action of VSRs, which should, in principle, inhibit both defensive and subversive effects of viRNAs (Figure 4A), implying that their deployment must be tightly regulated. Harnessing the dynamics of VSR expression and activity in space and time is surely a major challenge in plants and, perhaps, in animals, as related models could well apply to invertebrate infections.
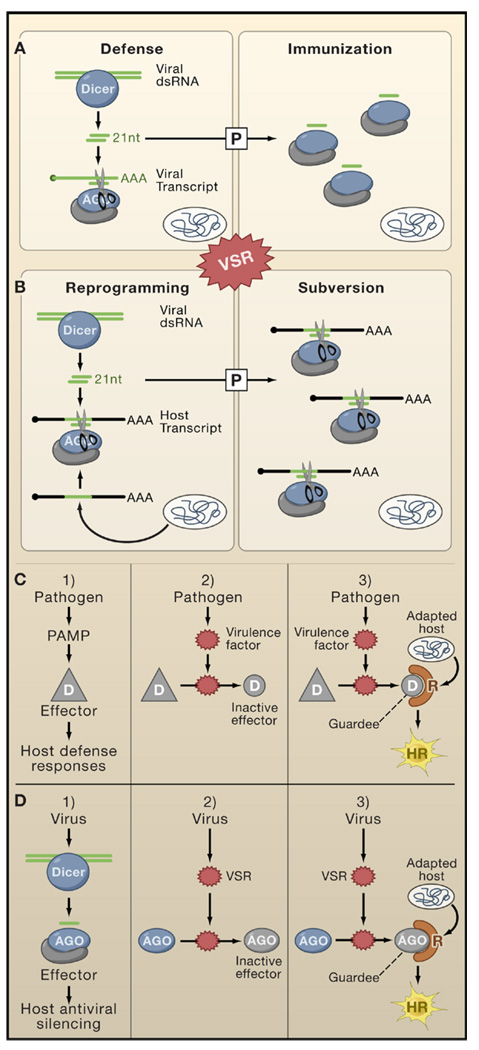
Figure 4. The Virus-Host Arms Race.
Complex and multilayered interactions exist between RNA silencing pathways, VSRs, and other plant immune pathways.
(A and B) This model depicts the possibility that viRNAs produced in virally-infected plants not only contribute to antiviral defense (upper panels) but might sometimes benefit viruses if viRNAs share sequence homologies with host transcripts (lower panels). The model also depicts the possibility that immunization and subversion by viRNAs might operate in cells ahead of the infection front. The extent of defense and subversion is influenced by the timing and level of VSR expression.
(C) This model illustrates the guard hypothesis, which proposes that suppression of PAMP-elicited basal defense responses in plants by pathogens’ effector proteins is detected by dedicated host-encoded R proteins, sometimes resulting in a hypersensitive response (HR).
(D) An adaptation of the guard hypothesis model explains how VSRs might elicit a hypersensitive response in specific plant ecotypes. The model entails that some effectors of antiviral silencing (e.g., AGO1) are modified by VSRs (such as P0 of poleroviruses; Figure 3C).
Mechanisms directed by viRNAs to turn off host genes in plants, mammals, and possibly invertebrates have implications for viral and host genome evolution. Single-nucleotide polymorphisms that prevent viRNA-directed repression of certain cellular transcripts might be under strong positive selection in the host genome. However, this might, in turn, favor the emergence of virus quasi-species carrying compensatory mutations that restore viRNA activities. Conversely, persistent virus infection might contribute to select random mutations that promote resistance of some cellular transcripts to some viRNAs. A similar rationale applies to host-encoded small RNAs interacting directly with viral RNA, either positively or negatively. The liver-specific miR-122 enhances HCV replication by binding to the 5′ end of the virion RNA, given that mutations of the viral miRNA-binding site reduce HCV titers (Jopling et al., 2005). Therefore, sequence polymorphisms in either cellular miR-122 or viral miR-122 target site could influence the susceptibility of specific individuals to specific HCV strains. Another aspect deserving attention is that repertoires and polymorphisms of cell-specific miRNAs might influence tissue permissiveness to certain viruses: the prevalence of miR-122 in the liver might contribute to the tropism of HCV for hepatocytes (Jopling et al., 2005).
Defense, Counterdefense, Countercounterdefense…
Components of plant and invertebrate antiviral silencing pathways must be under constant challenge by highly diverse VSRs and so should be continuously and rapidly evolving. Accordingly, in insects, the evolution rate of the antiviral defense factors Dcr2, R2D2, and Ago2 is considerably faster than that of their miRNA-pathway counterparts Dcr1, R3D1, and Ago1 (Obbard et al., 2006). Dcr2, R2D2, and Ago2 are evolving among the fastest 3% of all Drosophila proteins and display markedly reduced genetic diversity indicating recent selective sweep. These signatures of host-pathogens arms race strongly suggest that viruses shape host RNAi/antiviral functions in Drosophila and, presumably, in other invertebrates and plants. This probably explains the emergence of DCL2 as a surrogate for DCL4 in Arabidopsis (Figure 1) and the fluidity of VSR genes, presumably required to face the rapid changes in host antiviral proteins. This host-pathogens arms race could, therefore, result in paralogous VSRs sometimes having different modes of action, as for the AL2/AC2 proteins of begomo- and curtoviruses in the plant geminiviridae (Trinks et al., 2005; Wang et al., 2005).
Plant protein-based innate immune responses typically involve NBS-LRR genes, known as resistance (R) genes (Jones and Dangl, 2006). Their products, the R proteins, monitor or “guard” the integrity of specific host defense components called “guardees,” which are primary targets of pathogen’s virulence factors. Changes in guardees’ status usually result in R proteins triggering host defense reactions sometime culminating in a form of programmed cell death (called the hypersensitive response, HR), which is thought to restrict pathogen’s growth (Jones and Dangl, 2006; Figure 4C). VSRs are, by their very own nature, viral virulence factors, of which several are known to trigger R gene-dependent hypersensitive response in specific hosts; in at least one case, VSR mutations compromising silencing suppression also compromised induction of the hypersensitive response (Li et al., 1999; Ren et al., 2000). Therefore, some R genes may have evolved to specifically sense the damages incurred by VSRs to host antiviral silencing components (Figure 4D). Avoiding R gene recognition could constitute another strong selective pressure driving VSR gene evolution. Presumably, hosts could also directly neutralize VSRs through activities that degrade or relocate them into inappropriate subcellular compartments. The former probably explains the failure of specific alleles of the Cucumber mosaic virus-encoded VSR 2b to accumulate in Arabidopsis, possibly due to proteolysis (Zhang et al., 2006), whereas the latter is suggested by the nuclear relocation of the tombusviral P19 caused by ALY proteins, which interact with this VSR (Canto et al., 2006). These observations suggest that the varying efficacy of host-directed VSR suppression mechanisms and the polymorphism among VSR alleles may well contribute to the differences in viral susceptibility between ecotypes or between species.
ACKNOWLEDGMENTS
S.W.D. is supported by the National Institute of Allergy and Infectious Diseases (A1052447) and the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (2005-35319-15331). O.V. is funded by the European Union SIROCCO Integrated Project, the Schlumberger Foundation for Education and Research, and the French National Agency for Research.
REFERENCES
- Anandalakshmi R, Marathe R, Ge X, Herr JM, Mau C, Mallory A, Pruss G, Bowman L, Vance VB. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science. 2000;290:142–144. doi: 10.1126/science.290.5489.142. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis ARGO-NAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian XY, Rasheed MS, Seemanpillai MJ, Rezaian MA. Analysis of Silencing escape of Tomato leaf curl virus: an evaluation of the role of DNA methylation. Mol. Plant Microbe Interact. 2006;19:614–624. doi: 10.1094/MPMI-19-0614. [DOI] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Hohn T, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Canto T, Uhrig JF, Swanson M, Wright KM, MacFarlane SA. Translocation of Tomato bushy stunt virus P19 protein into the nucleus by ALY proteins compromises its silencing suppressor activity. J. Virol. 2006;80:9064–9072. doi: 10.1128/JVI.00953-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P, Vanitharani R, Fauquet CM. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. USA. 2005;102:10381–10386. doi: 10.1073/pnas.0504439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li WX, Xie D, Peng JR, Ding SW. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression. Plant Cell. 2004;16:1302–1313. doi: 10.1105/tpc.018986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. A marked end. Nat. Struct. Mol. Biol. 2007;14:259–260. doi: 10.1038/nsmb0407-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Viruses and microRNAs. Nat. Genet. 2006;38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Li F, Li WX, Ding SW. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007 doi: 10.1105/tpc.106.047449. Published online June 22, 2007. 0.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dunoyer P, Himber C, Voinnet O. Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 2006;38:258–263. doi: 10.1038/ng1722. [DOI] [PubMed] [Google Scholar]
- Ebhardt HA, Thi EP, Wang MB, Unrau PJ. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl. Acad. Sci. USA. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner BJ, Goh W, Kwang J. Dissection of the double-stranded RNA binding protein B2 from betanodavirus. J. Virol. 2007;81:5449–5459. doi: 10.1128/JVI.00009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7:1168–1175. doi: 10.1038/sj.embor.7400837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat. Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: A lesson in détente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Grishok A. RNAi mechanisms in Caenorhabditis elegans. FEBS Lett. 2005;579:5932–5939. doi: 10.1016/j.febslet.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Guo HS, Ding SW. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 2002;21:398–407. doi: 10.1093/emboj/21.3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2006;442:82–85. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- Gupta V, Huang X, Patel RC. The carboxy-terminal, M3 motifs of PACT and TRBP have opposite effects on PKR activity. Virology. 2003;315:283–291. doi: 10.1016/s0042-6822(03)00589-0. [DOI] [PubMed] [Google Scholar]
- Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Havelda Z, Hornyik C, Crescenzi A, Burgyan J. In situ characterization of Cymbidium Ringspot Tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J. Virol. 2003;77:6082–6086. doi: 10.1128/JVI.77.10.6082-6086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Pinzon I, Yelina NE, Schwach F, Studholme DJ, Baulcombe D, Dalmay T. SDE5, the putative homologue of a human mRNA export factor, is required for transgene silencing and accumulation of trans-acting endogenous siRNA. Plant J. 2007;50:140–148. doi: 10.1111/j.1365-313X.2007.03043.x. [DOI] [PubMed] [Google Scholar]
- Ho T, Pallett D, Rusholme R, Dalmay T, Wang H. A simplified method for cloning of short interfering RNAs from Brassica juncea infected with Turnip mosaic potyvirus and Turnip crinkle carmovirus. J. Virol. Methods. 2006;136:217–223. doi: 10.1016/j.jviromet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Imler JL, Ferrandon D, Royet J, Reichhart JM, Hetru C, Hoffmann JA. Toll-dependent and Toll-independent immune responses in Drosophila. J. Endotoxin Res. 2004;10:241–246. doi: 10.1179/096805104225005887. [DOI] [PubMed] [Google Scholar]
- Itaya A, Zhong X, Bundschuh R, Qi Y, Wang Y, Takeda R, Harris AR, Molina C, Nelson RS, Ding B. A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 2007;81:2980–2994. doi: 10.1128/JVI.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson KE. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. USA. 2004;101:17240–17245. doi: 10.1073/pnas.0406983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, Lopez-Moya JJ, Burgyan J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L, Szittya G, Silhavy D, Burgyan J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ding SW. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Li HW, Lucy AP, Guo HS, Li WX, Ji LH, Wong SM, Ding SW. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 1999;18:2683–2691. doi: 10.1093/emboj/18.10.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, García-Sastre A, Ball LA, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Folimonov A, Shintaku M, Li WX, Falk BW, Dawson WO, Ding SW. Three distinct suppressors of RNA silencing encoded by a 20 kb viral RNA genome. Proc. Natl. Acad. Sci. USA. 2004;101:15742–15747. doi: 10.1073/pnas.0404940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews REF. Plant Virology. Third Edition. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Moissiard G, Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl. Acad. Sci. USA. 2006;103:19593–19598. doi: 10.1073/pnas.0604627103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Moissiard G, Parizotto EA, Himber C, Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007;13:1268–1278. doi: 10.1261/rna.541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel J-B, Gordon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones J. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- Omarov RT, Ciomperlik JJ, Scholthof HB. RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc. Natl. Acad. Sci. USA. 2007;104:1714–1719. doi: 10.1073/pnas.0608117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007 doi: 10.1016/j.immuni.2007.05.014. in press. Published online July 5, 2007. 0.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C.elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Pantaleo V, Szittya G, Burgyan J. Molecular bases of viral RNA targeting by viral siRNA programmed. RISC. J. Virol. 2007;81:3797–3806. doi: 10.1128/JVI.02383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, Ziegler-Graff V. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Qu F, Ye X, Hou G, Sato S, Clemente TE, Morris TJ. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 2005;79:15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Qu F, Morris TJ. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell. 2000;12:1917–1926. doi: 10.1105/tpc.12.10.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robalino J, Bartlett T, Shepard E, Prior S, Jaramillo G, Scura E, Chapman RW, Gross PS, Browdy CL, Warr GW. Double-stranded RNA induces sequence-specific antiviral silencing in addition to nonspecific immunity in a marine shrimp: convergence of RNA interference and innate immunity in the invertebrate antiviral response? J. Virol. 2005;79:13561–13571. doi: 10.1128/JVI.79.21.13561-13571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers GC, van Wezel R, Zhang X, Hong Y, Nuss DL. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell. 2006;5:896–904. doi: 10.1128/EC.00373-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- Takeda A, Tsukuda M, Mizumoto H, Okamoto K, Kaido M, Mise K, Okuno T. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 2005;24:3147–3157. doi: 10.1038/sj.emboj.7600776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L. Slicer and the Argonautes. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barby P, Baillat V, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Trinks D, Rajeswaran R, Shivaprasad PV, Akbergenov R, Oakeley EJ, Veluthambi K, Hohn T, Pooggin MM. Suppression of silencing by geminivirus nuclear protein AC2 correlates with transactivation of host genes. J. Virol. 2005;79:2517–2527. doi: 10.1128/JVI.79.4.2517-2527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlirova M, Foy BD, Beaty BJ, Olson KE, Riddiford LM, Jindra M. Use of Sindbis virus-mediated RNA interference to demonstrate a conserved role of Broad-Complex in insect metamorphosis. Proc. Natl. Acad. Sci. USA. 2003;100:15607–15612. doi: 10.1073/pnas.2136837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanitharani R, Chellappan P, Pita JS, Fauquet CM. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 2004;78:9487–9498. doi: 10.1128/JVI.78.17.9487-9498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Non-cell autonomous RNA silencing. FEBS Lett. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 2005;79:7410–7418. doi: 10.1128/JVI.79.12.7410-7418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew RW, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA. 2004;101:6297–6302. doi: 10.1073/pnas.0304346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Patel DJ. RNA silencing suppressor p21 of Beet yellows virus forms an RNA binding octameric ring structure. Structure. 2005;13:1375–1384. doi: 10.1016/j.str.2005.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, JoshuaTor L, Mitani S, Simard MJ, Mello CC. Analysis of the C.elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Chapman EA, Yang Z, Carrington JC, Chen X. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in Arabidopsis. FEBS Lett. 2006;580:3117–3120. doi: 10.1016/j.febslet.2006.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Fan B, MacFarlane SA, Chen Z. Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant Microbe Interact. 2003;16:206–216. doi: 10.1094/MPMI.2003.16.3.206. [DOI] [PubMed] [Google Scholar]
- Zambon R, Vakharia VN, Wu L. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell. Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- Zamore PD. RNA silencing: genomic defence with a slice of pi. Nature. 2007;446:864–865. doi: 10.1038/446864a. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yuan Y, Pei Y, Lin S, Tuschl T, Patel DJ, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]