Abstract
Freshwater planarian flatworms possess uncanny regenerative capacities mediated by abundant and collectively totipotent adult stem cells. Key functions of these cells during regeneration and tissue homeostasis have been shown to depend on PIWI, a molecule required for Piwi-interacting RNA (piRNA) expression in planarians. Nevertheless, the full complement of piRNAs and microRNAs (miRNAs) in this organism has yet to be defined. Here we report on the large-scale cloning and sequencing of small RNAs from the planarian Schmidtea mediterranea, yielding altogether millions of sequenced, unique small RNAs. We show that piRNAs are in part organized in genomic clusters and that they share characteristic features with mammalian and fly piRNAs. We further identify 61 novel miRNA genes and thus double the number of known planarian miRNAs. Sequencing, as well as quantitative PCR of small RNAs, uncovered 10 miRNAs enriched in planarian stem cells. These miRNAs are down-regulated in animals in which stem cells have been abrogated by irradiation, and thus constitute miRNAs likely associated with specific stem-cell functions. Altogether, we present the first comprehensive small RNA analysis in animals belonging to the third animal superphylum, the Lophotrochozoa, and single out a number of miRNAs that may function in regeneration. Several of these miRNAs are deeply conserved in animals.
Keywords: microRNAs, miRNAs, piRNAs, regeneration, stem cells
Planarians have become a molecularly tractable model system in which to study regeneration, tissue homeostasis, and stem-cell biology (1). Planaria are free-living, triploblastic flatworms of the phylum Platyhelminthes, which is presently considered to belong to the superphylum Lophotrochozoa. Model systems for modern molecular and developmental biology have almost exclusively focused on the other 2 superphyla, i.e., the Deuterostomes (which includes vertebrates) and the Ecdysozoa (e.g., Caenorhabditis elegans and Drosophila melanogaster). Unlike these model systems, planarians possess remarkable regeneration abilities. Decapitation, for example, results in the complete regeneration of the head within 7 days after amputation. Such robust restoration of missing body parts is mediated by adult stem cells known as neoblasts (2). Of the thousands of known planarian species, Schmidtea mediterranea is arguably the species of choice for modern molecular biology and high-throughput, genome-wide approaches because it is diploid, it exists in sexual and asexual strains, and its genome has recently been sequenced and annotated (3). The size of its genome is roughly a third of the human genome, and ≈80% of the ≈20,000 annotated planarian genes have orthologs in humans. Moreover, by morphology alone, neoblasts and their immediate division progeny comprise ≈25% of all cells in the adult animal (4). In addition, RNAi screens have identified hundreds of genes specifically linked to planarian regeneration and stem-cell biology (5). Many of these genes are conserved in humans, and thus understanding planarian regeneration promises to yield important insights into human regeneration and stem cell biology.
In recent years, small, noncoding RNAs have emerged as essential players in almost all biological processes. Many different animal small-RNA species have by now been identified, although the biological functions of these species remain largely unclear (6, 7). Important exceptions are microRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs). miRNAs have been shown to play important roles in many differentiation processes, including regeneration (8), whereas at least one function of piRNAs has been shown to be in maintaining the integrity of the germ line (6). PIWI proteins are essential for the biogenesis and function of piRNAs, and they appear to have undergone an expansion in the planarian genome. We have identified at least 7 likely planarian PIWI genes, of which 3 (SMEDWI-1–3) have been in part functionally characterized (9, 10). For example, depletion of SMEDWI-2 has been shown to generate specific defects in stem-cell-mediated regeneration and homeostasis (9). Because neoblasts can give rise to germ-line cells in planaria, it is perhaps not surprising that at least SMEDWI-1 and SMEDWI-2 proteins are specifically expressed in neoblasts, and that depletion of SMEDWI-2 or SMEDWI-3 reduces piRNA production and both are required for neoblast function and regeneration (10).
Given the importance of miRNAs and piRNAs for planarian and stem-cell biology, it is essential to identify and classify small RNAs in S. mediterranea. Presently, 63 planarian miRNA genes encoding for 61 unique, mature miRNAs have been identified (11) and attempts have been made to describe their expression mainly by in situ hybridization of primary miRNA transcripts (12). However, mature miRNA expression can be highly regulated (13). Therefore, to determine the definitive spatial distribution of miRNAs, expression patterns of primary transcripts have to be complemented by mature miRNA expression data. Additionally, all known planarian miRNAs have been identified by classic cloning and Sanger sequencing, and it is highly likely that the true number of planarian miRNAs is much higher. A recent study (10) has further identified a few thousand piRNAs, which is also almost certainly a vast underestimate of the true number of planarian piRNAs (14).
We thus used massive, next-generation sequencing methods to define the full complement of small RNAs present in neoblasts, animals depleted of neoblasts, and whole animals. Altogether, we cloned, sequenced, mapped, and annotated millions of small RNAs. Extensive computational, qPCR, and Northern analyses allowed us to double the number of known planarian miRNAs, quantify their expression, and identify a number of mature miRNAs likely to be involved in stem-cell biology. Furthermore, we were able to study the expression, genomic organization, and biogenesis features of planarian piRNAs at a resolution orders of magnitude higher than any previous studies. Our dataset allowed us to compare planarian piRNA characteristics with known piRNA features in mammals and ecdysozoans. Altogether, our work brings the characterization and annotation of small RNAs in planarians to a depth that is at par with other model systems such as C. elegans.
Results
Comprehensive and Quantitative Deep Sequencing of Planarian Small RNAs.
To profile expression differences of small RNAs, we wished to compare neoblasts, intact animals, and animals devoid of neoblasts with each other. Therefore, RNA was obtained from the clonal asexual strain CIW4 of S. mediterranea from FACS-purified neoblasts, intact animals, and irradiated animals in which neoblasts were eliminated by radiation (1). Each of the 3 samples was sequenced with 2 different methods. Solexa (Illumina) technology was used to profile all species of small RNAs (size selection: 18–40 nt). Furthermore, we used the 454 Life Sciences (Roche) technology to specifically profile Dicer products (such as miRNAs) by using a more narrow size selection of 18–25 nt. By using a stringent mapping procedure (see SI Text), we matched a total of ≈4.2 million sequencing reads to ≈6.7 million loci in the planarian genome. Table 1 gives an overview of the 6 deep-sequencing datasets. We next assessed the samples' quality by 3 criteria: coverage, reproducibility, and accuracy of expression quantitation.
Table 1.
The 6 deep-sequencing datasets derived from untreated and irradiated planarians and isolated neoblasts
| Datasets | Sequencing platform | Sample type | Number of unique mapped reads | Number of mapped reads | Number of loci |
|---|---|---|---|---|---|
| 1 | 454 | Neoblast | 25,256 | 63,278 | 95,951 |
| 2 | 454 | Untreated | 27,461 | 93,412 | 104,769 |
| 3 | 454 | Irradiated | 21,163 | 61,391 | 85,051 |
| 4 | Solexa | Neoblast | 86,063 | 91,371 | 417,646 |
| 5 | Solexa | Untreated | 984,459 | 1,784,859 | 4,064,381 |
| 6 | Solexa | Irradiated | 767,496 | 2,050,669 | 3,018,709 |
| All | All | All | 1,507,162 | 4,144,980 | 6,700,894 |
Coverage.
To estimate the coverage of planarian small RNAs by the sequenced RNAs, we computed the overlap of our mapped reads with known planarian miRNAs and piRNAs. Previously identified miRNAs were detected by conventional cloning and sequencing small RNAs from S. mediterranea whole-body samples with a median miRNA count of 4 (11). We found all of these miRNAs in our pooled datasets, with a median of >9,000 counts (Table S1). Furthermore, our data contain the lowly expressed “star” miRNAs for 62 of the 63 miRNA genes. Another recent study reported ≈4,800 planarian piRNAs deep-sequenced from whole-body samples of planarians (10). We found 38% of these piRNAs in our data. Considering that animal piRNA populations are estimated to consist of hundreds of thousands of unique sequences (14), it is not surprising that our sequencing of piRNAs is not fully saturated.
Reproducibility.
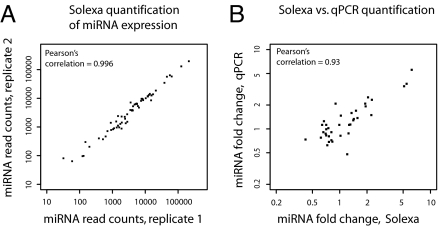
We compared 2 Solexa datasets obtained by sequencing biological replicate planarian samples. For each planarian miRNA in miRBase, we plotted the number of times the miRNA was sequenced in one sample vs. the other sample (Fig. 1A). The correlation was almost perfect (Pearson's correlation = 0.996), indicating high reproducibility.
Fig. 1.
Reproducible and quantitative sequencing of small RNAs. (A) Reproducibility of Solexa quantitation of miRNA expression. Each data point represents 1 miRNA. Samples are independent biological replicates of irradiated planarians. (B) Solexa vs. qPCR quantitation of miRNA fold-changes. Each data point represents 1 miRNA. Independent biological replicates were used for the Solexa and the qPCR quantitation.
Accuracy of Expression Quantitation.
We investigated whether our deep-sequencing data can accurately quantify differential miRNA expression. We measured expression fold-changes between intact and irradiated samples for 35 planarian miRNAs by using our Solexa data and quantitative PCR in samples from independent biological replicates (Taqman assay; Methods). We found a strong correlation between the deep-sequencing data and the qPCR measurements (Fig. 1B, Pearson's correlation = 0.93). We conclude that our data are comprehensive, reproducible, and can be used to quantify miRNA expression across samples.
Planarian Small RNAs Are Predominantly miRNAs and piRNAs.
We next identified the types of small RNAs present in planarians and quantified their expression in neoblast vs. whole-body extracts. We hypothesized that the comparison of neoblasts with an untreated whole-body sample should identify small RNA species up-regulated in the planarian adult stem cells. If such species are in fact specific to neoblasts, we would further expect them to have reduced expression in the irradiated whole-body sample compared with the samples from the intact, unirradiated animals. Moreover, these comparisons would allow us to detect artifacts, i.e., highly expressed small RNAs, that may have arisen as a result of cell dissociation and/or cell sorting.
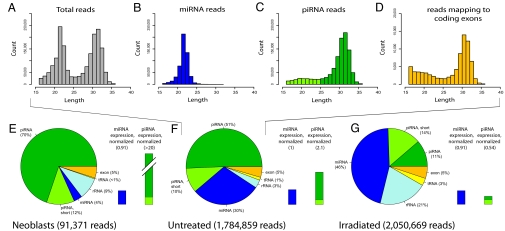
Small RNAs in the untreated sample showed a bimodal length distribution with 2 distinct peaks at nucleotides 22 and 32 (Fig. 2A). We first selected reads that mapped to known planarian miRNAs from miRBase as well as our novel miRNAs (see below). The length distribution of these reads had a single peak at nucleotide 22, typical for miRNAs (Fig. 2B). In fact, these 122 miRNAs (known and novel) account for the entire 22-nt peak in Fig. 2A, suggesting that few miRNAs remain to be discovered in S. mediterranea. When subtracting all reads mapping to annotated miRNAs, rRNAs and tRNAs, and coding exons, the length distribution forms a distinct peak at nucleotide 32 (Fig. 2C). We tentatively refer to these sequences as piRNAs, and will present further evidence for this classification in the next section on piRNAs. Reads mapping to annotated coding exons display a clear peak approximately at nucleotide 32 (Fig. 2D). However, this is likely an artifact caused by ambiguous read-mappings and the genome annotation (for discussion see SI Text and Table S2).
Fig. 2.
Small RNA contents in planarian neoblasts and whole-body samples. (A–D) Length profiles of Solexa reads from sequencing of the untreated whole-body sample. Only reads successfully mapped to the genome are considered. (A) All reads, (B) miRNA reads, (C) piRNA reads, and (D) reads mapping to coding exons are shown. Note that D is on a different scale. piRNA reads >25 nt (dark green) have features characteristic for piRNAs (see sections on piRNAs). These features were still present but weaker (70–80%, see SI Text) for the remaining piRNA reads (light green, length 17–25 nt). (E–G, pie charts) Contents of RNA species in the three Solexa datasets: (E) neoblasts, (F) untreated planarians, and (G) irradiated planarians. Dark green, piRNAs; light green, short piRNAs; dark blue, miRNAs; light blue, rRNA; yellow, tRNA; orange, mRNA. (E–G, bar graphs) miRNA (blue), piRNA (dark green), and short piRNA (light green) expression. The 454 data, produced by sequencing RNAs that were 18–25-nt long, were used to estimate miRNA expression, whereas the Solexa data were used for piRNA expression. Total miRNA and piRNA read counts were normalized to miR-71c. miRNA expression of the untreated sample was set to 1, and the other expression bars were scaled accordingly (numbers in parentheses above each bar). Note that piRNA expression in the neoblast sample is out of scale.
We next estimated the relative abundance of different classes of small RNAs across the 3 sample types (Fig. 2 E–G, pie charts). The intergenic piRNA fraction is predominant in sorted neoblasts (82%), intermediate in the untreated sample (61%), and low in the irradiated sample (25%). The increased fractions of rRNA and short piRNAs in the irradiated sample could be a result of degradation. In contrast, the miRNA fraction is low in the neoblast sample (4%), intermediate in the untreated sample (30%), and larger in the irradiated sample (46%).
Comparing the abundance of each class of small RNAs across different samples requires normalizing contents to a stably expressed endogenous control. We used miR-71c for library normalization because we observed that this miRNA is robustly and constantly expressed across our 3 samples based on a quantitative Taqman assay (SI Text and Fig. 3). By normalizing the total read counts of miRNAs and piRNAs to the read count of miR-71c, we were able to estimate the relative expression of small RNAs across samples (Fig. 2 E–G, bar graphs). Intergenic piRNAs have very high expression in neoblasts (>10-fold higher than in untreated whole-body planarians) and low expression in irradiated planarians (4-fold lower than in untreated planarians), consistent with the idea that piRNAs may be up-regulated in neoblasts and their division progeny, i.e., where PIWI proteins are specifically expressed (10). In contrast, total miRNA contents appeared roughly constant over the 3 samples, although the abundances of individual miRNAs varied. We independently repeated this analysis with 2 other miRNAs (miR-36 and miR-36c) that appeared roughly constant and obtained comparable results (see Table S3).
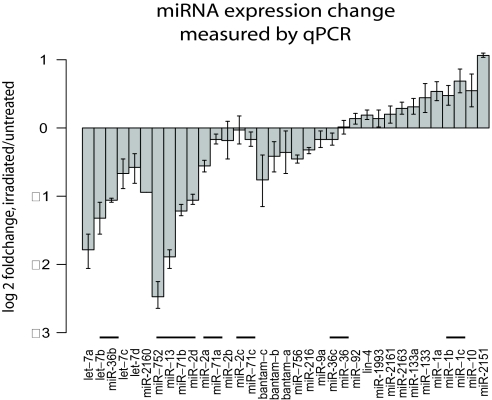
Fig. 3.
miRNA expression fold-changes measured by qPCR. Total RNA from untreated and irradiated animals was used to quantify expression fold-changes of 35 miRNAs by qPCR. Data are relative to expression detected for the ubiquitously expressed control ura4 (see Methods). Each miRNA is grouped with its family members. Horizontal bars indicate miRNA gene clusters.
Planarian piRNAs Share Key Features with Mammalian and Fly piRNAs.
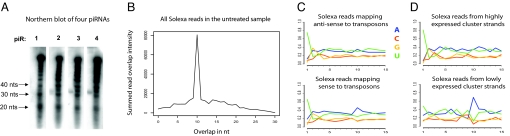
To characterize planarian piRNAs, we analyzed deep-sequencing reads that did not map to annotated miRNAs, rRNAs, tRNAs, or coding sequences (Fig. 2C). These reads display 2 of the defining features of mammalian and fly piRNAs (reviewed in ref. 14): a length distribution peaking approximately at nucleotide 30 and a diverse population (≈1.2 million unique sequences in our Solexa data). Northern blots validated size and expression of 3 planarian piRNAs (Fig. 4A).
Fig. 4.
Features of planarian piRNAs. (A) Northern blot analysis of 4 annotated piRNAs. Bands ≈32-nt long are visible for 3 of these piRNAs. (B) Summed overlap intensities for Solexa reads in the untreated sample. The horizontal axis is the length of overlap in nucleotides between the 5′ ends of reads mapping to the same genomic locus on opposite strands. The vertical axis shows the intensity of the overlap, summed over the entire dataset. (C and D) Sequence biases of piRNAs. The horizontal axis shows nucleotide positions from the 5′ end, the vertical axis represents nucleotide fractions. (E) piRNAs mapping antisense (Top) and sense (Bottom) to transposons. (F) piRNAs from highly (Top) and lowly (Bottom) expressed cluster strands.
Planarian piRNAs Display a Clear Tendency to Overlap by 10 Nucleotides.
The current model of biogenesis proposes that piRNAs are generated through iterative PIWI-mediated cleavage of transcripts with complementary sequence [the “ping-pong” amplification mechanism (15, 16)]. According to this model, piRNAs that map to opposite genomic strands tend to overlap by 10 nt. We investigated whether this signature is conserved in planarians. However, this is difficult because many piRNAs map to numerous genomic loci. For instance, if 2 reads map to the same 100 loci, their overlap would be counted 100 times. Thus, to avoid potentially inflated counts, we assigned “intensities” to mappings that were inverse to the number of mappings for the read. For example, a read mapping to 10 loci would be assigned an intensity of 0.1. Summing overlap intensities over each of our datasets, yielded major peaks with an overlap of exactly 10 nt in the neoblast and the untreated sample (Fig. 4B), whereas the peak for the irradiated sample was greatly reduced (Fig. S1).
Planarian Primary piRNAs Map Antisense to Transposons.
An important function of mammalian and fly piRNAs is to silence transposons. In mouse testes, PIWI proteins cleave transposon mRNA to generate primary piRNAs, with a uracil in the 5′ end (17). Primary piRNAs base pair with long transcripts that contain complementary sequence to cleave out secondary piRNAs, which thus typically have an adenosine at position 10. In fly testes, this is reversed. Primary piRNAs are cleaved from transcripts antisense to transposons, and the secondary piRNAs are cleaved from the transposon mRNA (15, 16).
We found that 32% of the planarian piRNAs map to annotated transposons (SI Text). piRNAs mapping antisense to transposons have a clear tendency for a beginning uracil and no other sequence biases (Fig. 4C), indicating that these are primary piRNAs. piRNAs mapping in the sense orientation to transposons have a bias toward a beginning uracil and an adenosine at position 10.
Planarian piRNAs Locate to Transposons as much as Mouse Prepachytene piRNAs.
Mammalian and fly piRNAs differ on the fraction of the population that is transcribed from transposons. Mouse pachytene piRNAs have no reported role in transposon silencing, and map to mouse transposons less than would be expected from the transposon genome coverage. In contrast, mouse prepachytene piRNAs and fly piRNAs have reported roles in transposon silencing (reviewed in ref. 18). These map to transposons as much (mouse prepachytene) or more (fly) than would be expected from the transposon genomic coverage.
Planarian transposons cover 31% of the genome and 32% of the piRNAs. These numbers resemble mouse prepachytene piRNAs. However, planarian piRNAs do display biases toward particular classes of transposons. For instance, Mariner elements, active in planarians (19), have 1.8 times more piRNAs mapping than would be expected by chance, whereas PiggyBac have half the number of mapping piRNAs as would be expected (Table S4). These findings are significant (P ≈ 0; see SI Text). piRNA transposon association changes little across the 3 samples.
Planarian piRNA Clusters Display Strand Expression Bias but Seem Not to Resemble Master Loci.
We observed that planarian piRNAs, similar to those of mammals and flies, tend to map to discrete regions. To annotate these piRNA clusters, we located 10-kb regions of the genome to which 100 or more long piRNAs can be unambiguously traced and where instances of 10-nt overlaps between such piRNAs occur. This yielded 119 piRNA cluster candidates to which 6% of all planarian piRNAs in the untreated sample can be traced (Table S5). These clusters are thus highly (and significantly; see SI Text) enriched in piRNAs, given that they only constitute about one thousandth of the planarian genome.
The majority (92%) of planarian piRNA clusters displayed a strong strand bias, with piRNA mapping intensities 10 times or higher on one strand (see Fig. S2). piRNAs originating from highly expressed cluster strands, like primary piRNAs in mouse and fly, have a strong bias for a 5′ uracil, whereas the ones from the lowly expressed cluster strands, like secondary piRNAs, have a strong tendency for an adenosine at position 10 (see Fig. 4D). Similar strand expression biases are observed in the fly “master loci”, which are piRNA clusters densely packed with nonfunctional transposons. However, we did not identify any master loci in the planarian genome, as none of the piRNA clusters contained large numbers of transposons.
Discovery and Validation of Novel Planarian miRNAs.
To discover novel miRNAs, we used miRDeep, an algorithm to detect and score Dicer hairpin products such as miRNAs in deep-sequencing data (20). Varying score cut-offs allow trade-offs between sensitivity and specificity. Sensitivity is computed as the fraction of known miRNAs recovered, whereas false positives are estimated by stringent statistical controls (20).
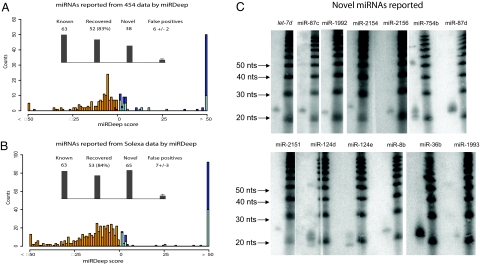
We separately searched the 454 and Solexa data (SI Text). With the default cut-off we recovered miRNAs with high sensitivity and specificity (Fig. 5 A and B). miRDeep identified 70 novel potential miRNAs, which were further curated (see SI Text). We thus report a subset of 61 high-confidence miRNAs (Table S6).
Fig. 5.
Novel planarian miRNAs. (A) miRNAs reported from 454 data by miRDeep. miRDeep scores are shown as a color-encoded histogram (score cut-off: 1). Known miRNAs, dark blue; novel miRNAs, light blue. Known miRNAs below the cut-off are plotted in red (false negatives). The number of false positives was estimated by miRDeep (20). (B) miRNAs reported from Solexa data by miRDeep; legend same as A. (C) Validation of novel miRNAs. 13 miRNAs were validated by Northern blot analysis, 11 by qPCR (Fig. 3). In some cases, miRNA precursors are also detected.
We subjected 20 miRNA candidates to Northern blot analysis and successfully validated 13 of them (see Fig. 5C). Candidates not observed by Northern blotting may be below detection threshold. In support of this, a more sensitive Taqman assay was used to validate 11 of 11 novel candidates tested (Fig. 3), 4 of which had also been validated by Northern blot analysis (Fig. 5C). In total, 20 novel candidates were validated.
The phylogenetic analysis of planarian miRNAs may be particularly informative as planarians are an outgroup relative to animal model systems used by the majority of researchers. miRNAs can be grouped into families based on sequence similarity at their 5′ end (7). Our novel miRNAs increase the number of planarian miRNA families from 37 to 79 (Fig. S3). The planarian miRNAs share 22 families with mammals and 33 with flies and with nematodes. Thus, we find planaria to resemble Ecdysozoa (flies, nematodes) more than mammals from the miRNA phylogeny. Interestingly, the majority (45 of 79) of planarian miRNA families do not show sequence similarity to known miRNAs. The presence of the miR-1992 family (21) combined with the absence of the miR-1994 family gives evidence to the hypothesis that flatworms are the sister group to the other lophotrochozoans.
More than a Dozen miRNAs Are Likely Linked to Neoblast Biology.
To identify miRNAs up-regulated in neoblasts, we calculated the miRNA expression fold-change between the untreated whole worm and isolated neoblast deep-sequencing datasets (SI Text and Table S7). 13 miRNAs were up-regulated by >2-fold in the neoblast sample. As an independent control, we used qPCR to profile the expression of these miRNAs in filtered cells enriched in neoblasts vs. untreated planarians. These were all up-regulated by >30% in the isolated neoblasts, whereas a number of other profiled miRNAs did not change.
To rule out that miRNA up-regulation may have been caused by cell dissociation or cell sorting, we used qPCR in independently obtained samples to calculate miRNA expression fold-changes between untreated and irradiated animals. We found 10 of 13 of the miRNAs of interest to be >25% down-regulated in the irradiated sample (see Table 2 and Fig. 3). These data suggest that a small subset of miRNAs is significantly up-regulated in neoblasts. Notably, miRNA genes comprised in clusters (miR-2d, miR-13, miR-71b, miR-752 and let-7b, miR-36b) as well as the miRNAs belonging to the same families (let-7, miR-2/miR-13) show a similar differential expression (Fig. 3).
Table 2.
Ten miRNAs up-regulated in neoblasts
| miRNAs | Fold change neo/untr (454) | Fold change untr/irr (qPCR) | Fold change untr/irr (Solexa) |
|---|---|---|---|
| let-7a | 2.4 | 3.3 | 5.1 |
| let-7b, miR-36b | 2.1, 3.8 | 2.5, 2.0 | 2.1, 0.9 |
| miR-2a | 3.7 | 1.4 | 1.0 |
| miR-2d, miR-13, miR-71b, miR-752 | 3.4, 4.9, 3.0, 7.0 | 2.0, 3.3, 2.5, 6.5 | 1.9, 5.5, 2.3, 6.2 |
| miR-756 | 2.0 | 1.4 | 1.6 |
| miR-2160 | 5.0 | 2.0 | 1.9 |
miRNAs listed in the same field locate to the same genomic cluster. Neo, neoblast sample; untr, untreated sample; irr, irradiated sample.
Interestingly, most of the up-regulated miRNAs in neoblasts belong to conserved families. The let-7 family has previously been associated with stem-cell identity. However, previous studies indicate that let-7 is down-regulated posttranscriptionally in stem cells (22). In flies, miR-2 and miR-13 target the proapoptotic genes grim, reaper, and sickle (23). We find that all 4 miRNAs in genomic cluster containing the planarian miR-2 and miR-13 are up-regulated in neoblasts, suggesting that these miRNAs are important for neoblast maintenance or neoblast-related function. Additionally, miRNAs that are typically expressed in specific somatic tissues such as miR-124 (brain tissues) and miR-1 and miR-133 (muscle tissues) were down-regulated in neoblasts.
Discussion
By using massive quantitative deep sequencing, we have annotated small RNA species in S. mediterranea. We have doubled the number of planarian miRNAs, and have validated a large fraction of our novel miRNAs. We find that the small RNA-length peak at nucleotide 22 disappears completely when miRNAs are removed (Fig. 2), suggesting that a diminishing number of miRNAs or other Dicer products remain to be discovered in S. mediterranea. We were also unable to detect evidence for phased processing of longer transcripts by Dicer. Moreover, we annotated more than one-million unique piRNA sequences that locate to genomic clusters. piRNAs have previously been well-described in Deuterostomes and ecdysozoans (15, 16, 24–26). We report ≈1.2 million unique planarian piRNAs locating to more than 100 genomic clusters, and thus give a first comprehensive description of piRNAs in lophotrochozoans. We find that piRNA features characteristic for piRNA biogenesis (sequence biases, 10-nt overlap) are shared in all 3 metazoan superphylae. Planarian piRNAs also share specific characteristics with either mammalian or fly piRNAs. Planarian primary piRNAs, like those in the fly, tend to map antisense to transposable elements, suggesting that planaria may defend their genome against transposons similarly to flies. However, from a different point of view, planarian piRNA biology resembles that of the mouse more than the fly. First, flatworm piRNAs associate with transposons as much as mouse prepachytene piRNAs. Second, the expression of planarian piRNAs is dispersed between numerous clusters, similar to what has recently been observed in the mouse (17). Third, we find no planarian clusters containing many transposon fragments akin to the characteristic fly master loci. Together, our data indicate that the piRNA pathway has undergone complex evolution.
We find that at least 10 miRNAs are up-regulated in neoblast samples. Deep sequencing and qPCR controls show that these are down-regulated in the irradiated samples depleted of neoblasts, indicating that they may be specific to neoblast biology. These miRNAs include all 4 miRNAs from a genomic cluster that contains miR-2 and miR-13, miRNAs known to inhibit proapoptotic genes in fly (23). We also find that at least 2 members (let-7a and let-7b) of the highly conserved let-7 family are up-regulated in neoblasts. Up-regulation of let-7 in neoblast samples was paralleled by let-7 down-regulation in irradiated samples. These findings are surprising because let-7 and its family members are known to be depleted in mammalian stem cells (22, 27) and have been shown in numerous species to repress cell proliferation and promote differentiation (reviewed in ref. 28). However, recent studies have shown that cells with the morphological appearance of neoblasts can be resolved into subtypes, and planarian stem cells may maintain proliferative activity after commitment (4, 29). Thus, high let-7 levels in neoblast samples may be derived from neoblasts that are exiting the stem-cell state and committing to a differentiation lineage. Further let-7 expression analyses, therefore, may help elucidate the specification of neoblast lineages.
Although planarian stem cells are collectively totipotent because they can give rise to both the somatic and germ lineages in the adult, our analyses indicate that planarians harbor only 1 miRNA (miR-92) known to be highly expressed in mammalian embryonic stem cells (30). However, we found no evidence that its 2 family members are up-regulated in neoblasts. Taken together, expression of miRNAs in planarian neoblasts share little if any similarity with mammalian embryonic stem cells, which may reflect both the adult nature of planarian stem cells as well as the inherent in vitro versus in vivo differences between these 2 populations of animal stem cells.
Finally, the small RNA profile of neoblasts resemble mouse and fly germ-line stem cells in being dominated by piRNAs. Because the genomic contents of germ-line cells and neoblasts are potentially immortal, both cell types need to strictly control their genome integrity during transmission to future generations, and particularly, to protect it against the uncontrolled propagation of mobile genetic elements. piRNAs have been shown selectively to silence transposons in the fly and mouse genomes (reviewed in ref. 18) and it is likely that piRNAs play such a role in planaria. Further studies are needed to determine whether planarian piRNAs also play a critical role in epigenetic silencing through DNA/chromatin methylation like their germ-line homologs (17).
Methods
Sample Preparation and Sequencing.
Planarians from the clonal, asexual CIW4 strain of S. mediterranea were starved for 1 week before all experiments. Planarian total RNA was isolated by using TRIzol (Invitrogen). Planarians for the irradiated samples were exposed to 60 Gy and RNA was extracted 8 days after irradiation. FACS sorting was performed as described in SI Text. Solexa and 454 sequencing was performed by using the manufacturer's protocol.
miRNA Identification.
Detection of novel miRNAs was performed as in ref. 20. For more details, see SI Text.
Northern Blot Analysis and qPCR.
Validation of miRNA and piRNA candidates was performed by Northern blot analysis (Table S8) as described previously (31). qPCR (Taqman miRNA custom assays, ABI) was used to quantify the expression fold-change of 35 miRNAs. cDNA was synthesized from 50 ng of total RNA from either irradiated or untreated animals. Samples without reverse transcriptase served as a negative control template. Each measurement was performed in triplicate. Two biological replicates were used. Threshold cycle values are relative to expression detected for the ubiquitously expressed control mRNA ura4 (SI Text). Relative expression of miRNAs is given as log2 of 2−ΔΔCt values.
Supplementary Material
Acknowledgments.
We thank Astrid Ferlinz from Applied Biosystems for the custom ABI Taqman primers. Sam Griffith-Jones facilitated the manuscript through a rapid assignment of miRBase names to the novel planarian miRNAs. Erik A. Sperling and Kevin J. Peterson gave valuable advice on miRNA phylogeny.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nln.nih.gov/geo (accession no. GSE16159).
This article contains supporting information online at www.pnas.org/cgi/content/full/0905222106/DCSupplemental.
References
- 1.Reddien PW, Sanchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 2.Randolph H. The regeneration of the tail in lumbriculus. J Morphol. 1892;7:317–344. [Google Scholar]
- 3.Robb SM, Ross E, Sanchez Alvarado A. SmedGD: The Schmidtea mediterranea genome database. Nucleic Acids Res. 2008;36:D599–D606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisenhoffer GT, Kang H, Sanchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Yin VP, et al. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 10.Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palakodeti D, Smielewska M, Graveley BR. MicroRNAs from the planarian Schmidtea mediterranea: A model system for stem cell biology. RNA. 2006;12:1640–1649. doi: 10.1261/rna.117206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Estevez C, Arseni V, Thambyrajah RS, Felix DA, Aboobaker AA. Diverse miRNA spatial expression patterns suggest important roles in homeostasis and regeneration in planarians. Int J Dev Biol. 2009;53:493–505. doi: 10.1387/ijdb.082825cg. [DOI] [PubMed] [Google Scholar]
- 13.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 14.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 15.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 16.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315(5818):1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 17.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Fernandez J, et al. High copy number of highly similar mariner-like transposons in planarian (Platyhelminthe): Evidence for a trans-phyla horizontal transfer. Mol Biol Evol. 1995;12:421–431. doi: 10.1093/oxfordjournals.molbev.a040217. [DOI] [PubMed] [Google Scholar]
- 20.Friedlander MR, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler BM, et al. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- 22.Thomson JM, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 25.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 26.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Higuchi S, et al. Characterization and categorization of fluorescence activated cell sorted planarian stem cells by ultrastructural analysis. Dev Growth Differ. 2007;49:571–581. doi: 10.1111/j.1440-169X.2007.00947.x. [DOI] [PubMed] [Google Scholar]
- 30.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.