Abstract
The subdivision of mesodermal cells into muscle and non-muscle cells is crucial to animal development. In the C. elegans postembryonic mesoderm, this subdivision is a result of an asymmetric cell division that leads to the formation of striated body wall muscles and non-muscle coelomocytes. Here we report that the Six homeodomain protein CEH-34 and its cofactor Eyes Absent, EYA-1, function synergistically to promote the non-muscle fate in cells also competent to form muscles. We further show that the asymmetric expression of ceh-34 and eya-1 is regulated by a combination of 1) mesodermal intrinsic factors MAB-5, HLH-1 and FOZI-1, 2) differential POP-1 (TCF/LEF) transcriptional activity along the anterior-posterior axis, and 3) coelomocyte competence-factor(s). These factors are conserved in both vertebrates and invertebrates, suggesting a conserved paradigm for mesoderm development in metazoans.
Keywords: ceh-34, eya-1, sys-1, pop-1, SIX homeodomain, Eyes absent, mesoderm, TCF, β-catenin, cell fate specification, M lineage, asymmetry, muscle, coelomocyte
INTRODUCTION
How distinct cell fates are acquired from multipotent progenitor cells is a fundamental question in developmental biology. We are interested in the mechanisms involved in distinguishing myogenic and non-myogenic fates in the mesoderm. The Six family of homeodomain proteins has been found to regulate cell fate specification in multiple tissue types, including in the mesoderm (for review, see Kawakami et al., 2000). The founding member of the Six family is the Drosophila sine oculis (so) gene (Cheyette et al., 1994; Serikaku and O’Tousa, 1994). The role of so is best characterized in the Drosophila eye, where it functions downstream of the Pax6 gene eyeless (ey) for proper eye development (Halder et al., 1998). so and its cofactor eyes absent (eya) function upstream of the transcription factor dachshund (dac) to regulate the expression of eye specification genes (Chen et al., 1997; Pignoni et al., 1997; Shen and Mardon, 1997; Halder et al., 1998). This Pax-Six-Eya-Dac network of regulation is also required for proper eye development in vertebrates (Hanson, 2001). Members of this network also function in other metazoan developmental processes such as mesodermal and sensory organ development (Heanue et al., 1999; Xu et al., 1999; Hanson, 2001; Clark et al., 2006)
C. elegans has four homologs of the Six family (CEH-32, CEH-33, CEH-34 and CEH-35/UNC-39) (Dozier et al., 2001). We have observed that knockdown of ceh-34 during postembryonic development results specifically in the loss of non-muscle cell types in the mesoderm. In this study we investigate how ceh-34 functions in the postembryonically-derived non-gonadal mesoderm, the M lineage (Sulston and Horvitz, 1977). During hermaphrodite larval development the pluripotent M mesoblast reproducibly produces three distinct cell types: 14 striated bodywall muscles (BWMs), 2 sex myoblasts (SMs) that subsequently give rise to the non-striated egg-laying muscles, and 2 non-muscle coelomocytes (CCs), which together with four other CCs generated during embryogenesis, act as macrophage-like cells (Sulston and Horvitz, 1977, Sulston et al., 1983). M lineage cell fate specification occurs in an asymmetric manner, as CCs are born dorsally and SMs ventrally (Fig. 1A). The LIN-12/Notch and TGFβ signaling pathways regulate proper asymmetry along the dorsal-ventral axis within the M lineage: the LIN-12/Notch pathway promotes ventral SM fates, while the C. elegans Schnurri homolog SMA-9 antagonizes the Sma/Mab TGFβ pathway to promote dorsal CC fates (Greenwald et al., 1983; Foehr et al., 2006; Foehr and Liu, 2008). Asymmetries also exist along the anterior-posterior axis in the M lineage, in which cell fate decisions are made between posterior BWMs and anterior CCs or SMs at the 16- and 18-M stages (Fig. 1A).
Figure 1. ceh-34 is required for CC fates.
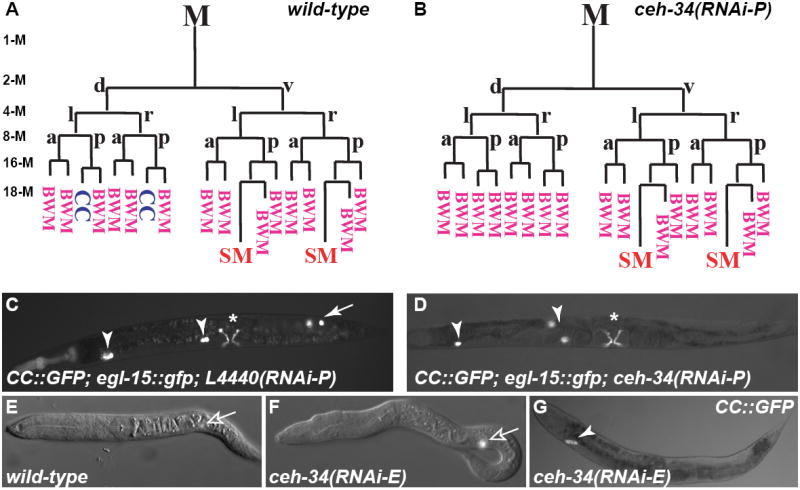
All images in Figures 1-4 are ventral/lateral views with anterior to the left, unless otherwise noted. (A,B) Early M lineage in wild-type (A) and ceh-34(RNAi-P) (B) animals. Stages of M lineage (1-M to 18-M) are indicated. (C,D) L4440-RNAi treated control (C) and ceh-34(RNAi-P) (D) adults. CCs are visualized using intrinsic CC∷gfp, with embryonic CCs labeled with arrowheads and M-derived CCs with arrows. Type I vulval muscles are visualized using egl-15∷gfp, denoted by asterisks. M-derived CCs are missing in ceh-34(RNAi) animals (D). (E-F) L1 larva of water-injected (E) or ceh-34 dsRNA (F) injected animals. The M mesoblast is indicated by expression of hlh-8∷gfp (open arrow). (G) ceh-34(RNAi-E) adult with only one pair of embryonic CCs (arrowhead). M, M mesoblast; d, dorsal; v, ventral; l, left; r, right; a, anterior; p, posterior; CC, coelomocyte; BWM, body wall muscle; SM, sex myoblast.
The Wnt/β-catenin asymmetry pathway plays a conserved role in multiple asymmetric fate specification events in C. elegans along the anterior-posterior or proximal-distal axes (for review, see Mizumoto and Sawa, 2007). Specifically, the β-catenin homolog SYS-1 and the TCF/LEF transcription factor POP-1 show reciprocal asymmetric nuclear distribution in multiple cell divisions, with SYS-1 being enriched in the posterior or distal nuclei and POP-1 enriched in the anterior or proximal nuclei (Lin et al., 1995; Lin et al., 1998; Herman, 2001; Siegfried and Kimble, 2002; Kidd et al., 2005; Huang et al., 2007; Phillips et al., 2007). POP-1 nuclear localization is further regulated by the LIT-1 kinase and another β-catenin WRM-1, which facilitate the nuclear export of POP-1 in an asymmetric manner (Lo et al., 2004; Takeshita and Sawa, 2005). Recently it has been shown that in the M lineage, WRM-1 localizes to the anterior cortex during anterior-posterior cell divisions but to the nuclei of posterior daughters afterwards (Takeshita and Sawa, 2005). However, a role of the Wnt/β-catenin asymmetry pathway in the M lineage has yet to be described.
A number of mesoderm-intrinsic transcription factors are required for specification of both muscle (BWM) and non-muscle (CC) fates derived from the M mesoblast. The single C. elegans MyoD family member HLH-1 functions redundantly with the Hox protein MAB-5 and another transcription factor, FOZI-1, to specify M-derived BWMs (Harfe et al., 1998a; Liu and Fire, 2000; Amin et al., 2007). Curiously, all of these factors are also expressed in and required for the specification of the M-derived non-muscle CCs (Harfe et al., 1998a; Liu and Fire, 2000; Amin et al., 2007). These observations suggest that other factor(s) must be required to differentiate between M-derived muscle and non-muscle fates.
Here, we describe the role of the Six2 family protein CEH-34 and its cofactor EYA-1 in proper specification of non-muscle CC fates. We propose a model in which ceh-34 and eya-1 expression and the subsequent specification of non-muscle cell fates from myogenic precursor cells are regulated in a combinatorial manner by the mesoderm-intrinsic factors HLH-1, FOZI-1 and MAB-5, by SMA-9 and LIN-12 along the dorsal-ventral axis, and by SYS-1 and POP-1 along the anterior-posterior axis.
MATERIALS AND METHODS
C. elegans strains
Strains were maintained and manipulated using standard conditions (Brenner, 1974). Analyses were performed at 20°C, unless otherwise noted.
The strains LW0683 [rrf-3(pk1426) II; ccIs4438 (intrinsic CC:::gfp)III; ayIs2(egl-15∷gfp) IV; ayIs6(hlh-8∷gfp) X] and LW1734 [jjIs1475(myo-3∷rfp) I; rrf-3(pk1426) II; ccIs4438(intrinsic CC:::gfp) III; ayIs2(egl-15∷gfp) IV; ayIs6(hlh-8∷gfp) X] were used to visualize M lineage cells in RNAi experiments. Intrinsic CC∷gfp is a twist-derived coelomocyte marker (Harfe et al., 1998b). Secreted CC∷gfp is another coelomocyte marker using a myo-3∷secreted GFP that is secreted from the body wall muscles and taken up by differentiated CCs (Harfe et al., 1998a). Additional M lineage specific reporters were as described in Kostas and Fire (Kostas and Fire, 2002). The M lineage was followed in live animals under a fluorescence stereomicroscope and a compound microscope. Other strains used in this work are:
LG I: eya-1(ok654) (Furuya et al., 2005), sys-1(q544) (Miskowski et al., 2001), pop-1(q645), pop-1(q624) (Siegfried and Kimble, 2002)
LG II: hlh-1(cc561ts) (Harfe et al., 1998a); rrf-3(pk1426) (Sijen et al., 2001)
LG III: dac-1(gk213), dac-1(gk211) (Colosimo et al., 2004);
LG V: ceh-34(tm3330) and ceh-34(tm3733) (gifts from Shohei Mitani, Tokyo Women’s Medical College, Tokyo, Japan), him-5(e1467) (Hodgkin et al., 1979)
Integrated transgenic lines:
sys-1p∷venus∷sys-1 (Phillips et al., 2007)
hlh-8p∷pop-1∷gfp (this work)
pop-1p∷pop-1∷gfp (Siegfried et al., 2004)
hlh-8p∷mRFP (Jiang et al., 2008)
myo-3∷nls∷rfp∷lacZ (this work)
Plasmid constructs and transgenic lines
Fragments spanning 3.9 kb of the ceh-34 promoter and the entire coding region or the promoter alone were PCR amplified from N2 genomic DNA using iProof™ High-Fidelity DNA Polymerase (Bio-Rad). We obtained a cDNA clone, yk209b2, which spans the entire ORF and the 3’UTR of ceh-34 (gift from Yuji Kohara, National Institute of Genetics, Japan). These PCR fragments and cDNA were used to generate the following ceh-34 reporter constructs: pNMA90: ceh-34p∷ceh-34 genomic ORF∷gfp∷unc-54 3’UTR; pNMA94: ceh-34p∷gfp∷ceh-34 cDNA∷ceh-34 3’UTR. Forced expression constructs: pNMA107: hlh-8p∷gfp∷ceh-34 cDNA∷ceh-34 3’ UTR; pNMA109: hlh-8p∷eya-1 genomic ORF∷unc-54 3’ UTR; pNMA88: hsp-16p∷ceh-34 cDNA∷ceh-34 3’UTR; pNMA110: hsp-16p∷eya-1 cDNA∷unc-54 3’UTR; Other reporter constructs: pJKL758: myo-3p∷NLS∷mRFP∷lacZ∷unc-54 3’UTR; pJKL601: hlh-8p∷pop-1∷gfp∷unc-54 3’ UTR. Plasmids for RNAi of ceh-34(11068D5), eya-1(11020D3), dac-1(11007F3), ceh-35(11062A7), pax-3(11038B3), ceh-33(11058H10) and egl-38(10018H7) were retrieved from the ORFeome-RNAi v1.1 library (Rual et al., 2004). sys-1(T23D8.9) and ceh-32(W05E10.3) RNAi plasmids were obtained from the RNAi library generated by Dr. Julie Ahringer and provided by Geneservice Ltd. The identities of all these RNAi clones were confirmed by sequencing. The eya-1 RNAi clone was subsequently used for additional cloning. pNMA49 (fozi-1 RNAi) and pNMA50 (mab-5 RNAi) were made by sub-cloning into L4440 (Timmons and Fire, 1998) full-length cDNAs for each gene from the plasmids pNMA24 (Amin et al., 2007) and p198 (Liu and Fire, 2000) respectively. pJKL833 (vab-3 RNAi), pJKL834 (pax-3 RNAi) and pJKL835 (pax-2 RNAi) were generated by subcloning into L4440 a PCR fragment corresponding to a genomic region for each gene. RNAi construct pSP28 (pop-1 RNAi) was a gift from David Eisenmann.
Transgenic lines were generated using the plasmids pRF4 (Mello et al., 1991) or LiuFD61 (mec-7p∷mRFP, gift from Sylvia Lee, Cornell University) as markers.
Heat-shock experiments
Transgenic animals harboring pNMA88 (hsp-16p∷ceh-34) and/or pNMA110 (hsp-16p∷eya-1) were subjected to periodic heat-shock at 37°C for 30 minutes followed by recovery for 3-4 hours at 20°C beginning at the 1-M stage until after M-derived CCs were visible using intrinsic CC∷gfp. Alternatively, animals were continuously heat-shocked at 30°C from embryogenesis until the 16-M stage. Both periodic and continuous heat-shock conditions yielded the same results. Non-transgenic heat-shocked animals were used as controls for heat-shock conditions.
RNAi
RNAi by injection (RNAi-E)
ceh-34 dsRNA was synthesized with the T7 RiboMax RNA Production System (Promega) using the ceh-34 RNAi plasmid as a template. To observe the effects of RNAi during embryonic development, dsRNA was further purified and injected into wild-type hermaphrodites of the reference strain LW0683, with water as an injection control. The progeny of injected animals were scored for phenotypes.
RNAi by ingestion (RNAi-P)
To observe the effects of RNAi during postembryonic development, we performed RNAi by ingestion with a synchronous population of L1 larvae. L1 animals synchronized by standard methods (Kamath and Ahringer, 2003) were plated in triplicate on HT115(DE3) bacteria expressing dsRNA for genes of interest. Bacteria for ingestion were prepared as described by Kamath and Ahringer (Kamath and Ahringer, 2003), using 4 mM IPTG to induce dsRNA production. RNAi-P was performed at 25°C and animals were scored for M lineage phenotypes or used for immunostaining 24-48 hours after plating.
Yeast two-hybrid assays
Two-hybrid analysis was performed using the protocol described by James et al. (James et al., 1996). Full length or portions of ceh-34 and eya-1 cDNAs were fused in frame with the GAL4 binding domain (pGBD-C1) and the GAL4 activation domain (pGAD-C1), respectively. Details of the plasmids are available upon request.
Immunofluorescence staining
Animal fixation, immunostaining, microscopy and image analysis were performed as described previously (Amin et al., 2007). Guinea pig anti-FOZI-1 (Amin et al., 2007; 1:200) and goat anti-GFP (Rockland Immunochemicals; 1:5000) antibodies were used. All secondary antibodies were from Jackson ImmunoResearch Laboratories and used in a dilution of 1:50 to 1:200.
RESULTS
ceh-34 is required for specifying the non-muscle CC fates in the M lineage
In an RNAi screen to identify transcription factors important for M lineage development (N.M.A., Z. Via and J.L., unpublished data), we found that RNAi knockdown of ceh-34 during postembryonic development (see Materials and Methods, referred to as RNAi-P) resulted in a loss of M-derived CCs (Table 1, Fig. 1B, D). We followed the M lineage in these animals using the hlh-8∷gfp reporter and αFOZI-1 immunostaining (Harfe et al., 1998b; Amin et al., 2007), and observed normal cleavage orientations and proliferation in the M lineage (Fig. 1A, B). However, at the 18-M stage, both M.drpa and M.dlpa were transformed from CCs to BWMs (as observed using myo-3∷rfp (Fig. 1A, B)) in 10 of 11 animals examined, while the remaining animal had only one CC transformed to a BWM (data not shown). Fate specification of SMs (visualized using hlh-8∷gfp) and their derivatives (visualized using egl-15∷gfp; Fig. 1B,D) was unaffected in ceh-34(RNAi-P) animals. While the lack of defect in the SM lineage could be due to incomplete knockdown of ceh-34 via RNAi-P, the CC to BWM fate transformation caused by ceh-34(RNAi-P) suggests that ceh-34 is required for specifying M-derived CC fates.
Table 1.
M lineage phenotypes of C. elegans Pax-Six-Eya-Dac mutants
| Gene family | Genotype | Number of Embryonic CCs | Number of M-derived CCs |
|---|---|---|---|
| wild-type | 4 (n>200) | 2 (n>200) | |
| ceh-34(RNAi-P) | 4 (n>200) | 0 (98.3%); 2 (1.7%) (n=362) |
|
| Six1/2 | ceh-34(RNAi-E) | 11.3% with 1st pair missing 16.7% with 2nd pair missing 14.1% with both pairs missing (n=538) |
0 (98.6%) 1-2 (1.4%) (n=425) |
| ceh-33(RNAi-P) | 4 (n>200) | 2 (n>200) | |
| Six3/6 | ceh-32(RNAi-P) | 4 (n>200) | 2 (n>200) |
| Six4/5 | ceh-35/unc-39 (RNAi-P) | 4 (n>200) | 2 (n>200) |
| Eyes absent | eya-1(ok654) | 9.5% with 1st or 2nd pair missing 87.7% with both pairs missing (n=203) |
0 (84.7%);1 (1.0%); 2 (14.3%) (n=203) |
| Pax1/9 | pax-1(RNAi-P) | 4 (n>200) | 2 (n>200) |
| Pax2/5/8 | pax-2(RNAi-P) | 4 (n>200) | 2 (n>200) |
| egl-38(RNAi-P) | 4 (n>200) | 2 (n>200) | |
| egl-38(n578) | 4 (n>200) | 2 (n>200) | |
| Pax3 | pax-3(RNAi-P) | 4 (n>200) | 2 (n>200) |
| Pax6 | vab-3(RNAi-P) | 4 (n>200) | 2 (n>200) |
| Dachshund | dac-1(gk211) | 4 (n>200) | 2 (n>200) |
| dac-1(gk213) | 4 (n>200) | 2 (n>200) | |
RNAi-P: RNAi was administered to synchronized L1 larvae via feeding throughout post-embryonic development.
RNAi-E: RNAi via injection into the gonads of gravid adults whose progeny were scored for phenotypes.
There are three other Six family members in C. elegans, ceh-32, ceh-33 and unc-39/ceh-35. ceh-32 is required for head morphogenesis and ceh-32(RNAi-E) leads to embryonic and larval lethality (Dozier et al., 2001). We found that ceh-32(RNAi-P) had no effect on the M lineage (Table 1). unc-39 has been reported to be required for the proper specification and migration of M, and subsequently, the proper development of the M lineage (Yanowitz et al., 2004). We found that unc-39(RNAi-P) gave no M lineage defects (Table 1), suggesting that the M lineage phenotypes observed in unc-39 mutants may reflect an earlier role of unc-39 in M cell specification. Neither ceh-33(RNAi-P) nor ceh-33(RNAi-E) gave any M lineage phenotypes (Table 1 and data not shown). Thus ceh-34 is the only Six family gene required for the specification of M-derived CC fate.
ceh-34 is essential for embryonic CC fates and larval viability
To further assess the role of ceh-34 during embryonic development, we examined two ceh-34 deletion mutants, tm3330 and tm3733 (Fig. 2A), and knocked down ceh-34 during embryogenesis by injecting ceh-34 dsRNA into wild-type animals (referred to as RNAi-E). tm3330 animals carry a 235bp deletion in the second intron and showed no M lineage defects (Table 1), suggesting this deletion does not compromise the function of ceh-34. In contrast, tm3733, which is predicted to truncate CEH-34 after amino acid 40, and ceh-34(RNAi-E), resulted in 100% (n>100) and 99% (n=2210) lethality, respectively. These animals arrest as three-fold embryos or L1 larvae with abnormal anterior morphology, but do specify the M mesoblast (Fig. 1F). These results suggest that ceh-34 is an essential gene required for embryonic and early larval development.
Figure 2. ceh-34 is expressed in the M-derived CC precursors.

(A) Representations of the ceh-34 translational reporter constructs: pNMA90 [ceh-34p∷ceh-34 genomic ORF∷gfp∷unc-54 3’UTR] and pNMA94 [ceh-34p∷gfp∷ceh-34 cDNA∷ceh-34 3’UTR]. The regions deleted in tm3330 and tm3733 are shown. (B-G): Representative images of live animals expressing ceh-34∷gfp (pNMA90) at different developmental stages. Two focal planes of a bean stage embryo (B,C, anterior to the left and dorsal up) and a 3-fold stage embryo (D,E). (F) A L1 larva and (G) an adult. Arrows in G denote BWMs. (H-O) The left side of a wild-type (H-K) or mab-5(RNAi-P) (L-O) L1 larva double labeled with CEH-34∷GFP (H,L) and anti-FOZI-1 antibody (I,M) at the 16-M stage. (J,N) Merged images of H and I, and L and M, respectively. CEH-34∷GFP was detected in M.dlpa (I) and M.drpa (not shown), both M-derived CC precursors. (K,O) Summary of ceh-34∷gfp expression in the M lineage, with ceh-34∷gfp-positive cells in green circles. All images are anterior to the left.
In the above RNAi-E experiments, eight animals escaped the embryonic/L1 arrest, but lacked some or all of the 4 embryonically-derived CCs (Fig. 1G). We repeated the ceh-34(RNAi-E) experiment and scored a larger number of animals that escaped the embryonic/L1 arrest using two independent reporters of the CC fate (see Materials and Methods). 42% (n=538) of these animals had a reduced number of embryonically-derived CCs (Table 1). These animals lacked a pair of CCs located in the head (the 1st pair), near the vulva (the 2nd pair) or both pairs. Thus, ceh-34 is also required for the proper development of the four embryonically-derived CCs.
ceh-34 is expressed in the CC precursor cells in the M lineage
To understand how ceh-34 functions during development, we generated two sets of transgenic lines with each of two translational ceh-34∷gfp fusion constructs, pNMA90 and pNMA94 (Fig. 2A). Both showed identical patterns of GFP expression and nuclear localization (Fig. 2). ceh-34∷gfp expression begins late during embryogenesis and persists in a few anterior BWM cells and other unidentified cells in the head throughout postembryonic development (Fig. 2B-G). A similar ceh-34 expression pattern has been independently observed by T. Hirose and H. R. Horvitz (personal communication).
To determine the expression pattern of ceh-34 in the M lineage, we performed double-labeling experiments using the ceh-34∷gfp fusions and hlh-8p∷rfp or αFOZI-1 staining to label M lineage cells (Harfe et al., 1998b; Amin et al., 2007). ceh-34∷gfp is transiently expressed in the M lineage, specifically in M.dlpa and M.drpa, before they terminally differentiate into CCs (Fig. 2H-K). ceh-34∷gfp expression is no longer detectable in these two cells after they differentiate into CCs. For simplicity, we will henceforth refer to the undifferentiated M.dlpa and M.drpa cells as M-derived CC precursor cells.
ceh-34 alone is not sufficient to specify CC fates
Because ceh-34 is expressed in the M-derived CC precursors and is required for specifying the M-derived CCs, we asked whether ceh-34 is sufficient to specify the CC fate. We forced ceh-34 expression in mixed-staged animals via a heat-shock inducible promoter (see Materials and Methods) and observed no effect on the number of cells expressing the intrinsic CC∷gfp marker. To more directly test whether ceh-34 is sufficient to specify the CC fate, we forced ceh-34 expression in all undifferentiated cells of the M lineage using the hlh-8 promoter (pNMA107), but did not detect any extra M-derived CCs (Fig. 3H). Thus, although ceh-34 is required for specifying the CC fate, it alone is not sufficient to induce other cells to adopt CC fates.
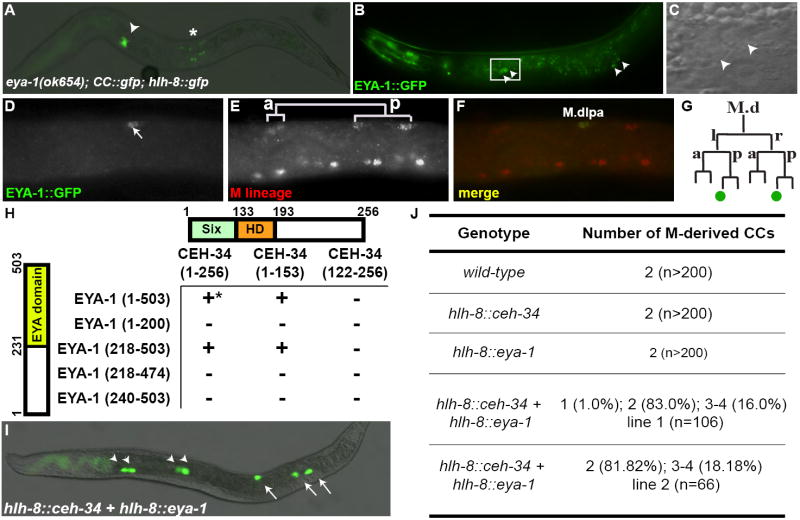
Figure 3. EYA-1 acts as a cofactor for CEH-34 in CC fate specification.
(A) An eya-1(ok654) L4 larva missing one pair of embryonic CCs and both M-derived CCs. Arrowhead denotes the anterior pair of embryonically-derived CCs. GFP positive cells near the asterisk are SM descendants labeled by hlh-8∷gfp. (B,C) A wild-type adult expressing eya-1∷gfp (B). GFP was detected in the four embryonically-derived CCs (arrowheads). (C) Magnified DIC view of the anterior pair of CCs in the box in (B). (D-F) The left side of an L1 larva double labeled with EYA-1∷GFP (D), anti-FOZI-1 antibody (E) at the 16-M stage. (F) A merged image of D and E. EYA-1∷GFP was detected in M.dlpa (F) and M.drpa (not shown). (G) Summary of eya-1∷gfp expression in the M lineage, with eya-1∷gfp-positive cells in green circles. (H) The Six domain of CEH-34 binds to the Eya domain of EYA-1 via the yeast two-hybrid assay. Respective amino acid residues are labeled next to schematics of each protein. *All interactions were tested in both directions, with the exception of GBD-EYA-1(1-503), which auto-activates reporter expression. Western blot analysis was performed to ensure proper protein expression. (I,J) Forced expression of both ceh-34 and eya-1 in the M lineage leads to ectopic CC fates. Panel I shows an example of such a worm. Arrowheads point to embryonically-derived CCs while arrows point to M-derived CCs.
EYA-1 is required for CC fate specification
In both Drosophila and vertebrates, Six homeodomain proteins have been shown to function together with other proteins, including Pax, Eya and Dac (Kawakami et al., 2000). C. elegans contains five Pax homologs, pax-1, pax-2, egl-38, pax-3, and vab-3/pax-6, and one homolog each for Eya and Dac, eya-1 and dac-1 (Hobert and Ruvkun, 1999; Dozier et al., 2001; Colosimo et al., 2004; Furuya et al., 2005). Mutants or RNAi-P against the Pax and Dac homologs do not cause any M lineage defects (Table 1), suggesting that none of the Pax genes on their own, nor the single Dachshund homolog, are essential in the postembryonic M lineage. However, the strong loss-of-function allele of eya-1, ok654 (Furuya et al., 2005), displayed M lineage phenotypes similar to ceh-34(RNAi-P): loss of M-derived CCs and variable loss of embryonic CCs (Table 1, Fig. 3A). Thus, both ceh-34 and eya-1 are required for specifying the embryonic and M-derived CCs.
EYA-1 acts as a cofactor for CEH-34 in CC fate specification
Six family proteins have been shown in the fruit fly and the mouse to use Eya as a cofactor to regulate gene expression (Pignoni et al., 1997; Ohto et al., 1999; Li et al., 2003). We tested whether EYA-1 can act as a cofactor for CEH-34 in the C. elegans M lineage. We found that eya-1∷gfp (Furuya et al., 2005) is expressed in a similar pattern to ceh-34∷gfp, but differs slightly from ceh-34∷gfp, which is transiently expressed in M-derived CC precursors; in contrast, eya-1∷gfp expression is detected in all six differentiated CCs throughout development (Fig. 3B-G). We also found that the physical interaction between Six and Eya proteins is likely conserved, as the Six domain of CEH-34 and the Eya domain of EYA-1 interact in yeast two-hybrid assays (Fig. 3H). Thus EYA-1 may serve as a cofactor for CEH-34 in CC fate specification.
To test whether CEH-34 and EYA-1 together can promote the CC fate, we forced the expression of each gene or both genes together using the hlh-8 promoter (Harfe et al., 1998b). Forced expression of either ceh-34 or eya-1 alone did not result in any ectopic CCs in the M lineage (Fig. 3J). However, when both genes were simultaneously expressed in the M lineage, we observed an increase in the number of M-derived CCs (Fig. 3I, J). Thus CEH-34 and EYA-1 can act together to specify ectopic CC fates. Taken together, our data are consistent with the role of EYA-1 as a cofactor for CEH-34 in non-muscle CC-specific transcription.
CEH-34 and EYA-1 act downstream of mesoderm-intrinsic transcription factors necessary for muscle and non-muscle fates
The expression of CEH-34 and EYA-1 in the M lineage is preceded by the expression of a number of transcription factors that are required for both BWM and CC fates. MAB-5, HLH-1 and FOZI-1 are expressed in and required for the CC and BWM precursor cells of the M lineage (Harfe et al., 1998a; Liu and Fire, 2000; Amin et al., 2007). We tested if these factors are required for the expression of both ceh-34 and eya-1. We found that mab-5(RNAi) and fozi-1(RNAi) animals lost expression of ceh-34∷gfp and eya-1∷gfp in the presumptive CCs (Fig. 2L-O; data not shown). Similarly, ceh-34∷gfp and eya-1∷gfp were not detected in the M lineage of the temperature sensitive hlh-1(cc561ts) mutants at the restrictive temperature (data not shown). Thus mab-5, hlh-1 and fozi-1 are required for the expression of ceh-34 and eya-1.
CEH-34 and EYA-1 act downstream of POP-1 and SYS-1, components of the Wnt/β-catenin asymmetry pathway
SYS-1 and POP-1 are asymmetrically distributed along the anterior-posterior axis in the M lineage
Since CEH-34 expression is limited to the anterior daughters of M.dlp and M.drp, we wanted to identify factors regulating this anterior expression. The Wnt/β-catenin asymmetry pathway is involved in multiple anterior-posterior fate decisions in C. elegans (Mizumoto and Sawa, 2007). In particular, the TCF/LEF homolog POP-1 is enriched in the anterior daughter, while the β-catenin homolog SYS-1 is enriched in the posterior daughter of an A-P cell division (Lin et al., 1995; Lin et al., 1998; Huang et al., 2007; Phillips et al., 2007). Could pop-1 and sys-1 play similar roles in A-P divisions in the M lineage?
We first examined the expression patterns of functional sys-1∷gfp and pop-1∷gfp reporters (Materials and Methods) within the early M lineage. Both sys-1∷gfp and pop-1∷gfp are present in the early M lineage at the 1-M stage and evenly distributed through the 4-M stage (Fig. 4A-D). As in other C. elegans lineages, SYS-1∷GFP is enriched in the posterior nucleus, while POP-1∷GFP is enriched in the anterior nucleus, after each subsequent cell division resulting in 8-, 16- and 18-M lineage descendants (Fig. 4A-H).
Figure 4. CEH-34 acts downstream of SYS-1 and POP-1 in the M lineage to specify CC fates.

(A-H) Localization of POP-1∷GFP (A) and SYS-1∷GFP (E) in the M lineage of wild-type animals. (B,F) M lineage cells marked by anti-FOZI-1 staining; (C,G) the corresponding merged images. POP-1∷GFP is enriched in the nuclei of anterior cells (A), while SYS-1∷GFP is enriched in the nuclei of posterior cells (E). (D,H) Summary of asymmetric localization patterns of POP-1∷GFP and SYS-1∷GFP, respectively, in the M lineage. Black circles represent enriched localization; grey circles represent lower GFP levels. (I) A sys-1(q544) L4 larva with extra M-derived CCs (arrow) and its corresponding lineage (J). Arrowheads denote embryonically-derived CCs. (K-R) Two sys-1(RNAi) larvae with ectopic ceh-34∷gfp expression. K-M are lateral views while O-Q are dorsal images. (K,O) ceh-34∷gfp expression; (L,P) anti-FOZI-1 staining (with dorsal M lineage cells labeled); (M,Q) corresponding merged images; (N,R) corresponding lineages for K-M and O-Q, respectively, with green circles representing ceh-34∷gfp. (S-Z) POPTOP∷mCherry reporter expression in the M lineage of wild-type (S-V) and sys-1(RNAi-P) (W-Z) animals. (S,W) mCherry signal; (T,X) M lineage cells (hlh-8∷gfp); (U,Y) corresponding merged images; (V,Z) Summary of mCherry (red circles) expression in the M lineage. Note the faint mCherry signal in M.dlp and strong mCherry signal in M.vlpa and M.vlpp (U) in the wild-type animal, and the absence of mCherry signal in the M lineage of the sys-1(RNAi-P) animal (Y).
Reduction of SYS-1 levels results in the presence of extra M-derived CCs and SMs
To test whether the asymmetric distribution of SYS-1 reflected a role for sys-1 in M lineage development, we examined M lineage development in strong loss-of-function mutant sys-1(q544) animals and sys-1(RNAi-P) animals. Both had an increase in the number of dorsal CCs and ventral SMs derived from the M lineage (Fig. 4I,J, Table 2). We used hlh-8∷gfp and αFOZI-1 staining to follow the M lineage and determine the cause of these phenotypes.
Table 2.
ceh-34 functions downstream of POP-1 and SYS-1 to specify CC fates
| A | |||||
|---|---|---|---|---|---|
| Genotype | Number of M-derived CCs (intrinsic CC:gfp) | ||||
| n | 0-1 | 2 | 3-4 | 5-7 | |
| wild-type | >200 | 0% | 100% | 0% | 0% |
| L4440 RNAi | 201 | 2.5% | 97.5% | 0% | 0% |
| sys-1(q544) | 112 | 0% | 10.7% | 58.9% | 30.4% |
| sys-1(RNAi-P) | 86 | 6.9% | 41.9% | 38.4% | 12.8% |
| sys-1(q544); ceh-34(RNAi-P) | 22 | 100% | 0% | 0% | 0% |
| pop-1(RNAi-P) | 67 | 65.7% | 20.9% | 13.4% | 0% |
| pop-1(q624) | 68 | 20.6% | 79.4% | 0% | 0% |
| pop-1(q645) | 49 | 2.0% | 8.2% | 59.2% | 30.6% |
| pop-1(q645); ceh-34(RNAi-P) | 53 | 100% | 0% | 0% | 0% |
| wrm-1(RNAi-P) | 96 | 13.5% | 51.0% | 33.4% | 2.1% |
| lit-1(RNAi-P) | 71 | 14.1% | 45.1% | 32.3% | 8.5% |
| B | |||||
| Genotype | Number of M-derived SMs (hlh-8∷gfp) | ||||
| n | 0-1 | 2 | 3-4 | ||
| wild-type | >200 | 0% | 100% | 0% | |
| L4440RNAi | >200 | 0% | 100% | 0% | |
| sys-1(q544) | 37 | 0% | 32.4% | 67.6% | |
| sys-1(RNAi-P) | 59 | 0% | 54.2% | 45.8% | |
| pop-1(RNAi-P) | 99 | 41.4% | 49.5% | 9.1% | |
| pop-1(q645) | 22 | 0% | 0% | 100% | |
| wrm-1(RNAi-P) | 45 | 0% | 22.2% | 77.8% | |
| lit-1(RNAi-P) | 47 | 8.5% | 46.8% | 44.7% | |
On the ventral side of sys-1(q544) animals, M.vlpp and/or M.vrpp underwent an extra division along the anterior-posterior axis, most often producing an SM and a BWM, much like their anterior sister cells, M.vlpa and M.vrpa (67.6%, n=37; Fig. 4J, Supplemental Fig. S1). We observed this behavior in 11 of 12 animals, with some variation in which cells generated the extra SMs (Supplemental Fig. S1). This variation may be due to residual SYS-1 in sys-1(q544) animals. Similar posterior-to-anterior fate transformations occurred in the ventral M lineage of sys-1(RNAi-P) animals (45.8%, n=59). These fate transformations at the 16-M stage are the cause for the extra SMs observed.
Unlike the ventral side, the extra CCs observed in sys-1(q544 or RNAi-P) animals on the dorsal side of the M lineage were not simply due to a posterior-to-anterior fate transformation. All 10 sys-1(q544) animals and 19 of 24 sys-1(RNAi-P) animals had 9-12 cells on the dorsal side of the M lineage (rather than the usual 8) due to an extra round of cell division by M.d(l/r)pp or M.d(l/r)pa or both (Supplemental Figs. S2 and S3). Thus sys-1 is required to suppress further proliferation of the daughters of M.d(l/r)p.
Increased cell proliferation alone does not account for all the extra CCs observed in sys-1(q544) and sys-1(RNAi-P) animals. In instances where M.d(l/r)pa and M.d(l/r)pp did not undergo an extra division, the BWM fate of M.d(l/r)pp was transformed to the CC fate of its anterior sister M.d(l/r)pa (Supplemental Fig. S3). As shown in Fig. 4 and Supplemental Fig. S2, descendants of M.d(l/r)pa and M.d(l/r)pp show a bias toward the CC fate in sys-1(q544) and sys-1(RNAi-P) animals. Thus sys-1 is required to suppress the CC fate in M.d(l/r)pp, the posterior sisters of the CC precursor cells M.d(l/r)pa.
sys-1 represses CC fates in the M lineage by negatively regulating ceh-34 and eya-1 expression
Because ceh-34 and eya-1 function to specify M-derived CCs, we asked whether the extra M-derived CCs in sys-1 mutant animals were due to inappropriate ceh-34 and eya-1 expression. We found that sys-1(RNAi-P) resulted in the ectopic expression of ceh-34∷gfp (Fig. 4K-R, Supplemental Fig. S3) and eya-1∷gfp (data not shown) within the M lineage in a pattern consistent with the transformations to the CC fate observed above.
To confirm that the extra CCs in sys-1 mutants are due to the ectopic expression of ceh-34, we performed ceh-34(RNAi-P) in a sys-1(q544) mutant background. As shown in Table 2, ceh-34(RNAi-P) resulted in a loss of all M lineage-derived CCs in sys-1(q544) animals. Thus sys-1 negatively regulates ceh-34 and eya-1 expression in the posterior sister cells of M-derived CCs and prevents those cells from adopting the CC fate.
pop-1 is required to activate ceh-34 expression to properly specify M-derived CCs
We examined the role of pop-1 in the M lineage by using two mutant alleles of pop-1, q645 and q624, and by performing pop-1(RNAi-P). q624 is a weak loss-of-function allele of pop-1 which inhibits DNA binding, while q645 carries a point mutation in the β-catenin interacting domain (Siegfried and Kimble, 2002). We detected a range of M lineage defects in pop-1(q624), pop-1(q645) and pop-1(RNAi-P) animals. Both pop-1(q624) and pop-1(RNAi-P) worms exhibit a loss of M-derived CCs (20.6%, n=68 for pop-1(q624) and 65.7%, n=67 for pop-1(RNAi-P)) and SMs (41.4%, n=99 for pop-1(RNAi-P)), an M lineage phenotype opposite to that of sys-1 mutants (Table 2). The loss of SMs was due to a fate transformation of M.v(l/r)pa to the fate of its posterior sister M.v(l/r)pp (Supplemental Fig. S4). The loss of CCs was due to a combination of under-proliferation of the dorsal M lineage and fate transformation of M.d(l/r)pa to the fate of its posterior sister M.d(l/r)pp (normally fated to become BWMs), and was accompanied by the loss of ceh-34∷gfp and eya-1∷gfp expression in the M lineage (Supplemental Fig. S5; data not shown). Thus pop-1 is required for specification of the M-derived CC fate by positively regulating proliferation and ceh-34 and eya-1 expression in M.dlpa and M.drpa.
Interestingly, pop-1(q645) animals displayed an M lineage phenotype that resembled that of sys-1 mutants. 89.8% (n=49) of q645 animals had extra M-derived CCs on the dorsal side and 100% of q645 (n=22) animals had extra SMs on the ventral side (Table 2). The extra SMs are due to a fate transformation of M.v(l/r)pp to M.v(l/r)pa (Supplemental Fig. S6). The ectopic CCs arise from the extra divisions of M.d(l/r)pa and M.d(l/r)pp and posterior-to-anterior fate transformations among the descendants of these two cells (data not shown). Furthermore, generation of extra CCs in q645 animals correlates with ectopic ceh-34∷gfp expression (data not shown) and depends on the presence of ceh-34, as ceh-34(RNAi-P) in pop-1(q645) mutants resulted in the loss of all M-derived CCs (Table 2). Since the q645 mutation is located in a conserved β-catenin binding domain of POP-1 (Siegfried and Kimble, 2002), the similarity in phenotypes of pop-1(q645) to sys-1 loss of function animals suggests that SYS-1 is required for the normal activity of POP-1 in M.d(l/r)pp and M.v(l/r)pp (see Discussion below).
POP-1 functions as a transcriptional activator in the posterior daughters of the M lineage
Previous studies have shown that a high SYS-1 to POP-1 ratio makes POP-1 a transcriptional activator (Kidd et al., 2005). We monitored a reporter of TCF/LEF activity, POPTOP-mCherry, in the M lineage (Green et al., 2008). Faint mCherry expression was detected in M.d(l/r)p and M.v(l/r)p, but not in their anterior counterparts, just before these cells divide (M.dlp in Fig. 4S-U). The mCherry signal remains visible in both the anterior and posterior descendants of M.d(l/r)p and M.v(l/r)p (M.vlp(a/p) in Fig. 4S-U). Faint mCherry expression was again detectable in the posterior descendants of M.d(l/r)a and M.v(l/r)a (data not shown). This pattern of mCherry expression was seen in 15 out of 16 animals examined; the overall expression pattern of mCherry is summarized in Fig. 4V. Taking into account the slow folding rate of mCherry (Shaner et al., 2005), which could account for the faint signals, and the potential persistence of mCherry in both daughters of a cell expressing the reporter, these results are consistent with activation of the POPTOP reporter in the posterior cells of the M lineage.
We asked whether the activation of the POPTOP reporter in the posterior cells requires SYS-1 by examining the expression of mCherry in sys-1(RNAi-P) animals. sys-1(RNAi-P) consistently led to an overall decrease of the mCherry signal in larvae (Fig. 4W-Y). Furthermore, in 8 out of 10 animals scored, sys-1(RNAi-P) led to a loss of mCherry expression in most, if not all, M lineage descendants (Fig. 4W-Z). Thus a high SYS-1 to POP-1 ratio in the posterior daughters of the M lineage likely converts POP-1 to a transcriptional activator.
DISCUSSION
An evolutionarily conserved Six-Eya cassette in mesodermal development
CEH-34 belongs to a highly conserved family of homeodomain proteins called the Six family. Previous studies have shown the Pax-Six-Eya-Dac network functions in multiple developmental processes in Drosophila and vertebrates (Heanue et al., 1999; Xu et al., 1999; Hanson, 2001; Clark et al., 2006). Mutations in Six1 and Eya1 in humans have also been shown to cause Brancio-oto-renal (BOR) syndrome (Kochhar et al., 2007). However, the composition of the Pax-Six-Eya-Dac network appears to vary in different developmental contexts. For example, ey and dac are coexpressed in the developing mushroom bodies of the Drosophila central nervous system, but eya and so are absent there (Kurusu et al., 2000; Martini et al., 2000; Noveen et al., 2000). Similarly, eya and dac function independently in Drosophila neuronal specification (Miguel-Aliaga et al., 2004). In C. elegans, the Pax6 homolog vab-3 is required for proper head morphogenesis and directly regulates the expression of the Six gene ceh-32 (Dozier et al., 2001). vab-3 also genetically interacts with eya-1 during embryonic morphogenesis, but mutants in the single Dac homolog dac-1 do not display any anterior morphogenesis defects (Colosimo et al., 2004; Furuya et al., 2005). Here we show that proper specification of the non-muscle coelomocytes in the C. elegans postembryonic mesoderm requires the functions of both ceh-34 and eya-1, but not dac-1 or any of the five Pax genes individually. Although we cannot rule out the possibility that the Pax genes may function redundantly in the M lineage, our data are consistent with the notion that not all members of the Pax-Six-Eya-Dac network always function together in different cell and tissue types. Similarly, CEH-34 and EYA-1 have been shown to both be required for the death of the MR motor neuron sister cell and to interact physically by T. Hirose and H. R. Horvitz (personal communication).
Six and Eya proteins bind to each other and function together in various developmental contexts, including the mesoderm. In Drosophila, Six4 and Eya function together for the proper patterning of the non-dorsal mesoderm (Clark et al., 2006; Liu et al., 2009). Six1 and Eya2 proteins in mouse function together to regulate the expression of myogenic regulatory factors involved in multiple aspects of skeletal myogenesis (Grifone et al., 2005). In this study, we showed that CEH-34 and EYA-1 function together to promote the non-muscle coelomocyte fate in the C. elegans postembryonic mesoderm. A number of conserved transcription factors function in mesoderm development throughout metazoans (Harfe et al., 1998; Evans, 1999; Fukushige et al., 2006). Except for the interaction module of the bHLH factor Twist and its E protein binding partner (Spicer et al., 1996; Harfe et al., 1998b), few other examples of protein interaction modules have been found to play a conserved role in mesoderm fate specification across metazoan species. Our findings suggest that Six-Eya protein interactions represent another evolutionarily conserved cassette essential for mesodermal development in metazoans.
The conservation of the Six-Eya protein complex is likely due to the distinct biochemical properties of these two proteins. In general, Six proteins can bind to DNA, but cannot activate transcription of downstream targets, suggesting a general repressive effect of Six proteins on their own (Li et al., 2003). Eya proteins are phosphatases that do not bind DNA directly, but function as co-activators of Six proteins and recruit additional co-activators (Li et al., 2003). Thus proper activation of downstream target genes requires the function of the Six-Eya protein complex. Even in cases where some Six proteins have intrinsic activation domains, activation of target genes via these proteins is only clearly evident in the presence of Eya (Kawakami et al., 1996; Spitz et al., 1998; Ohto et al., 1999).
CEH-34 and EYA-1 act downstream of the mesoderm-intrinsic transcription factors in the M lineage to specify non-muscle mesodermal fates in C. elegans
The mesoderm gives rise to a variety of muscle and non-muscle cell types. Previous studies in both vertebrates and invertebrates have identified a number of factors, including the myogenic regulatory factors (MRFs) that are critical for the specification of myogenic fates (Pownall et al., 2002). Much less is known about the mechanisms involved in the specification of non-myogenic mesodermal cells. HLH-1, the lone C. elegans MRF, is sufficient to induce BWM fates when ectopically expressed in the C. elegans early embryo, like its vertebrate counterparts (Fukushige and Krause, 2005). However, two of the cells expressing hlh-1 in the M lineage become specified as non-muscle CCs. Two other mesoderm-intrinsic transcription factors FOZI-1 and MAB-5, together with HLH-1, are required for specifying these two CCs as well as the M-derived BWMs (Harfe et al., 1998a; Liu and Fire, 2000; Amin et al., 2007). We have shown that ceh-34 and eya-1 expression in the CC precursors is regulated by hlh-1, fozi-1 and mab-5. Taken together, these observations suggest that HLH-1, FOZI-1 and MAB-5 make cells competent to become BWM and CC, and that ceh-34 and eya-1 are further required to specify non-muscle CCs from these bipotent precursors. This is not the first example in which cells expressing a MRF do not necessarily adopt muscle fates. Cells initially expressing the MRF Myf5 give rise to brown fat cells in addition to muscles (Seale et al., 2008). Once the brown adipose tissue is differentiated, Myf5 expression is no longer detectable. It will be interesting to see whether or not Myf5 is required for the specification of non-muscle brown fat cell fates.
CEH-34 and EYA-1 function downstream of the Wnt/β-catenin asymmetry pathway that regulates anterior-posterior asymmetry in the M lineage
The Wnt/β-catenin asymmetry pathway is involved in many anterior-posterior fate decisions during C. elegans development (Mizumoto and Sawa, 2007). Specifically, POP-1, the C. elegans TCF/LEF transcription factor, is enriched in the nucleus of the anterior daughter of anterior-posterior divisions, while the divergent β-catenin SYS-1 is enriched in the nucleus of the posterior daughter (Lin et al., 1998; Huang et al., 2007; Phillips et al., 2007). The ratio of high levels of SYS-1 to POP-1 in the posterior cell allows for binding of SYS-1 to POP-1, converting it from a repressor to an activator, while high concentrations of POP-1 in the anterior cell keep POP-1 as a repressor (Kidd et al., 2005; Liu et al., 2008). The asymmetric distribution of nuclear POP-1 and SYS-1 is crucial in the specification of anterior vs. posterior cell fates in the embryo (Lin et al., 1995; Lin et al., 1998; Huang et al., 2007). The reciprocal asymmetric distribution of POP-1 and SYS-1 in sister cells along the anterior-posterior axis is maintained during postembryonic development, but also in cells along the proximal-distal axis in the somatic gonad and the vulval precursor cells (Herman, 2001; Siegfried et al. 2002; Kidd et al., 2005; Phillips et al., 2007; Green et al., 2008). A reporter of POP-1 transcriptional activity (POPTOP) is activated in daughter cells of P5.p and P7.p which have high SYS-1 to POP-1 ratios (Green et al., 2008). Intriguingly, in the postembryonic lineages examined to date, loss-of-function mutants for both pop-1 and sys-1 give identical mutant phenotypes (Siegfried et al., 2004; Huang et al., 2007; Green et al., 2008).
We found that the reciprocal asymmetries of POP-1 and SYS-1 along the anterior-posterior axis are conserved in the M lineage. We also found that reducing the level of SYS-1 resulted in partially penetrant posterior-to-anterior fate transformations and ectopic expression of ceh-34 and eya-1 in posterior cells. Reduced levels of POP-1 give a reciprocal result: partially penetrant anterior-to-posterior fate transformations and loss of ceh-34 and eya-1 expression. In contrast to this phenotype, q645, a mutation in POP-1 that blocks its ability to bind SYS-1 (Siegfried and Kimble, 2002), causes a sys-1-like phenotype. RNAi of lit-1 or wrm-1, which blocks export of POP-1 from the posterior nuclei, also resulted in the loss of posterior fates (Table 2). Finally, a reporter for TCF/LEF function is activated in cells with high levels of SYS-1, and this activation is dependent on sys-1. These results can be reconciled by the model presented in Fig. 5.
Figure 5. A model for non-muscle CC fate specification in the M lineage.
(A) ceh-34 and eya-1 expression and subsequent specification of M-derived CC fate requires the combinatorial actions of three sets of factors: the M lineage intrinsic transcription factors including MAB-5, FOZI-1 and HLH-1 that are required for CC and BWM fate, the cell competence factor(s) X, and the repressive activity of POP-1 due to a low SYS-1 to POP-1 ratio. This low SYS-1 to POP-1 ratio leads to the repression of a repressor of ceh-34 and eya-1, which in turn results in the expression of ceh-34 and eya-1. (B) Combinatorial regulatory inputs leading to ceh-34 and eya-1 expression and CC fates. Solid lines in (A) and (B) do not represent direct regulation.
In this model, we propose that cells expressing the mesoderm-intrinsic transcription factors MAB-5, FOZI-1 and HLH-1 are fated to become either myogenic BWMs or non-myogenic CCs. Expression of ceh-34 and eya-1 promotes the CC fate. POP-1 functions as a repressor in the anterior daughters of each division by repressing an unknown transcriptional repressor of ceh-34 and eya-1 so that ceh-34 and eya-1 are expressed to specify the CC fate. However, not all cells expressing MAB-5, FOZI-1 and HLH-1 can respond to the POP-1 repressive activity. TCF/LEF proteins often act synergistically with other cell competence factors to affect gene expression in a cell-type specific manner (Barolo, 2006). We propose that another competence factor (Factor X) functions to distinguish the daughters of M.d(l/r)p from the daughters of M.d(l/r)a and acts as an additional activator of ceh-34 and eya-1. Thus, the expression of ceh-34 and eya-1 and the specification of the CC fate only happen because of a combination of three sets of activities: 1) the mesoderm-intrinsic transcription factors that specify CC and BWM fates, 2) the POP-1 repressive activity and 3) the competence Factor X. The existence of Factor X in regulating ceh-34 and eya-1 expression and for providing competence for cells to become CCs (Fig. 5B) is consistent with our observations that a) the defects in sys-1 or pop-1 mutants are only restricted to the daughters of M.d(l/r)p and M.v(l/r)p, and b) ectopic expression of ceh-34 and eya-1 throughout the M lineage did not convert all BWMs to CCs. Finally, the expression of ceh-34 and eya-1 and the specification of CCs on the dorsal side are also under the control of dorsal-ventral patterning mechanisms (data not shown) that involve the LIN-12/Notch and TGFβ (antagonized by SMA-9) signaling pathways (Fig. 5B). We envision that a similar model could be applied to the ventral M lineage for the specification of SMs.
This model states that a complex containing POP-1 and SYS-1 functions as a transcriptional activator, while POP-1 functions as a repressor. Consistent with this model, POP-1 repressor activity and ectopic CC fates in M.d(l/r)pp were achieved by 1) reducing SYS-1 levels, 2) increased nuclear levels of POP-1 by wrm-1(RNAi) or lit-1(RNAi) and 3) blocking POP-1 binding to SYS-1 as in the pop-1(q645) allele. Conversely, lowering the level of POP-1 led to a higher ratio of POP-1-SYS-1 complexes and resulted in the loss of CC fates in M.d(l/r)pa in pop-1(RNAi) and pop-1(q624) animals.
This model predicts that complete loss of POP-1 function in the M lineage would result in a failure to activate the repressor of ceh-34 and eya-1 in the M lineage. Our data support this, as some pop-1(RNAi-P) animals (Table 2, Supplemental Fig. S5), but not q624 animals, displayed an M lineage phenotype resembling that caused by sys-1 loss-of-function, including the presence of extra CCs and extra cells expressing ceh-34 and eya-1. This phenotype may reflect a complete knockdown of POP-1 in the daughters of M.d(l/r)p and M.v(l/r)p.
Supplementary Material
Acknowledgments
We thank the C. elegans Genetics Center, David Eisenmann, Judith Kimble, Morris Maduro, Brian Harfe, Yuji Kohara, Shohei Mitani, Paul Sternberg and Asako Sugimoto for strains and plasmids; Marisa Foehr, Ken Kemphues, Diane Morton and Chenxi Tian for helpful discussions and valuable comments on the manuscript. This work was supported by NIH R01 GM066953 (to J.L.). S.-E.L. was a Rawlings Cornell Presidential Research Scholar and T.L.C. was a Howard Hughes Undergraduate Research Scholar at Cornell University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amin NM, Hu K, Pruyne D, Terzic D, Bretscher A, Liu J. A Zn-finger/FH2-domain containing protein, FOZI-1, acts redundantly with CeMyoD to specify striated body wall muscle fates in the Caenorhabditis elegans postembryonic mesoderm. Development. 2007;1:19–29. doi: 10.1242/dev.02709. [DOI] [PubMed] [Google Scholar]
- Barolo S. Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene. 2006;57:7505–7511. doi: 10.1038/sj.onc.1210057. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;1:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;7:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;5:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Clark IB, Boyd J, Hamilton G, Finnegan DJ, Jarman AP. D-six4 plays a key role in patterning cell identities deriving from the Drosophila mesoderm. Dev Biol. 2006;1:220–231. doi: 10.1016/j.ydbio.2006.02.044. [DOI] [PubMed] [Google Scholar]
- Colosimo ME, Brown A, Mukhopadhyay S, Gabel C, Lanjuin AE, Samuel AD, Sengupta P. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr Biol. 2004;24:2245–2251. doi: 10.1016/j.cub.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Dozier C, Kagoshima H, Niklaus G, Cassata G, Burglin TR. The Caenorhabditis elegans Six/sine oculis class homeobox gene ceh-32 is required for head morphogenesis. Dev Biol. 2001;2:289–303. doi: 10.1006/dbio.2001.0325. [DOI] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008 doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foehr ML, Lindy AS, Fairbank RC, Amin NM, Xu M, Yanowitz J, Fire AZ, Liu J. An antagonistic role for the C. elegans Schnurri homolog SMA-9 in modulating TGFbeta signaling during mesodermal patterning. Development. 2006;15:2887–2896. doi: 10.1242/dev.02476. [DOI] [PubMed] [Google Scholar]
- Foehr ML, Liu J. Dorsoventral patterning of the C. elegans postembryonic mesoderm requires both LIN-12/Notch and TGFbeta signaling. Dev Biol. 2008;1:256–266. doi: 10.1016/j.ydbio.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM. Vertebrate Tinman Homologues and Cardiac Differentiation. Semin Cell Dev Biol. 1999;10:73–83. doi: 10.1006/scdb.1999.0282. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M. Defining the Transcriptional Redundancy of Early Bodywall Muscle Development in C. Elegans: Evidence for a Unified Theory of Animal Muscle Development. Genes Dev. 2006;20:3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Krause M. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development. 2005;8:1795–1805. doi: 10.1242/dev.01774. [DOI] [PubMed] [Google Scholar]
- Furuya M, Qadota H, Chisholm AD, Sugimoto A. The C. elegans eyes absent ortholog EYA-1 is required for tissue differentiation and plays partially redundant roles with PAX-6. Dev Biol. 2005;2:452–463. doi: 10.1016/j.ydbio.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Green JL, Inoue T, Sternberg PW. Opposing Wnt pathways orient cell polarity during organogenesis. Cell. 2008;4:646–656. doi: 10.1016/j.cell.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983;2:435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;9:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 2008;17:2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;12:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Hanson IM. Mammalian homologues of the Drosophila eye specification genes. Semin Cell Dev Biol. 2001;6:475–484. doi: 10.1006/scdb.2001.0271. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Branda CS, Krause M, Stern MJ, Fire A. MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development. 1998a;13:2479–2488. doi: 10.1242/dev.125.13.2479. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 1998b;16:2623–2635. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, Lassar AB, Tabin CJ. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;24:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman M. C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity. Development. 2001;4:581–590. doi: 10.1242/dev.128.4.581. [DOI] [PubMed] [Google Scholar]
- Hobert O, Ruvkun G. Pax genes in Caenorhabditis elegans: a new twist. Trends Genet. 1999;6:214–216. doi: 10.1016/s0168-9525(99)01731-x. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;1:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Shetty P, Robertson SM, Lin R. Binary cell fate specification during C. elegans embryogenesis driven by reiterated reciprocal asymmetry of TCF POP-1 and its coactivator beta-catenin SYS-1. Development. 2007;14:2685–2695. doi: 10.1242/dev.008268. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;4:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Shi H, Amin NM, Sultan I, Liu J. Mesodermal expression of the C. elegans HMX homolog mls-2 requires the PBC homolog CEH-20. Mech Dev. 2008;5-6:451–461. doi: 10.1016/j.mod.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;4:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Ohto H, Takizawa T, Saito T. Identification and expression of six family genes in mouse retina. FEBS Lett. 1996;2-3:259–263. doi: 10.1016/0014-5793(96)00899-x. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes--structure and function as transcription factors and their roles in development. Bioessays. 2000;7:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Kidd AR, 3rd, Miskowski JA, Siegfried KR, Sawa H, Kimble J. A beta-catenin identified by functional rather than sequence criteria and its role in Wnt/MAPK signaling. Cell. 2005;5:761–772. doi: 10.1016/j.cell.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Kochhar A, Fischer SM, Kimberling WJ, Smith RJ. Branchio-oto-renal syndrome. Am J Med Genet A. 2007;14:1671–1678. doi: 10.1002/ajmg.a.31561. [DOI] [PubMed] [Google Scholar]
- Kostas SA, Fire A. The T-box factor MLS-1 acts as a molecular switch during specification of nonstriated muscle in C. elegans. Genes Dev. 2002;2:257–269. doi: 10.1101/gad.923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu M, Nagao T, Walldorf U, Flister S, Gehring WJ, Furukubo-Tokunaga K. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and Dachshund genes. Proc Natl Acad Sci U S A. 2000;5:2140–2144. doi: 10.1073/pnas.040564497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Frasch M. Nuclear integration of positive Dpp signals, antagonistic Wg inputs and mesodermal competence factors during Drosophila visceral mesoderm induction. Development. 2005;6:1429–1442. doi: 10.1242/dev.01687. [DOI] [PubMed] [Google Scholar]
- Lee HH, Frasch M. Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development. 2000;24:5497–5508. doi: 10.1242/dev.127.24.5497. [DOI] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;6964:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. 1998;2:229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;4:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Liu J, Fire A. Overlapping roles of two Hox genes and the exd ortholog ceh-20 in diversification of the C. elegans postembryonic mesoderm. Development. 2000;23:5179–5190. doi: 10.1242/dev.127.23.5179. [DOI] [PubMed] [Google Scholar]
- Liu J, Phillips BT, Amaya MF, Kimble J, Xu W. The C. elegans SYS-1 protein is a bona fide beta-catenin. Dev Cell. 2008;5:751–761. doi: 10.1016/j.devcel.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YH, Jakobsen JS, Valentin G, Amarantos I, Gilmour DT, Furlong EE. A Systematic Analysis of Tinman Function Reveals Eya and JAK-STAT Signaling as Essential Regulators of Muscle Development. Dev Cell. 2009;16:280–291. doi: 10.1016/j.devcel.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Lo MC, Gay F, Odom R, Shi Y, Lin R. Phosphorylation by the beta-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell. 2004;1:95–106. doi: 10.1016/s0092-8674(04)00203-x. [DOI] [PubMed] [Google Scholar]
- Martini SR, Roman G, Meuser S, Mardon G, Davis RL. The retinal determination gene, dachshund, is required for mushroom body cell differentiation. Development. 2000;12:2663–2672. doi: 10.1242/dev.127.12.2663. [DOI] [PubMed] [Google Scholar]
- Mazieres J, He B, You L, Xu Z, Jablons DM. Wnt signaling in lung cancer. Cancer Lett. 2005;1:1–10. doi: 10.1016/j.canlet.2004.08.040. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;12:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel-Aliaga I, Allan DW, Thor S. Independent roles of the dachshund and eyes absent genes in BMP signaling, axon pathfinding and neuronal specification. Development. 2004;23:5837–5848. doi: 10.1242/dev.01447. [DOI] [PubMed] [Google Scholar]
- Miskowski J, Li Y, Kimble J. The sys-1 gene and sexual dimorphism during gonadogenesis in Caenorhabditis elegans. Dev Biol. 2001;1:61–73. doi: 10.1006/dbio.2000.9998. [DOI] [PubMed] [Google Scholar]
- Mizumoto K, Sawa H. Two betas or not two betas: regulation of asymmetric division by beta-catenin. Trends Cell Biol. 2007;10:465–473. doi: 10.1016/j.tcb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kim S, Ishidate T, Bei Y, Pang K, Shirayama M, Trzepacz C, Brownell DR, Mello CC. Wnt signaling drives WRM-1/beta-catenin asymmetries in early C. elegans embryos. Genes Dev. 2005;15:1749–1754. doi: 10.1101/gad.1323705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noveen A, Daniel A, Hartenstein V. Early development of the Drosophila mushroom body: the roles of eyeless and dachshund. Development. 2000;16:3475–3488. doi: 10.1242/dev.127.16.3475. [DOI] [PubMed] [Google Scholar]
- Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;10:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BT, Kidd AR, 3rd, King R, Hardin J, Kimble J. Reciprocal asymmetry of SYS-1/beta-catenin and POP-1/TCF controls asymmetric divisions in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;9:3231–3236. doi: 10.1073/pnas.0611507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;7:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;10B:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;7207:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serikaku MA, O’Tousa JE. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;4:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;12:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;1:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Kidd AR, 3rd, Chesney MA, Kimble J. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the Caenorhabditis elegans gonad. Genetics. 2004;1:171–186. doi: 10.1534/genetics.166.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Kimble J. POP-1 controls axis formation during early gonadogenesis in C. elegans. Development. 2002;2:443–453. doi: 10.1242/dev.129.2.443. [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;4:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of Myogenic bHLH and MEF2 Transcription Factors by the bHLH Protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- Spitz F, Demignon J, Porteu A, Kahn A, Concordet JP, Daegelen D, Maire P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci U S A. 1998;24:14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;1:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;1:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Sundaram M, Greenwald I. Genetic and phenotypic studies of hypomorphic lin-12 mutants in Caenorhabditis elegans. Genetics. 1993;3:755–763. doi: 10.1093/genetics/135.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita H, Sawa H. Asymmetric cortical and nuclear localizations of WRM-1/beta-catenin during asymmetric cell division in C. elegans. Genes Dev. 2005;15:1743–1748. doi: 10.1101/gad.1322805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;6705:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;2:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;1:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.