Abstract
Background
Intrapartum single-dose nevirapine decreases mother-to-child transmission of human immunodeficiency virus type 1 (HIV-1) but promotes nevirapine resistance. Although resistant viruses fade to undetectable levels in plasma, they may persist as stably integrated proviruses within the latent reservoir in resting CD4+ T cells, potentially complicating future treatment.
Methods
Blood samples were collected from 60 women from South Africa and Uganda >6 months after they had received single-dose nevirapine. To selectively analyze the stable latent form of HIV-1, resting CD4+ T cells were isolated and activated in the presence of reverse-transcriptase inhibitors and integrase inhibitors, which allows for the specific isolation of viruses produced by cells with stably integrated proviral DNA. These viruses were then analyzed for nevirapine resistance.
Results
Although only a small number of latently infected cells were present in each blood sample (mean, 162 cells), nevirapine resistance mutations (K103N and G190A) were detected in the latent reservoir of 4 (8%) of 50 evaluable women.
Conclusions
A single dose of nevirapine can establish antiretroviral resistance within the latent reservoir. This results in a potentially lifelong risk of reemergence of nevirapine-resistant virus and highlights the need for strategies to prevent transmission that do not compromise successful future treatment.
The most widely used intervention to prevent mother-to-child transmission of HIV-1 in resource-limited settings is a single dose of the nonnucleoside reverse-transcriptase inhibitor (NNRTI) nevirapine, administered to pregnant women at the onset of labor, followed by a dose of nevirapine administered to the infant ≤72 h after birth [1, 2]. Single-dose nevirapine decreases transmission by 41%−47% [1, 2]. However, the most sensitive assays available detected nevirapine-resistant virus in the plasma of up to 87% of mothers 6 − 8 weeks after treatment [3–6]. Although resistant virus typically fades to undetectable levels in the plasma within several months [5–7], the persistence of resistant virus in the plasma for up to 5 years has been reported [8]. The most common mutations selected by single-dose nevirapine include K103N, Y181C, and G190A [7]. These mutations also confer resistance to other NNRTIs. A major concern regarding single-dose nevirapine is that the first-line antiretroviral regimens in developing countries rely on an NNRTI along with 2 nucleoside reverse-transcriptase inhibitors. The presence of virus resistant to a key component of these regimens could lead to treatment failure.
Although the development and persistence of nevirapine-resistant virus in the plasma has been well studied [3–14], evidence of archived resistance in the latent reservoir for HIV-1 in resting CD4+ T cells is lacking. The latent reservoir is established after infection of activated CD4+ T cells and integration of proviral HIV-1 DNA into the host genome. A small fraction of HIV-1–infected, activated CD4+ T cells return to a resting state as memory cells. In these cells, HIV-1 enters a state of latency in which it is protected from cellular immunity and antiretroviral drugs [15, 16]. In these inherently long-lived resting memory CD4+ T cells, the integrated HIV-1 genome is preserved for the life of the cell. Activation of a latently infected memory cell can trigger the release of archived virus [17]. Among patients receiving highly active antiretroviral therapy (HAART) for whom HIV-1 viremia was suppressed to undetectable levels, the frequency of latently infected cells is stable [18]. Thus, the latent reservoir is a major barrier to curing HIV-1 infection.
It is unclear whether nevirapine-resistant virus can be permanently archived in the latent reservoir after a single dose. Analysis of this issue is complicated by the ongoing viral replication that continues in mothers after nevirapine has been cleared. In viremic patients, most of the HIV-1 DNA in resting CD4+ T cells is a labile, unintegrated form representing recent infection [19, 20], and standard methods thus cannot provide an accurate reflection of the stable latent reservoir in resting CD4+ T cells. To evaluate the presence of nevirapine-resistant virus in the latent reservoir of women who had received a single dose of this drug, we used a novel method to detect stably integrated HIV-1 in highly purified resting CD4+ T cells.
METHODS
Patient selection
We studied 60 women from Soweto, South Africa, and Rakai, Uganda, who had received single-dose nevirapine during labor to prevent mother-to-child transmission of HIV-1. None of the women had received other antiretroviral agents. Single-dose nevirapine was self-administered during labor, >6 months before enrollment. To ensure that a sufficient number of resting CD4+ T cells were available for analysis, enrollment criteria included a CD4+ T cell count >200 cells/mm3. Exclusion criteria included severe anemia and current pregnancy. Written informed consent was obtained from all participants.
Isolation of resting CD4+ T cells
Peripheral blood mono-nuclear cells and plasma were isolated from 60 mL of whole blood on site. Plasma samples were frozen at −80°C. Peripheral blood mononuclear cells were resuspended in 90% fetal calf serum and 10% dimethyl sulfoxide and slowly frozen to −80°C. Plasma and cell aliquots were shipped in liquid nitrogen to Johns Hopkins University, where they were stored at −80°C. Cells were thawed, with an average recovery of 39% (range, 10%−77%) after freezing and thawing. Resting CD4+ T cells were isolated by magnetic bead depletion and cell sorting, as described elsewhere [21]. Resulting cell populations generally contained >99% resting CD4+ T cells.
Culture assay for postintegration latent virus
Purified resting CD4+ T cells were cultured in the presence of 4 antiretroviral drugs [19], including 2 nucleoside reverse-transcriptase inhibitors (8.6 μmol/L of lamivudine and 470 μmol/L of tenofovir [PMPA]), 1 NNRTI (0.5 μmol/L of efavirenz), and 1 integrase inhibitor (4 μmol/L of L-870812; Merck [22, 23]). Resting cells were activated by addition of the T cell mitogen phytohemagglutinin (0.5 μg/mL) and irradiated peripheral blood mononuclear cells from an HIV-1–negative donor to stimulate virus production. Activated cells were cultured in RPMI containing 10% fetal calf serum, interleukin-2, and a cytokine-rich supernatant from activated T cells. Culture supernatants were collected daily, and fresh media and drugs were added to the culture.
Supernatant viral RNA extraction and complementary DNA (cDNA) synthesis
Viral loads for supernatant and plasma samples were performed with Roche Amplicor (version 1.5; Roche). Approximately 5000 copies of HIV-1 RNA were extracted using the QIAamp viral RNA isolation kit (Qiagen). For samples that contained <5000 copies of HIV-1 RNA, supernatants from ≥2 days were combined to standardize the quantity of RNA per sample. cDNA that included the entire reverse transcriptase gene was generated, and nested polymerase chain reaction (PCR) was performed, using 10 μL of cDNA to amplify a 700-bp fragment (for primer sequences, see the appendix, which appears only in the electronic edition of the Journal).
Analysis of plasma virus
Approximately 10,000 copies of plasma HIV-1 RNA were extracted for each sample. In 3 cases, 10,000 copies of plasma HIV-1 RNA could not be obtained owing to low viral load. In these cases, all plasma obtained was concentrated for analysis. However, because of the low amount of virus present, we were unsuccessful in our efforts to isolate viral RNA from these samples. The lowest plasma HIV RNA levels successfully used in LigAmp and allele-specific PCR assays were 825 and 1143 copies/mL, respectively. A 700-bp fragment of reverse transcriptase was amplified (for primer sequences, see the appendix) in a limiting dilution series to determine cDNA concentration. Two independent PCR products generated using 100 copies of cDNA were used for LigAmp analysis.
LigAmp analysis
LigAmp analysis for the presence of resistance mutations was performed as described elsewhere [4]. Briefly, mutation-specific oligonucleotides were designed for each mutation and each HIV-1 subtype (for LigAmp primer sequences, see the appendix). A ligation reaction was performed with sequence-specific primers, sample DNA template, and Pfu polymerase (Stratagene). The ligated product was detected by real-time PCR. Each sample was analyzed in duplicate, and a standard curve included in each experiment was used to determine the amount of resistant virus in each sample. The cutoff value for LigAmp analysis was set at 1% resistant virus. Because of sequence polymorphisms, 5 samples could not be analyzed for the Y181C mutation, and 4 samples could not be analyzed for the G190A mutation. In these samples, clonal sequence analysis was used, and no nevirapine resistance mutations were identified.
Allele-specific PCR
To confirm results of LigAmp analysis, real-time allele-specific PCR was performed to identify the K103N resistance mutation in plasma samples from South Africa; 0.5 mL of plasma was used for this analysis in accordance with the method described elsewhere [6]. No evidence of this mutation among plasma virus was found (data not shown). One discrepancy between allele-specific PCR and LigAmp analysis was noted for 1 woman (LT2049) in whom a low level of K103N-resistant virus was detected by LigAmp analysis but not by allele-specific PCR. This mutation was confirmed by clonal sequence analysis.
Clonal analysis
HIV-1 reverse transcriptase sequences from selected samples were analyzed via nested PCR amplification of cDNA. A 1.5-kb region of the HIV-1 pol gene was amplified with subtype-specific primers (for primer sequences, see the appendix). Sequences were determined either by direct sequencing of limiting dilution PCR products or after cloning of 10− 20-μL of cDNA into a vector. Identical sequences isolated from the same PCR reaction were removed, and error analysis was performed to exclude sequence variants generated by PCR error, as described elsewhere [24]. Between 3 and 102 clones were isolated to identify resistance mutations that could not be measured by LigAmp analysis owing to sequence variation in the primer binding region. Fewer clones were isolated when less viral RNA was available.
Phylogenetic analysis
Sequences were aligned using Bio-Edit software after removal of primer sequences. Phylogenetic model parameters were determined using the Akaike information criterion, as implemented in ModelTest (version 3.7) [25] with PAUP* software (version 4b10; Sinauer Associates). Phylogenetic trees were constructed with PhyML software (version 2.4.4), using the general time-reversible model, ModelTest-calculated parameters for proportion of invariant sites, and a shape parameter for modeling rate variation among sites. Tree images were generated using MEGA software (version 4) [26].
RESULTS
Patient characteristics
Between June 2005 and September 2006, 60 HIV-1–positive mothers with CD4+ T cell counts of >200 cells/mm3 who had received single-dose nevirapine during labor >6 months before sample collection were recruited (30 were from Soweto, and 30 were from Rakai). The mean time since receiving nevirapine was 28 months (range, 11−45 months). At the time of sampling, the mean plasma HIV-1 RNA level was 79,387 copies/mL, and the mean CD4+ T cell count was 556 cells/mm3 (table 1).
Table 1.
Characteristics of women from Uganda or South Africa who received single-dose nevirapine (Nvp).
| Characteristic | Ugandan group (n = 30) | South African group (n = 30) | Overall (n = 60) |
|---|---|---|---|
| Time between single-dose Nvp and sampling, months | 28 (15−42) | 27 (11−45) | 28 (11−45) |
| HIV-1 RNA level at sample collection, copies/mL | 118,922 (<50 to 1,080,903) | 39,852 (319−219,064) | 79,387 (<50 to 1,080,903) |
| CD4+ cell count, cells/mm3a | 579 (309−1293) | 534 (205−1019) | 556 (205−1293) |
| HIV-1 subtype | |||
| C | 1 | 29 | 30 |
| D | 23 | 1 | 24 |
| A | 5 | 0 | 5 |
| Not determined | 1 | 0 | 1 |
| Nvp-resistant virus in plasmab | |||
| Not detected | 0 | 17 | 17 |
| Detected | 1 | 13 | 14 |
| Not determined | 29 | 0 | 29 |
| Mutationc | |||
| K103N | 0 | 10 | . . . |
| Y188C | 0 | 3 | . . . |
| Y181C | 1 | 1 | . . . |
| G190A | 0 | 2 | . . . |
| K101E | 0 | 1 | . . . |
| V106A | 0 | 1 | . . . |
NOTE. Data are mean value (range) or number of participants.
CD4+ cell measurements were performed at variable time points before sample collection.
Blood samples for identification of Nvp resistance in plasma were obtained 6−8 weeks after single-dose Nvp administration.
Plasma virus was not tested for resistance in 29 of 30 women from Uganda. Several samples from South African women had >1 Nvp resistance mutation.
Isolation of virus from the latent reservoir
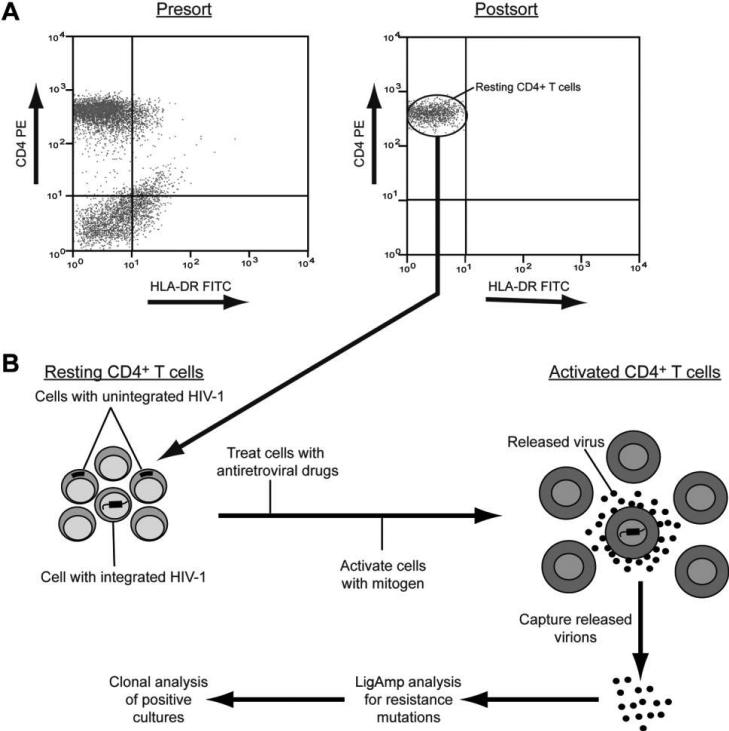
To determine whether nevirapine-resistant virus could enter and persist in the latent reservoir after treatment with single-dose nevirapine, a highly purified population of resting CD4+ T cells was isolated from each blood sample (figure 1A). This purification is important, because resting CD4+ T cells from blood do not produce virus without stimulation [19], and virus isolated from these cells after activation can therefore be assumed to have been latent. Stably integrated, latent viral genomes are present in only a very small fraction of these cells [15, 16]. Most of the infected resting CD4+ T cells from viremic patients harbor unintegrated forms of HIV-1 DNA [20]. These labile unintegrated viral sequences are present in resting CD4+ T cells owing to recent infection by circulating virus. They must be excluded from that analysis to visualize the stable latent reservoir of integrated HIV-1 proviruses. Therefore, resting CD4+ T cells were activated in the presence of 3 reverse-transcriptase inhibitors (lamivudine, tenofovir, and efavirenz) and an integrase inhibitor (L870812; Merck [22, 23]). These antiretroviral drugs prevent virus production by recently infected cells containing incomplete reverse transcripts or full-length linear unintegrated HIV-1 DNA. Only rare cells with stably integrated viral genomes are able to produce virus in this system [19] (figure 1B). Because of the low frequency of latently infected cells (<1 infected cell per 106 cells), only a small number of cells can be analyzed. Nevertheless, this method provides a reliable way to assess the latent reservoir in patients with ongoing viral replication. With use of this method, virus was successfully recovered from the latent reservoir for 50 of 60 women who received single-dose nevira-pine.
Figure 1.
Analysis of the latent reservoir in viremic patients. A, Representative flow cytometric analysis illustrating the isolation of a highly purified population of resting CD4+ T cells from peripheral blood mononuclear cells by magnetic bead depletion of unwanted populations (presort) and sorting for CD4+ HLA-DR− T cells (postsort). HLA-DR is a marker for activated CD4+ T cells. FITC, fluorescein isothiocyanate; PE, phycoerythrin. B, Culture assay for analysis of virus released from cells with HIV-1 that has stably integrated into the host cell genome. Highly purified resting CD4+ T cells are cultured in the presence of 3 reverse-transcriptase inhibitors and an integrase inhibitor. These drugs block the completion of reverse transcription and integration of HIV-1 that has not integrated into the host cell genome. Cells are then activated with a mitogen. Only cells with virus stably integrated into the host cell genome are able to release virus particles. Culture supernatants are collected daily and analyzed for the presence of virus containing nevirapine resistance mutations.
Nevirapine resistance in virus from the latent reservoir and plasma
A sensitive, sequence-specific assay, LigAmp [4], was used to screen virus released after activation of resting CD4+ T cells for the presence of 3 major nevirapine resistance mutations: K103N, Y181C, and G190A (table 2). Contemporaneous plasma virus was also analyzed for the presence of these mutations.
Table 2.
Identification of nevirapine-resistant HIV in the latent reservoir and/or plasma of persons from Uganda or South Africa >6 months after receipt of single-dose nevirapine.
| Participants, no. |
|||
|---|---|---|---|
| Reservoir and plasma findings | Ugandan group (n = 30) | South African group (n = 30) | Total (n = 60) |
| Latent reservoir | |||
| Virus isolated from cells of latent reservoir | 26 | 24 | 50 |
| Resistant virus present in cells of latent reservoir | 2a | 2 | 4 |
| Plasma | |||
| Plasma virus analyzed | 28 | 29 | 57 |
| Resistant virus present in plasma | 1a | 1 | 2 |
For patient LT1020, viral variants containing 2 different nevirapine resistance mutations were identified. One mutation was detected in both plasma virus and virus released from cells of the latent reservoir, whereas the other mutation was found only among viruses from cells of the latent reservoir.
Of 50 women for whom virus was isolated from the latent reservoir, nevirapine resistance mutations were identified in 4 (8%). At the same time point, only 2 (3.4%) of 58 women had nevirapine resistance mutations detectable in circulating plasma virus (3 women had plasma virus levels too low to be analyzed). One woman had 2 nevirapine-resistant variants detected, 1 that was present in both virus from the latent reservoir and virus from plasma and 1 isolated only from the latent reservoir. The K103N mutation was identified most frequently, followed by the G190A mutation. The Y181C resistance mutation was not identified among any samples (table 3). Of the 4 women with nevirapine-resistant virus identified in the latent reservoir, 2 had subtype C and 2 had subtype D virus. The same nevira-pine resistance mutation (K103N) identified in a previous analysis of plasma virus after single-dose nevirapine was identified among virus from the latent reservoir for 2 women in our study. The 3 remaining women in whom nevirapine resistance was identified in the latent reservoir or the plasma either had no resistance testing performed or showed no previous nevirapine resistance (table 3). These results show that a single dose of nevirapine can lead to stable archiving of resistant virus in at least some women.
Table 3.
Nevirapine (Nvp) resistance mutations detected in HIV from the latent reservoir and/or plasma of persons from Uganda or South Africa >6 months after receipt of single-dose nevirapine.
| HIV with Nvp resistance mutations |
||||||||
|---|---|---|---|---|---|---|---|---|
| Patient | Study site | HIV subtype | Interval between single-dose Nvp and sampling, months | CD4+ cell count, cells/mm3 | HIV-1 RNA load, copies/mL | Detected in plasma | Detected in latent reservoir | Identified in plasma before current study |
| LT1003 | Uganda | D | 37.5 | 336 | 129,546 | None | G190A | Not determined |
| LT1020 | Uganda | D | 23.2 | 309 | 244,545 | K103N | K103N, G190A | Not determined |
| LT2009 | South Africa | C | 15.3 | 286 | 219,064 | None | K103N | K103N |
| LT2028 | South Africa | D | 32.0 | 205 | 14,280 | None | K103N | K103N |
| LT2049 | South Africa | C | 25.6 | 291 | 32,339 | K103N | None | None |
In our study, the average number of resting CD4+ T cells isolated per patient was 1.62 × 106 cells. Of these, only ∼0.01% (∼162 cells) contained integrated HIV-1 DNA. The power to detect nevirapine-resistant virus in the latent reservoir is determined by the number of cells analyzed and the proportion of nevirapine-resistant virus present in the reservoir. With this level of sampling, we have >99.9% power to detect nevirapine-resistant virus in the latent reservoir if 10% of the latent reservoir contains nevirapine-resistant virus and 80.4% power if 1% of the reservoir contains resistant virus (figure 2, which appears only in the electronic edition of the Journal). However, at 0.1% and 0.01% rates of nevirapine-resistant virus, we have powers of only 15% and 1.6%, respectively (figure 2). Thus, we cannot exclude the possibility that resistant viruses were present in more women at these lower levels.
Figure 2.
Frequency of nevirapine-resistant virus detection among latently infected cells.
Phylogenetic analysis of virus from the latent reservoir
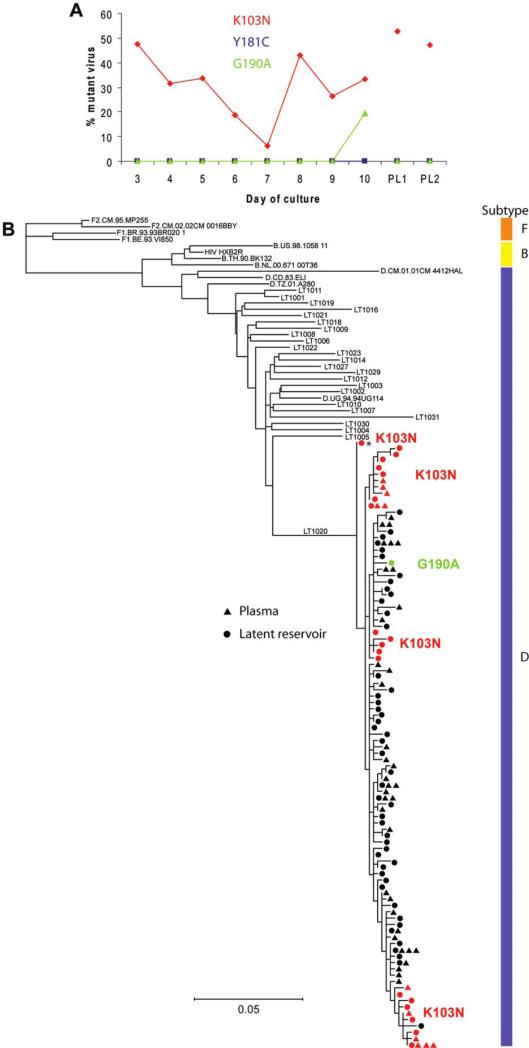
To understand the degree to which resistant virus had populated the latent reservoir and the relationship between the resistance mutations detected, viral sequencing and phylogenetic analysis were carried out in a subset of patients. For patient LT1020, LigAmp analysis indicated that virus isolated from the latent reservoir contained both the K103N and G190A resistance mutations (figure 3A). The variability in the amount of resistant virus isolated on different days of the culture assay may reflect the activation of different resting CD4+ T cells among the total population during the course of the assay. Sequences obtained for patient LT1020 were phylogenetically distinct from those obtained for all other patients with the same subtype (D) included in this study (figure 3B). Viruses containing the K103N resistance mutation were readily detected among viruses released from the latent reservoir and in plasma. Each sequence presented represents an independent viral sequence present in the patient. Therefore, the fraction of resistant viruses detected in our analysis indicates the degree of resistance present in plasma and the latent reservoir of this patient. Viruses with the K103N mutation composed 28% of the latent reservoir sequences analyzed and 23% of the plasma sequences analyzed. In addition, a single viral clone containing the G190A resistance mutation was identified among viruses released from the latent reservoir. This mutation was not detected in plasma virus. The K103N and G190A mutations were not present in the same viral genome, indicating that they arose independently in response to nevirapine.
Figure 3.
Phylogenetic analysis of nevirapine resistance. A, Identification of nevirapine resistance mutations in viruses released after in vitro activation of resting CD4+ T cells and in viruses from the plasma of patient LT1020. The percentage of mutant virus present for each of 8 days of cell culture is indicated for the K103N (red diamonds), Y181C (blue squares), and G190A (green triangles) mutations. The level of mutant virus present in plasma at the same time point was measured in 2 individual polymerase chain reaction reactions, labeled PL1 and PL2. B, Phylogenetic tree from patient LT1020 with sequences from virus released from the latent reservoir (circles) and plasma virus (triangles). Viral sequences containing the K103N resistance mutation are shown in red. The viral sequence containing the G190A resistance mutation is shown in green. The asterisk indicates a possible ancestral K103N-containing sequence. Representative plasma viral sequences from other study patients with the same subtype (D) are included (all sequence names beginning with LT). Subtype B (including HXB2R) and D reference sequences obtained from the Los Alamos National Laboratory sequence database are also included. The tree is rooted on subtype F reference sequences obtained from the Los Alamos sequence database.
Four clusters of viruses containing the K103N resistance mutation were revealed by phylogenetic analysis. Of these 4 clusters, 2 were detected in plasma and the latent reservoir, but the remaining 2 were found only in the reservoir. For resistant viruses that persisted in both compartments, it appears that the fitness cost of the K103N mutation was sufficiently low such that mutant viruses could continue to replicate over a prolonged period. One K103N-containing viral sequence appears to be ancestral to all other viral sequences isolated from patient LT1020 (asterisk in figure 3B), indicating that this patient could have been infected by virus containing the K103N resistance mutation. However, this is unlikely because of the low frequency at which antiretroviral therapy was used in Rakai at the time of her infection. The location of this viral sequence on the phylogenetic tree can also be explained by the presence of different sequence polymorphisms that result in its ancestral placement on the tree. One wild-type viral sequence clusters with a group of K103N-containing viral sequences, suggesting that it is a revertant sequence (located at the bottom of the phylogenetic tree). This wild-type viral sequence has 3 unique polymorphisms that distinguish it from the rest of the sequences in this clade, 1 of which resulted in the disappearance of the K103N mutation. This is the most parsimonious tree construction, although other more complex scenarios that do not indicate this reversion are plausible.
DISCUSSION
We have shown that nevirapine-resistant HIV-1 arising after a single dose of nevirapine can be archived in the latent reservoir of resting CD4+ T cells. Resistant virus was detected in the latent reservoir in 8% of study participants an average of 27 months after receipt of a single dose of nevirapine. Resistant viruses were detected in the plasma of 3.4% of study participants at the same time point. The persistent replication of these resistant viruses long after receipt of single-dose nevirapine likely indicates that the fitness of the resistant virus is similar to that of wild-type virus from the same patient. Archived nevirapine-resistant virus is of concern because it may reemerge and manifest itself clinically if these women later begin HAART regimens that include NNRTIs.
The methods used in this study allow for specific analysis of the small number of resting CD4+ T cells that compose the stable latent reservoir for HIV-1. Nevirapine-resistant virus is generally detected transiently in the plasma of women after single-dose nevirapine, providing a limited chance for this virus to enter the latent reservoir. Repopulation of the latent reservoir is more likely when the resistant viruses have fitness similar to that of wild-type viruses and replicate for prolonged periods (as demonstrated here for patient LT1020). It is likely that latently infected cells with nevirapine-resistant virus are present in recipients of single-dose nevirapine over a wide range of frequencies. The level of nevirapine-resistant virus present in the latent reservoir may be low for most women. Our results identified nevirapine-resistant virus in a minority of study participants. However, because the frequency of latently infected cells is low, only a small number of latently infected cells are present in each blood sample, and our method does not have the power to detect resistant virus present at levels of ≤0.1%. These factors suggest that archived nevirapine resistance may be more prevalent than identified in our study. The small fraction of the latent reservoir assayed in this study may also account for the presence of nevirapine-resistant virus among plasma virus, but not among virus from the latent reservoir in patient LT2049 (table 3). It is also possible that this nevirapine-resistant virus identified in plasma arose from a reservoir for HIV-1 other than resting CD4+ T cells.
Previous studies have identified nevirapine resistance mutations among peripheral blood mononuclear cells after single-dose nevirapine [6, 27–29]. Most HIV-1 found in peripheral blood mononuclear cells of viremic patients represents unintegrated virus in recently infected cells. Therefore, those studies fail to provide information regarding the composition of the stable latent reservoir, which can be analyzed only by using special methods, such as the one described here.
Recent studies have shown that the time between receipt of single-dose nevirapine and initiation of antiretroviral treatment can affect virologic suppression [30, 31]. Women who began nevirapine-based antiretroviral regimens ≤6 months after receiving single-dose nevirapine (plus zidovudine from week 34 of gestation through delivery) had higher rates of virologic failure than women without previous exposure to nevirapine [30]. Clearly, some treatment failures observed in women who began combination therapy soon after receipt of single-dose nevira-pine result from the active replication of resistant virus when combination therapy is started. Our results suggest that resistant virus can persist in the latent reservoir even when it is no longer detectable in the plasma. Although this archived resistance does not lead to immediate treatment failure, its eventual reemergence may cause an increased rate of treatment failure in the long term. Available outcome studies with follow-up periods of 6−24 months may not adequately describe the long-term risk of nevirapine-based antiretroviral regimens in women treated with single-dose nevirapine. Further studies with longer follow-up periods are needed to evaluate the effectiveness of HAART in women treated with single-dose nevirapine.
Because of the high incidence of resistance after receipt of single-dose nevirapine [3–14], strategies have been investigated that incorporate either a short course or a single dose of a nucleoside reverse transcriptase inhibitor to supplement single-dose nevirapine. Preliminary studies have shown that this strategy reduces but does not eliminate the development of NNRTI resistance [32–35]. However, these regimens are not available or feasible in all settings, and, for a large number of women, single-dose nevirapine remains the only option for preventing mother-to-child transmission of HIV-1 infection.
In summary, this study proves the principle that archiving of resistant virus can occur after receipt of a single dose of nevira-pine. Nevirapine-resistant virus was identified in the latent reservoir of 4 women 15−37 months after exposure to nevirapine. This archiving could potentially create lifelong resistance to a key agent in the first-line HAART regimen recommended by the World Health Organization and could have important consequences for the choice of HAART regimens in women exposed to NNRTIs to prevent mother-to-child transmission of HIV-1.
Acknowledgments
Dativa Mukasa and Jane Kasozi (Rakai Health Sciences Program, Rakai, Uganda) and Hao Zhang (Johns Hopkins School of Public Health, Baltimore, Maryland) contributed to this work. Women in Soweto, South Africa were recruited for this study by Lynette Modise.
Financial support: HIV Prevention Trials Network, sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Child Health and Human Development, National Institute on Drug Abuse, National Institute of Mental Health, and Office of AIDS Research, National Institutes of Health (NIH), Department of Health and Human Services (grants U01-AI-46745, U01-AI-48054, and U01-AI-068613 to S.H.E.); International Maternal Pediatric Adolescent AIDS Clinical Trials Network; NIAID (grant U01-AI-068632 to S.H.E.); NIH (grant K01-AI071754 to B.L.); Division of Intramural Research, NIAID (T.C.Q.); Howard Hughes Medical Institute and NIH (grant AI43222 to R.F.S.).
Footnotes
Potential conflicts of interest: N.M. manages a grant from US President's Emergency Plan for AIDS Relief that provides antiretroviral treatment. S.H.E. is a coinventor of the LigAmp assay, for which Johns Hopkins University has filed a patent application with the US Patent and Trademark Office; the inventors may receive royalty payments if the patent is awarded and licensed. All other authors: none reported.
Presented in part: 15th Conference on Retroviruses and Opportunistic Infections, Boston, Massachusetts, 3−6 February 2008 (poster 643).
References
- 1.Guay LA, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 2.Jackson JB, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: 18-month follow-up of the HIVNET 012 randomised trial. Lancet. 2003;362:859–68. doi: 10.1016/S0140-6736(03)14341-3. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JA, Li JF, Morris L, et al. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192:16–23. doi: 10.1086/430741. [DOI] [PubMed] [Google Scholar]
- 4.Flys TS, Chen S, Jones DC, et al. Quantitative analysis of HIV-1 variants with the K103N resistance mutation after single-dose nevirapine in women with HIV-1 subtypes A, C, and D. J Acquir Immune Defic Syndr. 2006;42:610–3. doi: 10.1097/01.qai.0000221686.67810.20. [DOI] [PubMed] [Google Scholar]
- 5.Palmer S, Boltz V, Martinson N, et al. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad SciUSA. 2006;103:7094–9. doi: 10.1073/pnas.0602033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loubser S, Balfe P, Sherman G, Hammer S, Kuhn L, Morris L. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. AIDS. 2006;20:995–1002. doi: 10.1097/01.aids.0000222071.60620.1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eshleman SH, Mracna M, Guay LA, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012). AIDS. 2001;15:1951–7. doi: 10.1097/00002030-200110190-00006. [DOI] [PubMed] [Google Scholar]
- 8.Flys TS, Donnell D, Mwatha A, et al. Persistence of K103N-containing HIV-1 variants after single-dose nevirapine for prevention of HIV-1 mother-to-child transmission. J Infect Dis. 2007;195:711–5. doi: 10.1086/511433. [DOI] [PubMed] [Google Scholar]
- 9.Eshleman SH, Becker-Pergola G, Deseyve M, et al. Impact of human immunodeficiency virus type 1 (HIV-1) subtype on women receiving single-dose nevirapine prophylaxis to prevent HIV-1 vertical transmission (HIV Network for Prevention Trials 012 Study). J Infect Dis. 2001;184:914–7. doi: 10.1086/323153. [DOI] [PubMed] [Google Scholar]
- 10.Flys T, Nissley DV, Claasen CW, et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevira-pine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J Infect Dis. 2005;192:24–9. doi: 10.1086/430742. [DOI] [PubMed] [Google Scholar]
- 11.Eshleman SH, Hoover DR, Chen S, et al. Nevirapine (NVP) resistance in women with HIV-1 subtype C, compared with subtypes A and D, after the administration of single-dose NVP. J Infect Dis. 2005;192:30–6. doi: 10.1086/430764. [DOI] [PubMed] [Google Scholar]
- 12.Eshleman SH, Guay LA, Mwatha A, et al. Characterization of nevirapine resistance mutations in women with subtype A vs. D HIV-1 6−8 weeks after single-dose nevirapine (HIVNET 012). J Acquir Immune Defic Syndr. 2004;35:126–30. doi: 10.1097/00126334-200402010-00004. [DOI] [PubMed] [Google Scholar]
- 13.Eshleman SH, Guay LA, Wang J, et al. Distinct patterns of emergence and fading of K103N and Y181C in women with subtype A vs. D after single-dose nevirapine: HIVNET 012. J Acquir Immune Defic Syndr. 2005;40:24–9. doi: 10.1097/01.qai.0000174656.71276.d6. [DOI] [PubMed] [Google Scholar]
- 14.Jackson JB, Becker-Pergola G, Guay LA, et al. Identification of the K103N resistance mutation in Ugandan women receiving nevirapine to prevent HIV-1 vertical transmission. AIDS. 2000;14:F111–5. doi: 10.1097/00002030-200007280-00001. [DOI] [PubMed] [Google Scholar]
- 15.Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–8. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 16.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1:1284–90. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 17.Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–7. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 18.Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–8. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 19.Monie D, Simmons RP, Nettles RE, et al. A novel assay allows genotyping of the latent reservoir for human immunodeficiency virus type 1 in the resting CD4+ T cells of viremic patients. J Virol. 2005;79:5185–202. doi: 10.1128/JVI.79.8.5185-5202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–60. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 22.Pandey KK, Bera S, Zahm J, et al. Inhibition of human immunodeficiency virus type 1 concerted integration by strand transfer inhibitors which recognize a transient structural intermediate. J Virol. 2007;81:12189–99. doi: 10.1128/JVI.02863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazuda DJ, Young SD, Guare JP, et al. Integrase inhibitors and cellular immunity suppress retroviral replication in rhesus macaques. Science. 2004;305:528–32. doi: 10.1126/science.1098632. [DOI] [PubMed] [Google Scholar]
- 24.Bailey JR, Sedaghat AR, Kieffer T, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–57. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, Dudley J, Nei M, et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Zhou Y, Alcock C, et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol. 2004;78:1718–29. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMahon MA, Jilek BL, Brennan TP, et al. The HBV drug entecavir: effects on HIV-1 replication and resistance. N Engl J Med. 2007;356:2614–21. doi: 10.1056/NEJMoa067710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaix ML, Ekouevi DK, Peytavin G, et al. Impact of nevirapine (NVP) plasma concentration on selection of resistant virus in mothers who received single-dose NVP to prevent perinatal human immunodeficiency virus type 1 transmission and persistence of resistant virus in their infected children. Antimicrob Agents Chemother. 2007;51:896–901. doi: 10.1128/AAC.00910-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lockman S, Shapiro RL, Smeaton LM, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med. 2007;356:135–47. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- 31.Chi BH, Sinkala M, Stringer EM, et al. Early clinical and immune response to NNRTI-based antiretroviral therapy among women with prior exposure to single-dose nevirapine. AIDS. 2007;21:957–64. doi: 10.1097/QAD.0b013e32810996b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIntyre JA, Martinson N, Gray GE, et al. Addition of short course combivir (CBV) to single-dose viramune (sdNVP) for the prevention of mother to child transmission (pMTCT) of HIV-1 can significantly decrease the subsequent development of maternal and paediatric NNRTI-resistant virus [abstract TuFo0504].. Program and abstracts of the International AIDS Society Meeting; Brazil. 24−27 July 2005; Rio de Janeiro; [Google Scholar]
- 33.Chaix ML, Ekouevi DK, Rouet F, et al. Low risk of nevirapine resistance mutations in the prevention of mother-to-child transmission of HIV-1: Agence Nationale de Recherches sur le SIDA Ditrame Plus, Abidjan, Cote D'Ivoire. J Infect Dis. 2006;193:482–7. doi: 10.1086/499966. [DOI] [PubMed] [Google Scholar]
- 34.Arrive E, Newell ML, Ekouevi DK, et al. Prevalence of resistance to nevirapine in mothers and children after single-dose exposure to prevent vertical transmission of HIV-1: a meta-analysis. Int J Epidemiol. 2007;36:1009–21. doi: 10.1093/ije/dym104. [DOI] [PubMed] [Google Scholar]
- 35.Chi BH, Sinkala M, Mbewe F, et al. Single-dose tenofovir and emtricitabine for reduction of viral resistance to nonnucleoside reverse transcriptase inhibitor drugs in women given intrapartum nevirapine for peri-natal HIV prevention: an open-label randomised trial. Lancet. 2007;370:1698–705. doi: 10.1016/S0140-6736(07)61605-5. [DOI] [PubMed] [Google Scholar]