Summary
The budding yeast, Saccharomyces cerevisiae, has emerged as an archetype of eukaryotic cell biology. Here we show that S. cerevisiae is also a model for the evolution of cooperative behavior by revisiting flocculation, a self-adherence phenotype lacking in most laboratory strains. Expression of the gene FLO1 in the laboratory strain S288C restores flocculation, an altered physiological state, reminiscent of bacterial biofilms. Flocculation protects the FLO1-expressing cells from multiple stresses, including antimicrobials and ethanol. Furthermore, FLO1+ cells avoid exploitation by non-expressing flo1 cells by self/non-self recognition: FLO1+ cells preferentially stick to one another, regardless of genetic relatedness across the rest of the genome. Flocculation, therefore, is driven by one of a few known “green beard genes”, which direct cooperation towards other carriers of the same gene. Moreover, FLO1 is highly variable among strains both in expression and in sequence, suggesting that flocculation in S. cerevisiae is a dynamic, rapidly-evolving social trait.
Keywords: Flocculation, FLO1, drug resistance, evolution, ethanol, selfish gene, Darwin, Dawkins, Hamilton, green beard gene
Introduction
Since Darwin, evolutionary biologists have been troubled by cooperative behavior. Darwin systematically identified the phenomena that were the greatest challenge to his ideas. Cooperation was, and remains (Pennisi, 2005), one of these: “If it could be proved that any part of the structure of any one species had been formed for the exclusive good of another species, it would annihilate my theory, for such could not have been produced through natural selection.” (Darwin 1859). Cooperation is a problem for evolution by natural selection because individuals are predicted to act in a way that maximizes their personal reproduction. Costly behaviors that invest in a common good, therefore, are expected to be disrupted by so-called “cheaters” that save on the cost of cooperation but reap in the benefits of the investment of others. Such cheaters will be fitter than cooperators and take over the population, ultimately resulting in the loss of the cooperative behavior.
Why then, do organisms frequently evolve behaviors that help others? For example, honeybee workers labor their whole life without reproducing, birds make alarm calls and humans often help one another. This fundamental question has received considerable attention over the last 50 years with the development of the field of sociobiology. Following the work of Hamilton (1964), it is now widely accepted that cooperative behaviors evolve because they directly help the actor alongside any recipients, or they help individuals who share more alleles with the actor than predicted by chance (genetic relatedness), or both (Dawkins, 1976; Hamilton, 1964; Queller, 1984; West et al., 2006). In extreme cases, therefore, cooperators can successfully transmit their genes by helping another individual that carries these alleles, as occurs when near-sterile honeybee workers help their mother to reproduce. Typically, it is assumed that the correlation in genotype among individuals is generated by family, as is the case for sister workers in the social insects. However, Hamilton also engaged in a thought experiment, in which he proposed that cooperation is also possible if a single gene that drives the tendency to cooperate can also preferentially directs cooperation to other carriers of the gene. Such a (hypothetical) gene was later named a “green beard gene” by Dawkins, the green beard being the recognizable “tag” that enables organisms to direct their interactions to other carriers of the gene (Dawkins, 1976; Hamilton, 1964). Hamilton predicted that green beard genes would be extremely rare owing to the requirement that altruism and recognition be performed by a single gene, a prediction that seems correct in social animals (Keller and Ross, 1998; Krieger and Ross, 2002).
Social animals have been well studied, but sociobiology has tended to overlook the fact that many microbes form groups. This is now changing with the realization that microbes offer particular advantages to sociobiology, including the ability to study the genetics of social traits in a system where culture and learning have minimal impact (Foster et al., 2007). Considerable attention has being paid to developmentally-sophisticated species, like the slime mold Dictyostelium discoideum, which appears to have a green beard gene that has swept through the population to fixation (Queller et al., 2003). While fascinating, however, such species are probably exceptional in their social sophistication, and many other microbes live in groups that seem to require cooperation. Notable among these are large surface-attached groups, known as biofilms, (d'Enfert, 2006; Hall-Stoodley et al., 2004; Palkova, 2004). Biofilms have received enormous attention from microbiologist owing to their resistance to stress and antimicrobials (d'Enfert, 2006; Hall-Stoodley et al., 2004) but little from sociobiology. Other aggregation phenotypes, often overlooked by microbiologists and sociobiologists alike, occur in one of the most familiar and tractable of microbes, the budding yeast, Saccharomyces cerevisiae. Several studies have begun to uncover S. cerevisiae’s remarkable capacity to form pseudohyphae and multicellular “mats” on low-density agar (Gimeno et al., 1993; Palkova and Vachova, 2006; Reynolds and Fink, 2001). Another multicellular form has been known for hundreds of years in the brewing industry. Brewers make effective use of the tendency of their yeast strains to adhere to each other to form large clumps or “flocs” consisting of thousands of cells that rapidly sediment from the medium. This process, known as “flocculation”, is routinely used in today’s beer production as a simple and cost-effective method to remove flocs of yeast cells from beer after fermentation.
The molecular mechanism underlying adhesion and flocculation is relatively simple. Flocculating cells express specific cell-surface proteins encoded by the FLO genes. Each FLO gene encodes a slightly different cell-surface protein capable of forming lectin-like bonds with mannan oligosaccharide chains that make up the outermost layer of the S. cerevisiae cell wall. In this way, the Flo adhesins make cells adhere to each other, resulting in the formation of flocs (for a review, see (Verstrepen and Klis, 2006)). However, while the basic molecular mechanisms are known, many fundamental questions about the physiological role and dynamics of flocculation remain unanswered. Flocculation has received relatively little scientific attention because commonly used laboratory yeasts do not flocculate. Records of the pioneering yeast geneticists show how feral strains were specifically crossed and selected to obtain S. cerevisiae strains with reduced cell-cell and cell-surface adhesion characteristics, making them more suited for laboratory use (Mortimer and Johnston, 1986). Thus, interesting open questions are: why do yeast cells flocculate? Is flocculation a true cooperative trait with associated benefits and costs, and if so, how does flocculation fit the theory of social evolution?
Here, we use the nonflocculent laboratory strain S288C and its flocculating feral ancestors to investigate the physiology, biological function, and evolution of flocculation. Our results indicate that flocculation is a cooperative protection mechanism that shields cells from stressful environments, under the control of one key gene FLO1. Moreover, we show that FLO1 provides a built-in mechanism to direct cooperation towards other FLO1 carries, and protect against potential cheater strains. The ability of a single gene to both generate cooperation and solve the problem of cheaters makes FLO1 a green beard gene. Moreover, FLO1 displays considerable expression and sequence variability in natural populations, suggesting that FLO1 continues to rapidly evolve in nature.
Results
Strong flocculation in ancestors of the laboratory strain S288C is linked to expression of a single gene, FLO1
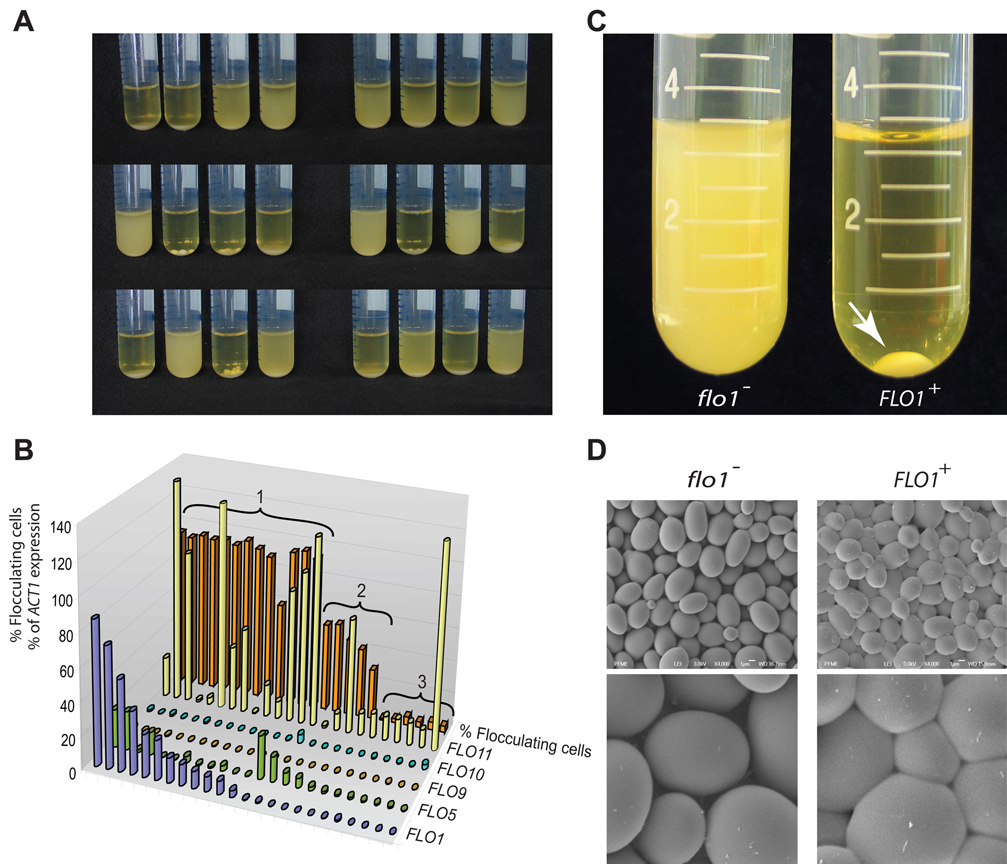
Since none of the flocculation (FLO) genes are transcriptionally active in the commonly used laboratory strain S288C, it is unknown which of the different FLO genes (if any) is responsible for the flocculation of feral strains. To investigate which FLO genes play a role in natural S. cerevisiae flocculation, we turned our attention to the ancestor of S288C, the feral strain EM93 (Mortimer and Johnston, 1986). In contrast to its domesticated sibling, EM93 and its haploid derivatives show extensive flocculation (Fig. 1A). We measured the expression of the 5 known FLO genes and correlated these levels to the rate of flocculation in 24 haploid EM93 strains (Fig. 1A and B). The results show that the EM93 strains show an extraordinary range of flocculation, from extremely strong to almost non-existent. Moreover, strong flocculation was tightly correlated with expression of one specific FLO gene, FLO1.
Figure 1. FLO1 confers strong flocculation in S. cerevisiae.
A. Haploid derivatives of the feral strain EM93 show a wide variation in flocculation behavior when grown in rich YPD medium. Some strains flocculate strongly, so that all cells clump together and sink to the bottom of the tube, while others show virtually no flocculation, leaving all cells in suspension. B. Flocculation and expression of the five known flocculation (FLO) genes was quantified in 24 haploid EM93 strains. Strains were divided into three groups. Group 1 shows expression of FLO1 (at least 1% of ACT1 levels), Group 2 shows expression of FLO5, but not FLO1, and Group 3 does not show expression of either FLO1 or FLO5. Strains from Group 1 generally showing strong flocculation (>85%), Group 2 intermediate flocculation (20–50%) and Group 3 no flocculation (< 5%). C. The FLO1 gene of the nonflocculent laboratory strain S288C was brought under the transcriptional control of the inducible GAL1 promoter (KV210). When this strain is grown in YPGal medium, FLO1 is expressed, resulting in strong flocculation (arrow). A control strain (KV22) containing the same resistance marker gene, but not the promoter, does not show flocculation. D. Scanning electron microscopy of centrifuged pellets of nonflocculent (KV22) and flocculent (KV210) S288C cells shows that the flocculent cells stick together to form a densely packed three dimensional structure with little intercellular space. By contrast, the (centrifuged) nonflocculent cells behave as stacked independent spheres with clear gaps between the cells.
To confirm that FLO1 expression leads to strong flocculation, we brought the genomic FLO1 copy of the non-flocculent S288C laboratory strain under transcriptional control of the inducible GAL1 promoter. Induction of FLO1 in galactose-containing medium leads to strong flocculation, closely resembling the phenotype observed in those EM93 strains that express FLO1. FLO1-expressing S288C cells aggregate to form spherical flocs of 5–8 mm in diameter (Fig. 1C). Scanning electron microscopy of the flocs shows that the yeast cells are packed extremely densely within the floc. The cell walls seem to bind strongly to each other, resulting in a remarkable three-dimensional tiling pattern of deformed yeast cells with virtually no intercellular space (Fig. 1D).
Flocculation confers stress resistance
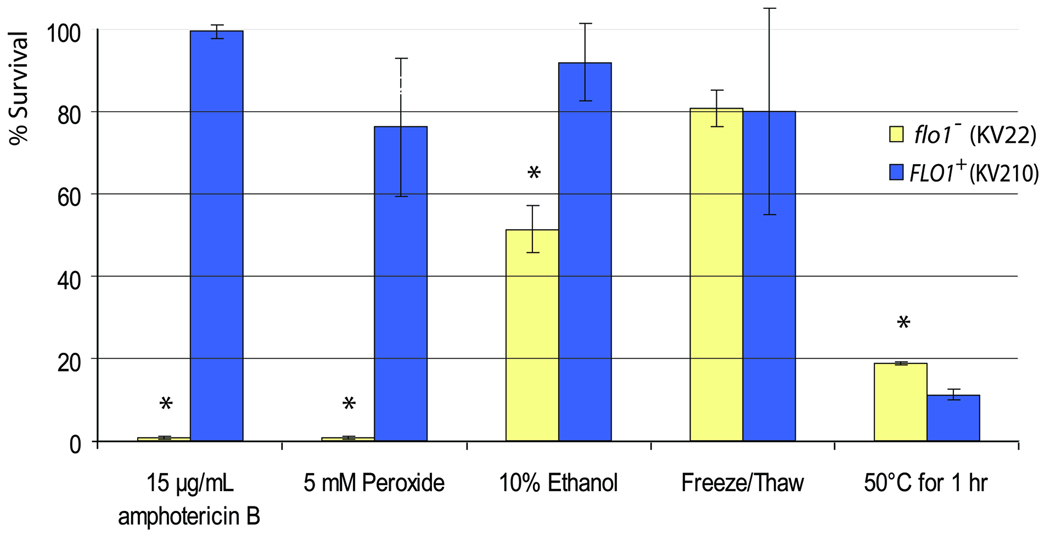
Using the wild-type (flo1−) and FLO1-expressing laboratory strains, we asked whether FLO1-induced flocculation confers resistance to environmental and chemical stress, as observed in microbial biofilms. Flocculent and non-flocculent cultures were subjected to freeze/thaw cycles, heat shock, oxidative stress, ethanol, and addition of amphotericin B, and cell survival was measured using standard Colony Forming Units (CFU) counts. Amphotericin B is a natural antifungal agent produced by Streptomyces nodosus, a soil bacterium that uses amphotericin production to inhibit the growth of competing fungi (Trejo and Bennett, 1963). The compound is one of the most commonly used drugs to fight pathogenic fungi such as Candida albicans. The number of cells surviving the stress treatment was 2-fold greater for flocculent cells subjected to ethanol stress, and more than 100-fold for treatment with peroxide and amphotericin B. No significant differences were found for the freeze/thaw stress. In the case of heat treatments, the flocculent cultures were slightly less resistant than planktonic flo1− cells (Fig. 2). This sensitivity to heat might be a consequence of changes in membrane lipids and sterols in flocculating cells (see further).
Figure 2. Flocculation confers resistance to certain stresses.
S. cerevisiae cells with (KV210) and without (KV22) FLO1 expression were subjected to various stress treatments, after which the percentage of surviving cells was determined. Asterisks indicate statistically significant differences between flocculent and nonflocculent cultures (α = 0.05); error bars correspond to standard deviation.
We noted that the stresses to which the flocculating cells are most resistant are all chemical stresses. For these stresses to have an effect, it is essential that molecules can physically reach the cells. Given that flocs are such a densely packed structure with virtually no intercellular space (Fig. 1D), we hypothesized that cells on the inside of flocs might be physically shielded from the chemicals in the growth medium. To test this hypothesis, we first performed survival assays with increasing concentrations of amphotericin B. The results show that the majority of flocculating cells survive treatments with as much as 100 µg ml−1 of amphotericin B (i.e. 100-fold higher than the minimum inhibitory concentration for non-flocculent cells) (Supplemental Fig. 1).
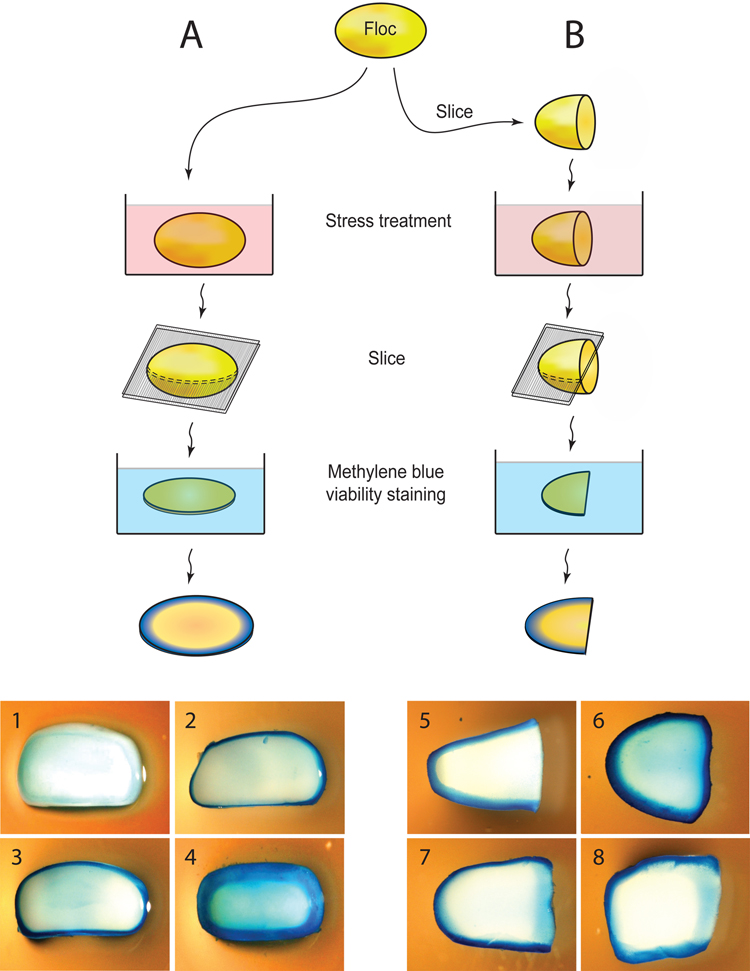
To further confirm that cells embedded within flocs are indeed physically shielded, we carried out viability assays on cross-sections of flocs before and after very severe stresses (70% ethanol or 100 µg ml−1 amphotericin B) (Fig. 3A). We hypothesized that a cell cannot survive these extreme concentrations if it comes in direct contact with these substances. Hence, if cells survive, this is likely because the chemicals were not able to reach the cells. The results indicate that after a short stress treatment, only cells at the outer edge of the floc are affected. Longer treatments result in a wider band of dead cells, but the inner cells remain unaffected (Fig. 3A). These results agree with a model in which chemicals can only very slowly penetrate flocs because the inner cells are physically shielded from the environment by the outer cell layers. However, this does not exclude the possibility that flocculating cells may also become inherently more resistant to stresses.
Figure 3. Flocculating cells are physically shielded from the external milieu.
A. Integral flocs were submerged in medium containing lethal levels of amphotericin B or ethanol. The flocs were subsequently sliced into thin sections and stained for viability using methylene blue, a dye that stains dead cells blue, while live cells remain white. (1) Control (no ethanol or amphotericin B); (2.) 100 µg ml−1 amphotericin B for 45 minutes; (3) 70% ethanol for 1 minute; (4) 70% ethanol for 45 minutes. B. Flocs were first cut in half before they were subjected to a stress treatment. (5) Control, with 45 min. amphotericin B treatment prior to slicing and staining the floc (note that this control was not sliced in half prior to treatment); (6) sliced before treating with 70% ethanol; (7) sliced before treatment with 100 µg ml−1 amphotericin B; (8) sliced before treatment with 15% ethanol. Note how for these intermediate conditions (7 and 8), cells that were originally situated in the heart of the floc (right-hand edge of specimens) are stained less than cell at the original periphery, but more than cells that remained shielded within the floc during stress treatment.
To investigate if cells embedded within flocs become inherently more resistant to stress, a variation of the former stress-survival assay was used. This time, flocs were sliced in half before stress treatment, so that the cells that were originally situated near the core of the floc now come in direct contact with the stress agent (Fig. 3B). After the stress treatment, a slice of the floc was stained for cell viability. The results indicate that cells in the core of the floc are more resistant then cells on the outside (Fig. 3B). This suggests that in addition to being physically shielded from toxins, cells embedded within a floc induce physiological changes that promote stress resistance.
Cells within flocs display a characteristic gene expression pattern
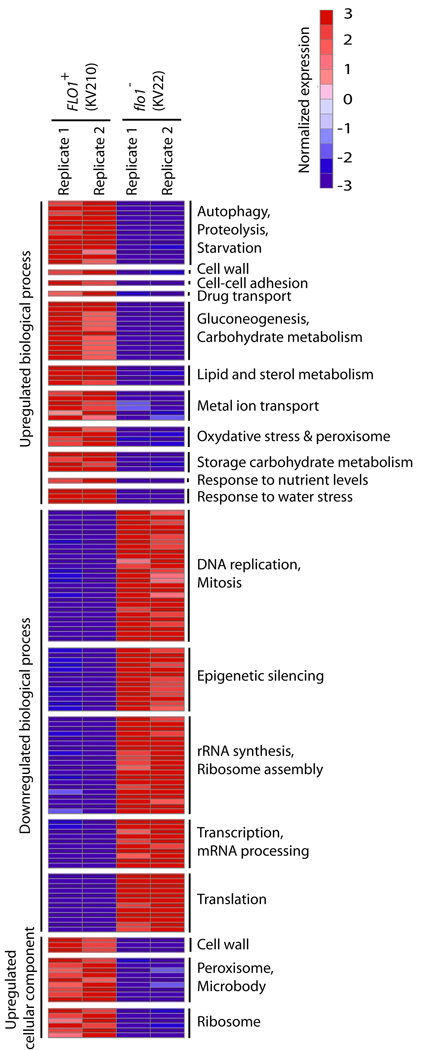
In order to investigate potential molecular mechanisms for the increased resistance to toxins, we performed genome-wide transcriptome analysis. The results indicate that cells embedded within flocs show considerable differences in gene expression compared to non-flocculating cells (Fig. 4 and Supplemental Fig. 2). The observed changes fall in several broad categories. First, decreased expression of genes involved in mitosis and protein synthesis (including ribosomal genes) indicate that the cells arrest or at least slow their growth, which was also confirmed experimentally (see further). The lack of growth might be associated with the limited availability of nutrients, as indicated by an upregulation of starvation genes as well as genes involved in gluconeogenesis and autophagy. Flocculating cells also upregulate a plethora of genes involved in stress resistance as well as multidrug transporters. Last but not least, genes involved in cell wall, lipid and sterol metabolism are also upregulated.
Figure 4. Flocculating and non-flocculating cells show differential expression of several gene clusters.
Genes were grouped into standard Gene Ontology (GO) sets. All GO gene sets that differ significantly between flocculating and non-flocculating cells are shown (rows). These sets are grouped together based on a higher-order category (labels in the right). For each gene set, the median expression of the leading-edge genes is shown. Expression was normalized by mean centering and unit scaling prior to visualization. Red and blue respectively represent induction and repression as compared to average across all experiments. See materials and methods and supplemental data online for more details.
The most highly upregulated gene in yeast flocs is DAN1. DAN1 encodes a cell-wall protein involved in sterol uptake and is typically upregulated in anaerobic cells, presumably in an attempt to take up sterols from the medium to complement for the arrested cellular synthesis of sterols, which is dependent on oxygen (Alimardani et al., 2004). In addition, several other genes linked to anaerobic growth, such as the TIR and PAU family, are induced in flocs (Supplemental spreadsheet).
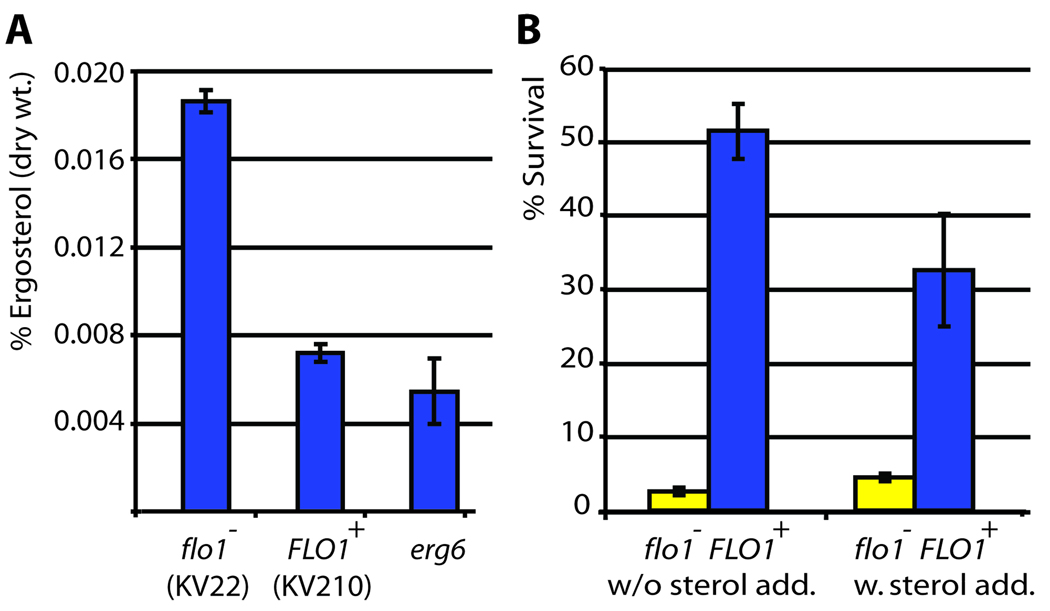
To investigate if the upregulated genes are crucial for floc formation and stress resistance, we deleted 10 of the most highly upregulated and interesting candidate genes. None of the deletion mutants showed significant changes in flocculation-dependent stress resistance (Supplemental Fig. 3). However, the particularly strong upregulation of DAN1 in flocculating cells pointed us at a potential mechanism for the increased resistance to amphotericin B. Upregulation of DAN1 suggests that cells embedded in flocs experience sterol deprivation. Interestingly, ergosterol is the target of amphotericin B (Ghannoum and Rice, 1999). The results in Figure 5 confirm that flocculating cells show a 60% reduction in ergosterol levels compared to planktonic cells, possibly as a consequence of a lack of oxygen inside the flocs, which is needed for sterol synthesis. To test if low sterol levels contribute to amphotericin resistance of flocculating cells, we supplemented flocculating and non-flocculating cultures with 20 ng ml−1 ergosterol and analyzed survival rates after treatment with amphotericin B (Fig. 5B). While sterol addition slightly increased survival rates of the planktonic cells (presumably because some of the amphotericin was sequestered by free ergosterol in the medium), the flocculating cultures show a marked 40% decrease in survival rates. Hence, apart from the limited penetration of the antifungal drug into the core of the yeast floc, resistance is further enhanced by the lower levels of ergosterol, the target of amphotericin B, in flocculating cells.
Figure 5. Lack of sterols in flocculating cells contributes to amphotericin B resistance.
Ergosterol levels of planktonic cells (KV22), flocculating cells (KV210) and cells defective in ergosterol sythesis (erg6 deletion) were measured. A. Calculated sterol levels. Both KV210 and the erg6 mutant show statistically significant lower ergosterol levels compared to the WT control (α = 0.05). B. Survival of nonflocculent (KV22) and flocculent (KV210) strains after amphotericin B treatment. In a first assay (left), no sterols were added to the growth medium. For the second assay, ergosterol was added to the growth medium (final concentration 20 µg ml−1). Flocculating cells survive significantly less when sterols are added to the medium; while nonflocculent cells survived slightly better. All pairwise differences within and between treatments are statistically significant (α = 0.05), error bars indicate standard deviation.
Natural flocculation in feral strains also correlates with drug resistance
All experiments above were carried out by comparing two S. cerevisiae strains, one in which FLO1 is transcriptionally silent, and one in which FLO1 is overexpressed. While this “clean” set-up has several advantages to determine the effect of FLO1 expression, there is also the risk of artifacts. We therefore investigated if flocculation also correlates with stress resistance in a set of naturally flocculating feral strains. We used the same set of 24 haploid EM93 strains as mentioned above and measured survival rates after amphotericin B treatment. The results show that flocculation indeed correlates with survival, with a Pearson correlation coefficient R2 of 0.60 (p < 10−5) (Supplemental Fig. 4). Given that the various EM93 strains are not isogenic, it is remarkable that more than half of the variation in stress resistance in these strains is linked to flocculation.
Variability in FLO1 generates variability in stress resistance
We and others have previously reported that flocculation is an extremely unstable phenotype. Closely related S. cerevisiae strains often show very distinct flocculation phenotypes (see for example Fig. 1), and even within one population, flocculation is often not stable (for a review, see (Verstrepen and Klis, 2006)). At least part of this variability is due to an unstable tandem repeat sequence in the FLO1 gene. The number of repeated DNA units varies at rates that are at least 100 fold greater than the average (point) mutation rates. In general, an increased number of repeats leads to stronger flocculation. To investigate the consequence of repeat variation on flocculation-mediated stress resistance, we overexpressed a series of FLO1 alleles with an increasing number of repeats. The results (Supplemental Fig, 5) show that stress resistance increases with increasing numbers of tandem repeats in the FLO1 gene.
Flocculation is under quorum-sensing regulation
Flocculation is a social trait that depends on multiple cells cooperating at one time. We therefore investigated the effect of known quorum-sensing molecules on the flocculation behavior of the diploid EM93 strain (Fig. 6), which does not show flocculation when grown in standard rich growth medium (YPD). However, given that some of the haploid segregants show strong constitutive flocculation (see Fig. 1), the EM93 diploid must have a functional set of flocculation genes, and its lack of flocculation is most likely due to transcriptional silencing.
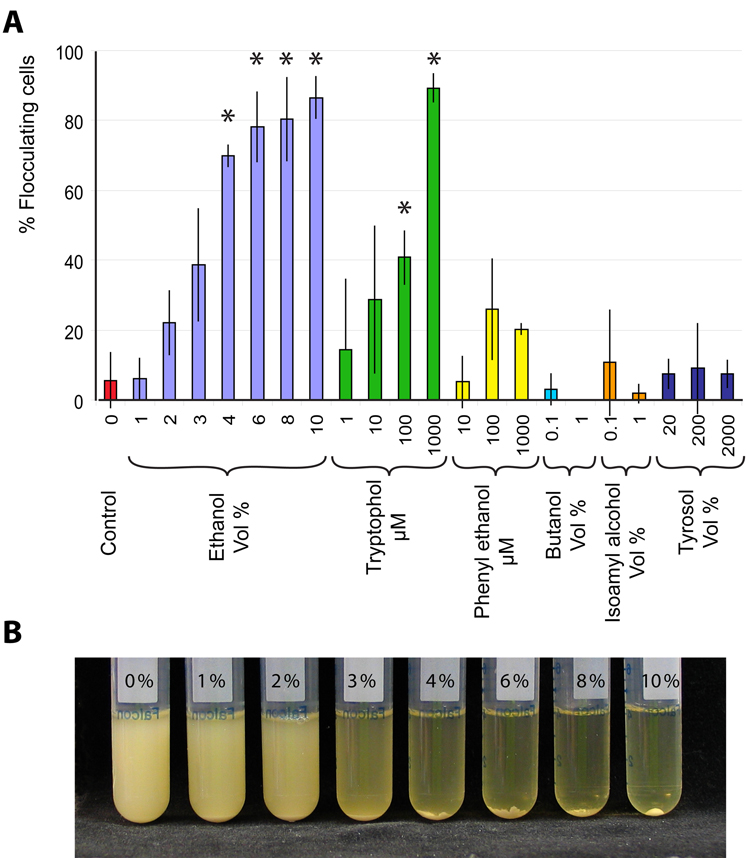
Figure 6. Ethanol and the quorum sensing molecule tryptophol induce flocculation in the feral EM93 strain.
Cultures of S. cerevisiae EM93 were grown in standard rich medium (YPD) with or without the addition of various known quorum sensing molecules and analogues thereof. A. Measurement of flocculation. Error bars represent standard deviation, asterisks indicate statistically significant differences from the control (i.e. no addition of any agent) (α = 0.05). B. Increasing flocculation of EM93 cultures observed with increasing concentration of ethanol.
Adding the known quorum sensing molecule tryptophol (Chen and Fink, 2006) induced strong flocculation in EM93. By contrast, phenyl ethanol, butanol and isoamyl alcohol, three other secondary metabolites that have been shown to act as intercellular signaling molecules (Lorenz et al., 2000), did not induce flocculation, nor did tyrosol, a known quorum-sensing agent of the fungal pathogen Candida albicans (Chen et al., 2004). Interestingly, addition of physiological concentrations of ethanol (6–10 Vol%) also induced very strong flocculation. This prompted us to ask if ethanol produced by a population of fermenting yeast cells is sufficient to trigger flocculation. We therefore grew EM93 cells in increasing sugar concentrations, which results in a corresponding increase in ethanol formation. As shown in Supplemental Fig. 6, increasing the sugar concentration from 2 to 22 % (w/v) results in a gradual increase in flocculation. While we cannot be sure what causes this flocculation, these results suggest that ethanol can function as a quorum-sensing molecule in S. cerevisiae, perhaps in combination with the other known molecules, such as tryptophol.
Expression of FLO1 comes at a fitness cost
Our data show that entering a floc can confer benefits to the inhabitants through resistance to various stress factors. Yet, not all feral yeast strains display strong flocculation. To understand this natural diversity, we investigated the costs and benefits of flocculation. To estimate the cost of FLO1 expression, we measured the relative fitness of FLO1-expressing S288C cells compared to wild-type S288C cells that do not express FLO1. Under normal growth conditions, FLO1-induced flocculation slowed growth rate more than 4-fold as compared to the non-flocculent strain. Similar differences were observed between naturally flocculating and non-flocculating EM93 strains. This enormous fitness cost is perhaps not surprising given that cells embedded in flocs only have limited access to nutrients and oxygen. We also investigated the cost of FLO1 expression without flocculation, using a FLO1-overexpressing strain grown in medium containing mannose, which prevents flocculation by competitively inhibiting interactions between Flo1 and the mannose in cell walls of adjacent cells (Kobayashi et al., 1998). Even when flocculation was completely inhibited by mannose addition, cells expressing FLO1 still show a significant fitness defect because of the expression of FLO1 (Fig 7A). The same is true when we compared a FLO1-expressing EM93 strain to the fitness of the same strain in which FLO1 was deleted. The FLO1-expressing strains show a slight fitness defect, illustrating the cost of FLO1 expression in a natural system (Fig. 7A).
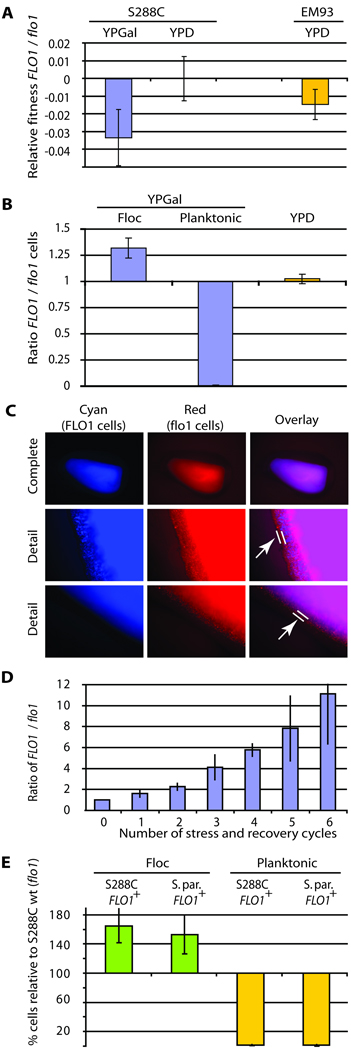
Figure 7. FLO1 is a Green Beard gene.
A. FLO1 expression comes at a fitness cost. Strain KV210 (FLO1 driven by the inducible GAL1 promoter) shows a significant (3%) fitness defect compared to strain KV22 (flo1−) when grown in YPGal medium, but not in YPD (where FLO1 is not induced, control). A significant (1.5%) fitness defect was also observed when a naturally flocculating FLO1+ EM93 strain was compared to its flo1 null mutant, demonstrating that natural expression of FLO1 also has a fitness cost. For all these experiments, the medium was supplemented with mannose (to block flocculation and only measure the cost associated with FLO1 expression, not flocculation; see text). Error bars represent 95% confidence intervals. B. FLO1 cells preferentially aggregate together. Cultures were inoculated with equal proportions of flocculating (KV210) and non-flocculating (KV22) cells. After induction of flocculation, the relative proportion of flocculating and non-flocculating cells in the planktonic and flocculating cell populations was measured, revealing an unequal distribution, with flocculating cells preferentially embedded within flocs, and the majority of nonflocculent “cheater” cells in the planktonic phase (P < 0.01). In YPD medium (no FLO1 expression, no flocculation), the fractions of both cell types remain equal. Error bars represent 95% confidence intervals. C. Fluorescence microscopy of flocs obtained from mixed cultures shows that flocs consist of perfectly mixed flocculent cells (KV210, Cyan) and non-flocculent “cheater” cells (KV22, Red). However, the outermost layer of the floc is almost exclusively made up out of nonflocculent cells (arrow). D. Mixed cultures of FLO1-expressing and flo1− cheater cells subjected to consecutive cycles of stress treatments show a gradual increase in FLO1-expressing cells. Cultures were inoculated with equal proportions of flocculating (KV210) and non-flocculating (flo1− KV22) cells. After 20 hours of growth in YPGal medium (to induce FLO1 expression in KV210), the mixed cultures were subjected to a 4h amphotericin treatment. After the treatment, the dead planktonic cells were removed. The remaining cells were washed, deflocculated and used to re-inoculate a fresh YPGal culture, which was again subjected to stress treatment after 20 h of growth. In these 20 hours, FLO1+ cells went through an average of 8.9 cell doublings, while flo1− cheaters divided about 9.4 times. After each cycle, the ratio of FLO1-expressing cells to flo1− cheater cells increases, indicating that these conditions strongly select for FLO1-expressing cells. A similar trend was observed when naturally flocculating (FLO1-expressing) and non-flocculating (FLO1-silent) EM93 strains were used (not shown). Error bars represent standard deviations. E. S. paradoxus and S. cerevisiae cells expressing FLO1 co-flocculate and exclude S. cerevisiae flo1− “cheater” cells. Three strains were co-cultivated: S. cerevisiae S288C (flo1−); S. cerevisiae KV210 (FLO1+) and a recombinant S. paradoxus strain that expresses the S. cerevisiae FLO1 gene. The graph shows the relative enrichment in flocs (green) and depletion in the planktonic fraction (yellow) of the two flocculent strains relative to the nonflocculent S288C wild-type cells (p < 0.01). FLO1-expressing S. cerevisiae and S. paradoxus cells co-flocculate and exclude S. cerevisiae cells that do not express FLO1, despite the closer genetic relatedness of the two S. cerevisiae strains. Error bars represent 95% confidence intervals.
FLO1–expressing cells preferentially interact with other FLO1-expressing cells
The cost of FLO1 expression raises the question of if and how flocculation is immune to cheater strains, which do not invest in expressing FLO1 but enter flocs and receive the protective benefits. In order to investigate this question, FLO1-expressing and non-flocculating flo1 S288C cells (“cheater” cells) were mixed in a 1:1 ratio and flocculation was induced by shifting the cells to galactose medium. After 16 hours of growth, flocs were separated from planktonic cells, and the number of cells of each of the two strains in both fractions was counted. The results (Fig. 7B) show that, while cells of each strain are found in both fractions, FLO1-expressing cells are significantly enriched in the flocs, and almost completely absent from the planktonic fraction (less than 1% of FLO1-expressing cells are planktonic). These ratios are more dramatic when one keeps in mind that as a consequence of the different growth rates, there are about 6-fold more nonflocculent cells than flocculent cells present in the culture. Hence, FLO1-expressing cells are able to direct their cooperative behavior (the formation of protective flocs) to other FLO1-expressing cells. These results are not an artifact of using the GAL1 promoter to induce FLO1 expression. We repeated the experiment with naturally flocculating and non-flocculent EM93 strains. A similar depletion of FLO1-expressing cells in the planktonic fraction (less than 2%) and enrichment of flocculating cells in flocs was observed (68% flocculent, 32% non-flocculent), demonstrating that the preferential embedding of FLO1 cells within flocs also occurs in naturally flocculating feral strains.
To investigate whether FLO1-expressing cells and the minority of flo1 “cheater” cells that are entrapped into the flocs are homogeneously mixed inside flocs, we expressed a different fluorescent protein in each cell type and investigated the fluorescence pattern in sliced flocs. The results show that the cheater cells are not found in separate clusters within the floc, but instead mix with FLO1-expressing cells, except for the outermost layer of the flocs, which almost completely exists of non-flocculating cells (Fig. 7C). It is interesting to note that cells in this outside layer are not protected from the outside environment, but do contribute to the protection of the inner cells. Hence, the first line of defense in the floc actually ends up being provided by flo1− “cheater” cells.
The unequal distribution of FLO1-expressing cells and flo1 “cheaters” in flocs versus the planktonic fraction is also reflected in the survival rates upon stress treatment. When mixed cultures of FLO1-expressing and flo1− cheater cells are repeatedly subjected to stress treatments (with 20 h recovery growth in-between treatments), the proportion if flo1− cheater cells gradually decreases (Fig. 7D). This is because the vast majority of flo1− cells are found in the planktonic fraction. Survival rates in this fraction are much lower than in the floc, which is enriched in FLO1-expressing cells. Similar results were obtained with naturally flocculating EM93 strains (not shown). FLO1 expression, therefore, confers a significant fitness advantage under some stress conditions. Moreover, the preferential interactions between FLO1-expressing cells are sufficient to gradually eliminate flo1 cheater cells.
Together, these results indicate the FLO1 expression leads to cooperative behavior that is preferentially directed towards other cells expressing FLO1 and effectively prevents flo1 cheater cells from reaping in the benefits of this cooperation without contributing to the cost of FLO1 expression. To investigate if this really only depends on the expression of a single gene, and not of any other genes in the genome, we used the strongest possible test: FLO1 was ectopically expressed in a different species, Saccharomyces paradoxus. The S. paradoxus genome does not contain a FLO1 ortholog, and S. paradoxus does not flocculate. However, ectopic expression of the S. cerevisiae FLO1 does confer strong flocculation in S. paradoxus. When either one of the FLO1-expressing species is mixed with a non-flocculent “cheater” strain of the other species, the floc is enriched in the FLO1-expressing organism, while the planktonic fraction is enriched in the other with the ratio’s being equal to those obtained using only flocculent and nonflocculent S. cerevisiae cells (not shown). When equal numbers of flocculating cells of S. cerevisiae and S. paradoxus are cultured together, virtually all cells aggregate in flocs that consist of equal proportions of both species (not shown). In a last set of experiments, three strains were cultured together: a nonflocculent S. cerevisiae strain S288C (KV22), a FLO1-expressing flocculent S. cerevisiae S288C strain (KV210) and a FLO1-expressing flocculent S. paradoxus strain. This results in yeast flocs that contain equal proportions of the flocculating S. cerevisiae and S. paradoxus strains, while the vast majority of nonflocculating S. cerevisiae cells are excluded from the floc and grow as planktonic cells (Fig. 7E). Together, these results show that flocculation and the exclusion of cheater cells occurs independently of the genetic background and depends solely on the presence of FLO1.
Discussion
Flocculation is a protective social response
This study shows how expression of a single dominant gene, FLO1, results in a multicellular phenotype in yeast. Upon expression of FLO1, cells adhere to each other to form tight flocs consisting of thousands of cells. As diffusion in this structure is severely impaired, the outer cells protect the inner cells from harmful compounds in the medium, something that cannot be achieved by single cells alone. Investing in the production of costly Flo adhesins is only useful when there is a sufficient concentration of other cells to form a floc. This may be why flocculation is regulated by the known quorum sensing molecule tryptophol, as well as by the primary metabolite ethanol. Together, these results reveal a complex and tightly-regulated social behavior in S. cerevisiae.
Similarities and differences between flocculation and biofilm formation
The protection provided to inner cells by the floc is reminiscent of microbial biofilms, where the tight structure of cells and extracellular material may shield inner cells from harmful compounds, including drugs. The floc's resistance to stress may be due not only to shielding but also to altered gene expression in cells embedded in flocs. Floc cells show upregulation of anaerobic and starvation genes, as well as genes encoding multidrug transporters, whereas genes involved in mitosis as well as ribosomal genes are downregulated. This transcription profile again bears resemblance to gene expression patterns in biofilms and S. cerevisiae mats: complex biofilm-like structures that form on low-density agar plates (Reynolds, 2006; Reynolds and Fink, 2001). Moreover, as is the case in biofilms and mats, deletion of the individual upregulated genes seems not to affect cellular stress resistance (d'Enfert, 2006; Reynolds, 2006). This could mean that the altered gene expression does not contribute to resistance or that the genes are partially redundant, as is the case in biofilms (d'Enfert, 2006).
The differences in gene expression between planktonic and flocculating cells are likely to be a consequence of the altered microenvironment inside flocs. The limited diffusion means that flocs are likely to be depleted of nutrients and oxygen, whereas waste products accumulate. The altered microenvironment in flocs also results in a deficit of ergosterol, a primary target of several antifungal drugs. Again, similar observations have been made for biofilms in pathogenic fungi, which also display reduced sterol levels that are at least partially responsible for their increased resistance to commonly used antifungal drugs (d'Enfert, 2006). Differential gene expression in general, and the different sterol composition in particular, may also explain why flocculating cells are not more resistant to all stresses, and for example show greater sensitivity to heat stress (Figure 2). It is interesting to speculate that the observed increase in stress resistance might not be limited to flocs. Adhesin genes like FLO1 may for example also be active in yeast colonies, providing cells growing on solid substrates with increased resistance.
FLO1 as a selfish green beard gene
Flocculation is a social trait that can confer both benefits (i.e. protection from stress) and costs (i.e. slower growth due to the burden of FLO1 expression). This leads to a central question of sociobiology: how can flocculation have evolved when cheater cells that do not express FLO1 could exploit cells that do? Our results indicate that FLO1 is a bona fide example of a green beard gene that confers both a social trait and a built-in mechanism for the preferential treatment of other FLO1-expressing cells. Expression of FLO1 entails a significant fitness cost, making it a target for flo1 “cheaters” who could invade flocs and benefit from the protective social structure without investing in it by carrying the fitness cost of FLO1 expression. In fact, we sometimes observed the emergence of non-flocculent cells in cultures inoculated with FLO1-expressing cells (not shown), which might indicate that cheaters develop relatively frequently. However, FLO1-expressing cells preferentially form flocs with other FLO1-expressing cells, limiting the frequency of flo1 cheater cells in the protective group.
The explanation for the preferential treatment of FLO1-expressing cells is likely to be mechanistically simple. Cells expressing Flo1 proteins can form reciprocal (two-way) attachments, which are stronger than the one-way interactions between a flocculent and a nonflocculent cell. Moreover, non-flocculating flo1 cells may be trapped at the surface of the “sticky” clump of FLO1-expressing cells, which explains why flocs are surrounded by a thin layer of nonflocculent cells (Fig. 7C). What may seem trivial from a mechanistic point of view, however, provides an attribute that has not been observed in most eukaryotes: the direct recognition of, and cooperation with, individuals that share the same genotype at a single locus.
There are only a few candidate examples of green beard genes to date. One case is the Gp-9 locus in Solenopsis invicta (red fire ant), where workers selectively kill egg-laying queens homozygous for one specific allele of the locus (Keller and Ross, 1998; Krieger and Ross, 2002). Here the “gene” is likely to represent several linked genes and the behavior involves spitefully killing individuals that do not bear the locus, rather than conferring cooperation among bearers of the green beard locus (Foster et al., 2001). A second example of possible green beard-like recognition is the case of cooperation among color morphs in side-blotched lizards, although the traits appear to be encoded by multiple loci across the genome (Sinervo et al., 2006). The best example of a green beard gene to date, and the most similar to FLO1, is the csaA gene from the social amoeba Dictyostelium discoideum (Queller et al., 2003). This gene encodes an adhesin vital for the formation of multicellular fruiting bodies on soil. Cells that do not bear a functional copy of csaA are depleted in the fruiting body and thus have a reduced chance of producing spores that can overcome a period of stress.
Importantly, the two species, S. cerevisiae and D. discoideum, are from different kingdoms and evolved both the escape response and green beard system independently. This convergent evolution in two distantly-related clades suggests that green beard recognition may emerge as a major phenotype in the sociobiology of microbes. Along with the similarities between these S. cerevisiae and D. discoideum adhesion genes, however, there also are major differences. For example, D. discoideum uses direct homophilic binding of the green beard protein, while the S. cerevisiae functions by binding to a distinct component of the cell wall. Most importantly though, csaA displays little or no within-species variability (Mehdiabadi, N., Queller, D. C, and Strassmann, J. E. personal comm.). That is, one allele of the gene has fixed and no longer plays a direct role in the evolutionary dynamics of D. discoideum. By contrast, the FLO genes are a very dynamic and variable system. Firstly, FLO expression levels vary strongly among strains, resulting in a great diversity of flocculation levels (Fig 1). Moreover, FLO genes are subjected to chromatin silencing and show stochastic silencing and desilencing (Halme et al., 2004). Finally, the FLO genes also show instability at the nucleotide sequence level because of the presence of an unstable internal tandem repeat region located in the coding DNA. Recombination events between these internal repeat units generate novel alleles that confer different flocculation characteristics (Verstrepen et al., 2005; Verstrepen et al., 2004).
While it remains to be confirmed exactly how this FLO variability combines to affect the evolution of yeast social interactions, it is clear that the FLO genes represent a highly dynamic system. The exceptional variability may result from ongoing competition between strains that cooperate and defect during flocculation, especially since FLO1 expression confers a significant fitness burden and does not completely avoid exploitation by cheating flo1 cells. The occurrence of multiple alleles of FLO genes also raises the possibility of fine discrimination among different allotypes (commonly referred to as the existence of multiple “colors” of beards). However, given the adhesion mechanism, with Flo proteins recognizing mannose residues that may be independent of the specific allele of expressed FLO gene, it seems likely that this effect will be weak at best.
A key conclusion from our work is that genetic identity at a single locus (FLO1) is more important for a social phenotype (flocculation) than genetic identity between organisms across the rest of their genomes. Activation or inactivation of FLO1 in S. cerevisiae, while leaving all other genes intact, induces or abolishes flocculation. Similarly, insertion of FLO1 into a different species (S. paradoxus) that normally lacks FLO1 causes strong flocculation that closely resembles the flocculation phenotype observed in FLO1-expressing S. cerevisiae cells. Most convincing is that co-culturing the FLO1-expressing cells of S. cerevisiae and S. paradoxus results in flocs that contain equal numbers of each species, whereas non-flocculent S. cerevisiae cells are underrepresented in the flocs. This shows that genetic identity at the FLO1 locus is more important for the social phenotype than genetic identity because the two genomes differ significantly. This system then epitomizes the notion of the selfish gene that can, at least temporarily, act to increase its own frequency irrespective of evolutionary interests of other genes in the genome, an idea popularized in Dawkin’s “The selfish gene” (Dawkins, 1976). The example of FLO1 is particularly telling because it counters the common misconception that selfish genes always result in selfish organisms: FLO1 is a “selfish” green beard gene that drives an act of remarkable cooperation.
Experimental Procedures
Microbial strains, growth conditions and molecular techniques
All yeast strains used are listed in Supplemental Table 1. EM93 haploid tetrads were derived from the feral diploid strain EM93 (Mortimer and Johnston, 1986). Yeast cultures were grown as described before (Sherman et al., 1991). YPGal medium contained 2% raffinose, 2% galactose, 2% peptone and 1% yeast extract. Ergosterol-enriched cultures were prepared by adding 60 µL of ergosterol stock solution (1 mg ml−1 ergosterol in 50:50 v/v Tergitol NP-40) and 30 µL of Tween 80 to 3 mL YPGal before inoculation. Flocs were disrupted in a 200 mM EDTA solution. Flocculation and ergosterol levels were measured as described previously (d'Hautcourt and Smart, 1999) (Arthington-Skaggs et al., 2002). Real-time PCR using the ABI 7500 system (Applied Biosystems) was carried out as recommended by the supplier. All oligonucleotides are listed in Supplemental Table 2 online. Cells were counted using the number of Colony Forming Units (CFU). To count two or more strains in mixed cultures, each strain was labeled with different resistance markers to enable discrimination on selective media. Alternatively, strains were labeled with fluorescent markers and counted using flow cytometry. Constitutively expressed fluorescent tags were derived from plasmids pSR240 or pKT139 (EUROSCARF), and inserted in BY4741 under the control of TDH3 promoter. In a second step, the constructs were amplified and inserted in an intergenic region of chromosome II (coordinates 343239–343409 in S288C). To obtain a flocculating S. paradoxus, DNA template from KV210 was used to amplify the KAN-GAL1p::FLO1 region which was used for transformation. S. paradoxus wild-type and the flocculating derivative.
Stress resistance assays
Survival testing is described in Fig. 3. In brief, cultures were subjected to the stress treatment, and subsequently centrifuged (4 min. at 3000 × g), washed and deflocculated using 250 mM EDTA, diluted and plated onto nonselective YPD agar to determine the number of colony forming units (CFU). To ensure that slight differences in culture medium or treatment could not account for the observed differences, this whole procedure was repeated with one alteration. In this case, 0.5 ml of non-flocculent cells was added to the test tube containing the flocculent culture, so that an equal number of flocculent and non-flocculent cells were present in the same medium during stress treatment. No differences were found between these two methods.
Microscopy
Intact flocs or floc slices were treated as indicated in 3 ml YPGal for 45 minutes at room temperature. Flocs and once-sliced flocs were washed with YPGal and then sliced to create ellipses or half-ellipses of approximately 1 mm thickness. Floc slices were stained in 1 ml of 0.1% methylene blue (Sigma-Aldrich) in YPGal for 1 minute. Stained specimens were washed with YPGal, and examined using a Zeiss Discovery V12 stereoscope with epifluorescence kit. Electron microscopy, was performed as described previously (Beauvais et al., 2007). Briefly, flocs of KV210 or centrifuged KV22 cultures were frozen using a Gatan alto 2500 cryostage and cryopreparation chamber and observed using a Jeol JSM-6700F apparatus.
Gene array analysis
Gene array analysis was performed using Affymetrix S98 chips as recommended by the producer. Gene Set Enrichment Analysis (GSEA) as described previously (Subramanian et al., 2005). For details, refer to the supplemental text.
Fitness measurements
Relative Malthusian fitness was determined as described before (Thompson et al., 2006). For details, please refer to the supplemental text.
Flow cytometry
Deflocculated samples were analyzed using a LSRII (Becton Dickson) flow cytometer. Fluorescent intensities were examined using a 488 nm excitation and a 530 ± 30 nm emission wavelength filter to detect YFP-tagged cells and a 561 nm excitation and 620 ± 20 nm emission wavelength filter to detect RFP tagged cells. Results were analyzed using FlowJo software (Treestar Inc.).
Supplementary Material
Acknowledgments
We apologize for the omission of relevant references due to space restrictions. We thank 3 reviewers, B. Stern, B. Aertsen, S. Alonzo, M. Legendre, A. Schier, A. Regev, R. Losick and A. Murray for their suggestions. KJV and KRF are supported by NIH NIGMS grant 5P50GM068763, and KJV also by the Human Frontier Science Program Award HFSP RGY79/2007, an Odysseus fellowship of the Flemish Fund for Scientific Research (FWO) and VIB. JPL is supported by contract LSHB-CT-2004-511952 FUNGWALL. GRF is supported by NIH grant GM40266. NP is a fellow of the Belgian American Educational Foundation (B.A.E.F.). M.D.V. is a fellow of the Ford Foundation and the B.A.E.F. S.S. and C.Y. thank the Harvard College Research Program, PRISE and the Bauer Internship Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Cited references
- Alimardani P, Regnacq M, Moreau-Vauzelle C, Ferreira T, Rossignol T, Blondin B, Berges T. SUT1-promoted sterol uptake involves the ABC transporter Aus1 and the mannoprotein Dan1 whose synergistic action is sufficient for this process. Biochem J. 2004;381:195–202. doi: 10.1042/BJ20040297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthington-Skaggs BA, Lee-Yang W, Ciblak MA, Frade JP, Brandt ME, Hajjeh RA, Harrison LH, Sofair AN, Warnock DW. Comparison of visual and spectrophotometric methods of broth microdilution MIC end point determination and evaluation of a sterol quantitation method for in vitro susceptibility testing of fluconazole and itraconazole against trailing and nontrailing Candida isolates. Antimicrob Agents Chemother. 2002;46:2477–2481. doi: 10.1128/AAC.46.8.2477-2481.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais A, Schmidt C, Guadagnini S, Roux P, Perret E, Henry C, Paris S, Mallet A, Prevost MC, Latge JP. An extracellular matrix glues together the aerial-grown hyphae of Aspergillus fumigatus. Cell Microbiol. 2007;9:1588–1600. doi: 10.1111/j.1462-5822.2007.00895.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Fink GR. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 2006;20:1150–1161. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fujita M, Feng Q, Clardy J, Fink GR. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc Natl Acad Sci U S A. 2004;101:5048–5052. doi: 10.1073/pnas.0401416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Enfert C. Biofilms and their role in the resistance of pathogenic Candida to antifungal agents. Current drug targets. 2006;7:465–470. doi: 10.2174/138945006776359458. [DOI] [PubMed] [Google Scholar]
- d'Hautcourt O, Smart K. Measurement of brewing yeast flocculation. J Am Soc Brew Chem. 1999;57:123–128. [Google Scholar]
- Dawkins R. The selfish gene. Oxford, UK: Oxford University Press; 1976. [Google Scholar]
- Foster KR, Parkinson K, Thompson CR. What can microbial genetics teach sociobiology? Trends Genet. 2007;23:74–80. doi: 10.1016/j.tig.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KR, Wenseleers T, Ratnieks FLW. Spite: Hamilton’s unproven theory. Annales Zooogici Fennici. 2001;38:229–238. [Google Scholar]
- Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12:501–517. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink G. Characterization of Saccharomyces cerevisiae pseudohyphal growth. In: Bossche HV, editor. Dimorphic fungi in biology and medicine. New York: Plenum Press; 1993. [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles CA, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. The genetical evolution of social behavior II. J Theor Biol. 1964;7:17–52. doi: 10.1016/0022-5193(64)90039-6. [DOI] [PubMed] [Google Scholar]
- Keller L, Ross KG. Selfish genes: a green beard in the red fire ant. Nature. 1998;394:573–574. [Google Scholar]
- Kobayashi O, Hayashi N, Kuroki R, Sone H. Region of Flo1 proteins responsible for sugar recognition. J Bacteriol. 1998;180:6503–6510. doi: 10.1128/jb.180.24.6503-6510.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger MJ, Ross KG. Identification of a major gene regulating complex social behavior. Science. 2002;295:328–332. doi: 10.1126/science.1065247. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Cutler NS, Heitman J. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Genealogy of principal strains of the Yeast Genetic Stock Center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palkova Z. Multicellular microorganisms: laboratory versus nature. EMBO Rep. 2004;5:470–476. doi: 10.1038/sj.embor.7400145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palkova Z, Vachova L. Life within a community: benefit to yeast long-term survival. Fems Microbiol Rev. 2006;30:806–824. doi: 10.1111/j.1574-6976.2006.00034.x. [DOI] [PubMed] [Google Scholar]
- Pennisi E. How did cooperative behavior evolve? Science. 2005;309:93. doi: 10.1126/science.309.5731.93. [DOI] [PubMed] [Google Scholar]
- Queller DC. Kin selection and frequency dependence: a game-theoretic approach. Biol J Linn Soc. 1984;23:133–143. [Google Scholar]
- Queller DC, Ponte E, Bozzaro S, Strassmann JE. Single-gene greenbeard effects in the social amoeba Dictyostelium discoideum. Science. 2003;299:105–106. doi: 10.1126/science.1077742. [DOI] [PubMed] [Google Scholar]
- Reynolds TB. The Opi1p transcription factor affects expression of FLO11, mat formation, and invasive growth in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:1266–1275. doi: 10.1128/EC.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds TB, Fink GR. Bakers' yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. [Google Scholar]
- Sinervo B, Chaine A, Clobert J, Calsbeek R, Hazard L, Lancaster L, McAdam AG, Alonzo S, Corrigan G, Hochberg ME. Self-recognition, color signals, and cycles of greenbeard mutualism and altruism. Proc Natl Acad Sci U S A. 2006;103:7372–7377. doi: 10.1073/pnas.0510260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, Desai MM, Murray AW. Ploidy controls the success of mutators and nature of mutations during budding yeast evolution. Curr Biol. 2006;16:1581–1590. doi: 10.1016/j.cub.2006.06.070. [DOI] [PubMed] [Google Scholar]
- Trejo WH, Bennett RE. Streptomyces nodosus sp. n., the amphotericin-producing organism. J Bacteriol. 1963;85:436–439. doi: 10.1128/jb.85.2.436-439.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Klis FM. Flocculation, adhesion and biofilm formation in yeasts. Mol Microbiol. 2006;60:5–15. doi: 10.1111/j.1365-2958.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- Verstrepen KJ, Reynolds TB, Fink GR. Origins of variation in the fungal cell surface. Nat Rev Microbiol. 2004;2:533–540. doi: 10.1038/nrmicro927. [DOI] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.