Abstract
Neural crest cells (NCCs) can adopt different neuronal fates. In NCCs, neurogenin-2 promotes sensory specification but does not specify different subclasses of sensory neurons. Understanding the gene cascades that direct Trk gene activation may reveal mechanisms generating sensory diversity, because different Trks are expressed in different sensory neuron subpopulations. Here we show in chick and mouse that the Runt transcription factor Runx1 promotes axonal growth, is selectively expressed in neural crest-derived TrkA+ sensory neurons and mediates TrkA transactivation in migratory NCCs. Inhibition of Runt activity depletes TrkA expression and leads to neuronal death. Moreover, Runx1 overexpression is incompatible with multipotency in the migratory neural crest but does not induce expression of pan-neuronal genes. Instead, Runx1-induced neuronal differentiation depends on an existing neurogenin2 proneural gene program. Our data show that Runx1 directs, in a context-dependent manner, key aspects of the establishment of the TrkA+ nociceptive subclass of neurons.
Information from external and internal environments is conveyed through peripheral sensory neurons terminating in specialized structures termed sensory receptors located in the skin, muscles and organs of the body. The somatic sensory system comprises different perceptual modalities. Neurons of the dorsal root ganglia (DRG) mediate tactile sensation elicited by mechanoreceptive stimuli, limb proprioceptive sensation elicited by displacement and the static tension of muscles, thermal sensation and pain. Thus, DRG sensory neurons are essential for receiving and relaying information about many types of external stimuli to the brain and are also responsible for conveying visceral pain and proprioceptive information. This can be accomplished through the presence of different subclasses of DRG neurons that convey different perceptual modalities. Thus, DRG neurons have different molecular properties and central termination points within the spinal cord1. Much remains to be understood about how sensory neuron subclasses are created during embryogenesis.
During development, neuroepithelial cells in the dorsal neural tube undergo conversion into NCCs, which in turn generate DRG neurons. Several neural crest-specifying transcription factors control cell shape and cell adhesion through downstream mediators. They thereby initiate delamination from the neural tube, which is followed by migration and differentiation2. Premigratory and migratory NCCs have stem cell-like properties. They are multipotent and competent to generate neurons, glia and melanocytes, as well as smooth muscle cells. Expression of the high-mobility group transcription factor Sox10 is necessary to maintain their multipotency3. NCCs leave the neural tube starting at embryonic day 9 (E9) in mice4 and migrate in chain-like structures5 to form the DRG in a ventral-to-dorsal order. Within the condensed ganglion, sensory progenitors continue to proliferate and the different subclasses of DRG neurons are born (that is, they exit the cell cycle) at different developmental stages. The large-diameter mechanoreceptive neurons are born first, showing a peak of birth in the cervical region at E10.5 in mice, whereas E11.5 is the peak for birth of small-diameter nociceptors6.
The early neurons survive cell-autonomously and the acquisition of trophic factor dependency for survival is coordinated with the arrival of their axons in the target field7. Different subclasses of sensory neurons depend on different neurotrophic factors for their survival. This dependency is reflected by expression of TrkA (encoded by Ntrk1) and TrkC (encoded by Ntrk3) in subpopulations of the embryonic DRG8. Mice carrying null mutations in nerve growth factor beta (encoded by Ngfb) or its receptor TrkA show a specific loss of virtually all nociceptors9,10, whereas neurotrophin-3 (Ntf3) and Ntrk3 null mice lack limb proprioceptive neurons11. The pro-apoptotic protein Bax regulates survival of neurotrophin-dependent peripheral neurons. In Bax-/- and in compound Ngfb-/- Bax-/- or Ntf3-/- Bax-/- mice, excessive cell death due to trophic factor deprivation is eliminated12,13. In surviving neurons, however, the lack of NGF signaling leads to severely diminished nociceptive marker expression, including neuropeptides and ion channels12,14, showing that NGF signaling not only promotes neuronal survival, but is also required for full phenotypic maturation of nociceptors. In a recent study14, a strain of mice was generated in which Ntrk3 genetically replaces Ntrk1 (that is, TrkC replaces TrkA protein). In these mice, some presumptive TrkA-expressing neurons adopted a proprioceptive phenotype. These results show that neurotrophin signaling executes cytochemical expression and functional connectivity characteristic of different sensory neuron subclasses and thereby has an instructive role in sensory subclass specification. Identification of transcription factors controlling Trk expression could therefore provide a link between a general sensory neuron program executed by the neurogenin (Neurog1 and Neurog2) family of proneural genes and the establishment of different sensory neuron subclasses. However, little is known about the genetic control of Trk receptor expression.
In mammals, there are three members of the runt-domain family of transcription factors, Runx1, 2 and 3. They have been shown to have crucial roles in specifying cell fate in bones, hematopoietic stem cells, cartilage and the gastrointestinal tract15. Runx3 is selectively expressed in the TrkC population of sensory neurons; in null mutant mice, TrkC is at the onset of expression greatly reduced and disappears a few days later. Furthermore, these mice show decreased expression of proprioceptive markers and axonal projection deficits; their proprioceptors ultimately perish due to lack of trophic support16,17. Runx1 is also expressed in a subpopulation of DRG neurons during embryogenesis18, but little is known about its function in sensory neuron development.
Here we show that Runx1 is expressed in the TrkA population of DRG neurons. In this population, Runx1 acts as a transcriptional activator and is necessary first for specific aspects of differentiation and later for survival of nociceptors. Its activities depend on an Ngn2 proneural gene program. One direct role of Runx1 in the differentiation of the nociceptor subclass is to induce TrkA expression. In the absence of Runx1, TrkA is not expressed and the neurons die by apoptosis.
RESULTS
Developmental expression of Runx1 in mice and chick DRG
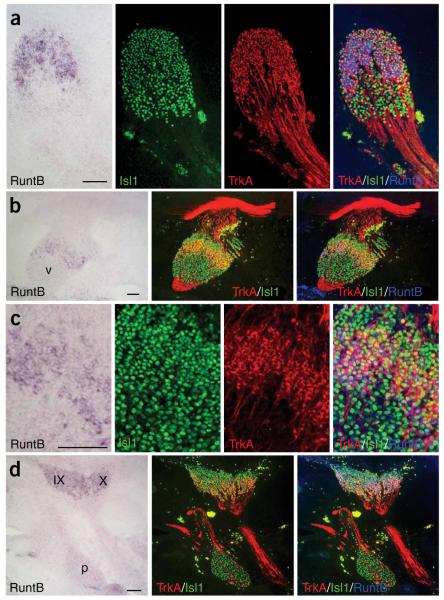
We first investigated Runx1 expression and its chick homolog RuntB during the development of cranial and spinal sensory neurons. In situ hybridized sections for RuntB were double immunostained for Isl1 to detect all sensory neurons and TrkA for identification of nociceptive neurons. TrkA expression in dorsal root, trigeminal and superior/jugular ganglia was seen in the subpopulation of neurons expressing RuntB (Fig. 1a-d). Among placode-derived sensory neurons, RuntB was present in facial and vestibuloacoustic but not the petrose ganglion and TrkA was not expressed in any of these placode-derived ganglia (Fig. 1d, Supplementary Fig. 1 online and data not shown). Thus, RuntB expression colocalizes with TrkA in neural crest-derived sensory neurons. Runx1 expression appeared later than Ngn2 in mouse and chick DRG during development (Supplementary Fig. 1). Ngn2 was expressed in migratory NCCs and later in some cells of the DRG. At E10.5 and E11.5 in mice, Runx1 expression was observed in cells populating the DRG, but not at earlier stages in the migratory NCCs, suggesting that expression appears after initiation of neurogenesis. The Ngn2 expression pattern is similar in chicks and mice19. In chicks, RuntB was not present in migratory NCCs at stage HH19 (ref. 20) and its expression started in DRG at HH24 in few cells populating the DRG. At stage HH29, a robust expression of RuntB was observed in a dorsomedial subpopulation of DRG neurons. The increase of RuntB between HH24 and HH29 correlated with a threefold increase in DRG neurons expressing TrkA (data not shown). Taken together, these results suggest that Runx1 may act in concert and/or sequentially with Ngn2, and its expression is compatible with a role in the specification of nociceptive neurons in vertebrates.
Figure 1.
RuntB is expressed in the TrkA+ subpopulation of neural crest-derived trunk and cranial sensory neurons. In situ hybridization for RuntB and immunohistochemical stainings for TrkA (red) and Isl1 (green) are shown (a) in the same sagittal sections of DRG, (b,c) in the trigeminal ganglion (V) and (d) in the superior (IX), jugular (X) and petrosal (p) ganglia at stage HH29 of the chick embryo. Merged images of RuntB In situ hybridization (blue pseudocolor) and immunohistochemical stainings for TrkA and Isl1 show an overlapping expression of RuntB and TrkA in trigeminal, superior, jugular and dorsal root ganglia but not in the petrosal ganglion. Scale bars, 100 μm.
Runx1b promotes survival of Ngn2-fated bNCSCs
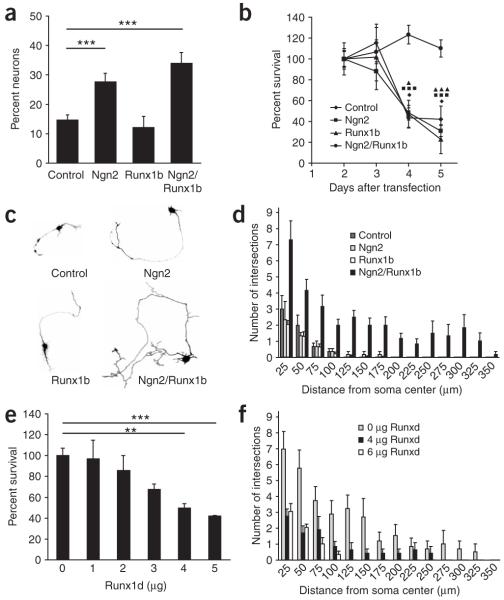
To investigate the role of Runx1 in neural progenitors, we examined its function in p75+/nestin+ boundary cap neural crest stem cells (bNCSCs)21. In these assays, the percentage of neurons that differentiate all the way into sensory neurons is very low21. We therefore used bNCSCs to examine the effects of Runx1 on the differentiation and survival of neural progenitors into immature βIII-Tubulin+ neurons. The cells expressed very low levels of Ngn2 and Runx1 endogenously (Supplementary Fig. 2 online). Ngn2, the full-length Runx1 (Runx1b) or a combination of both were overexpressed and the number of neurons was counted 3 d later. As previously shown22, Ngn2 induced a neuronal phenotype with neurites (27.7 ± 2.9% of Ngn2-transfected cells showed a neuronal phenotype, versus 14.6 ± 1.8% in control samples; P < 0,001; Fig. 2a). Runx1b alone did not lead to any increase in the number of neurons. Similarly, coexpression of Runx1b with Ngn2 did not increase the number of neurons compared to Ngn2 alone (Fig. 2a). These data indicate that Runx1b has no neurogenic effect on bNCSCs. Four and five days after transfection, cells expressing Ngn2 or Runx1b alone died similar to control cells (Fig. 2b). Unexpectedly, such a loss was not observed at any of the analyzed time points for Ngn2/Runx1b-overexpressing neurons (Fig. 2b). The survival was cell autonomous, since removal of trophic support from the culture did not affect survival (Supplementary Fig. 2). We conclude that Runx1b depends on Ngn2 for its activities in bNCSCs.
Figure 2.
Runx1b promotes survival and axonal growth of Ngn2-fated bNCSCs. (a) Percentage of neurons 3 d after differentiation with Ngn2, Runx1b or both. Ngn2 exerted a neurogenic effect, whereas Runx1 did not. (b) Camera lucida drawings of representative neurons observed 3 d after differentiation in control and with Ngn2, Runx1 and Ngn2/Runx1. Note that cotransfection with Ngn2 and Runx1 led to a marked increase in neurite growth. (c) Percentage survival of neurons at 3, 4 and 5 d after differentiation of bNCSCs. Neurons in cultures receiving only GFP plasmid (control), or after Ngn2 or Runx1b transfection, died massively between 3 and 5 days, while Ngn2/Runx1 coexpressing neurons survived. (d) Quantification by Sholl analysis of neurite length and number of branching of neurons 3 d after differentiation with Ngn2, Runx1 and Ngn2/Runx1. Cotransfection with Ngn2 and Runx1 significantly increased both neurite length and branching at all distances. (e,f) Runx1 transactivation domain mediates its survival and differentiation effects. (e) Percent survival of neurons 3 d after differentiation with Ngn2/Runx1 and increasing concentrations of Runx1d (a competitive form of Runx1b blocking the activity of the full protein) compared to survival after transfection with Ngn2/Runx1 only. Runx1d reversed the survival effect of Runx1b on Ngn2-fated bNCSCs in a dose-dependent manner. (f) Quantification of neurite length and number of branches by Sholl analysis of neurons 3 d after differentiation with Ngn2/Runx1b and increasing concentrations of Runx1d. Runx1d reversed the growth effect of Runx1b on Ngn2-fated bNCSCs. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Student’s t-test. Error bars = s.e.m.
Runx1b promotes axonal growth of Ngn2-fated bNCSCs
To analyze whether the survival effect of Runx1b on Ngn2-fated cells correlated with differentiation in vitro, we studied the morphology of cells expressing different combinations of transcription factors. The number of neurites derived from a single neuron was counted at 25-μm intervals using concentric rings to the cell body (Sholl analysis)23. Neurite length and branching markedly increased in neurons coexpressing Ngn2 and Runx1 (Fig. 2c,d). These results indicate that Runx1b stimulates axonal outgrowth of Ngn2-fated neurons.
In vitro effects of Runx1 require its transactivation domain
Runx1b encodes a protein of 452 amino acids, including the Runt domain and a carboxy (C)-terminal domain capable of DNA binding and transactivation, respectively. Runx1d encodes an endogenous isoform of 243 amino acids that binds to DNA but is not capable of transactivation24-26. Runx1d interferes with the activity of all three mammalian Runx proteins due to its higher affinity for the consensus DNA binding site25. Transfection of Runx1d alone did not induce survival or axonal growth. To examine if the survival and differentiation effects of Runx1 depend on the transactivation domain, we cotransfected cells with Ngn2 and Runx1b together with increasing concentrations of Runx1d. Measurement of survival showed that Runx1b-induced survival was reversed in a dose-dependent manner (Fig. 2e). Furthermore, Runx1d prevented the Runx1b-dependent increase in both neurite length and branching (Fig. 2f). Thus, both survival and axonal growth elicited by Runx1b depend on DNA binding and an intact transactivation domain.
Runt activities are required for sensory neurons in vivo
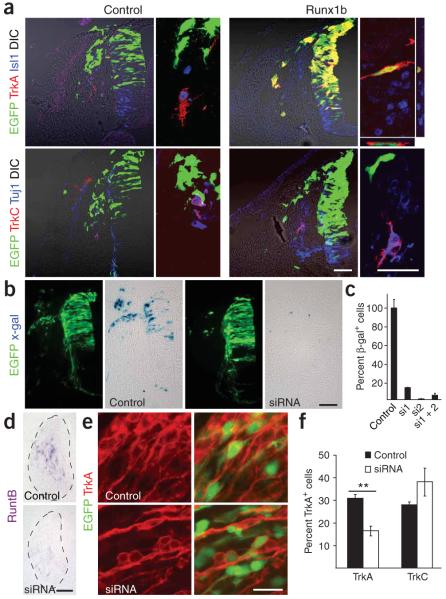
Because Runx1 null mutant mice die at E11.5 (refs. 27,28), they could not be used to study the role of Runx1 in the DRG. To study the function of runt domain transcription factors in development of peripheral sensory neurons, we developed an in vivo model in which genes were introduced into the premigratory neural crest of the chick neural tube. Fluorescent cells were identified in migratory and post-migratory NCCs, including the DRG, after electroporation with an expression vector for enhanced green fluorescent protein (EGFP) (Supplementary Fig. 3 online). Co-electroporation of EGFP and red fluorescent protein (RFP) revealed that more than 95% of the transfected cells received both vectors (data not shown). Runx1d and EGFP were introduced into the chick and embryos were analyzed 96 h after gene delivery (HH29). In control embryos expressing EGFP alone, positive cells appeared throughout the whole ganglion and differentiated into TrkA-, TrkB- and TrkC-expressing cells (Fig. 3a). Embryos overexpressing Runx1d exhibited a significantly smaller total number of EGFP+ cells in the DRG compared with controls. This loss occurred in the populations of neurons expressing TrkA, TrkB and TrkC and specifically in the cells receiving the DNA constructs (the EGFP+ cells), since TrkA-, TrkB- and TrkC-expressing neurons in the untransfected cells (or EGFP- cells), remained unchanged, as did the overall size of the DRG. The remaining EGFP+ cells in the Runx1d-transfected cells showed a mesenchymal-like morphology and were located in the very dorsal part of the DRG (Fig. 3b). A similar cell population was also seen in control animals. These cells also failed to express general neuronal markers such as βIII tubulin (Tuj1) and NeuN (Fig. 3 and Supplementary Fig. 4 online).
Figure 3.
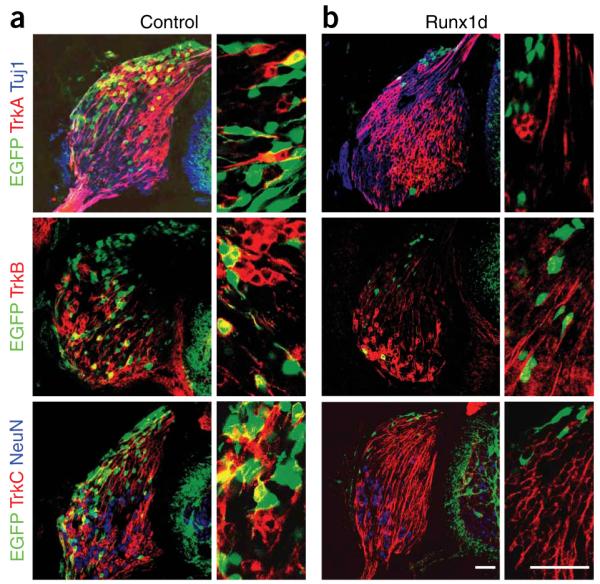
Blocking Runt activity is incompatible with establishment of neural crest-derived sensory neurons. Micrographs of stage HH29 DRG of (a) control and (b) Runx1d-overexpressing chick embryos stained for TrkA, TrkB or TrkC in red, the early neuronal marker Tuj1 (βIII tubulin) or NeuN in blue and corresponding high-magnification images where only the green and red channels are shown. Note in panel a that EGFP was present in TrkA+, TrkB+ and TrkC+ neurons, whereas animals overexpressing Runx1d (in panel b) showed a marked loss of EGFP-expressing cells. The remaining cells, mostly residing in the very dorsal part of the DRG, did not express TrkA, TrkB or TrkC. Scale bars, 50 μm.
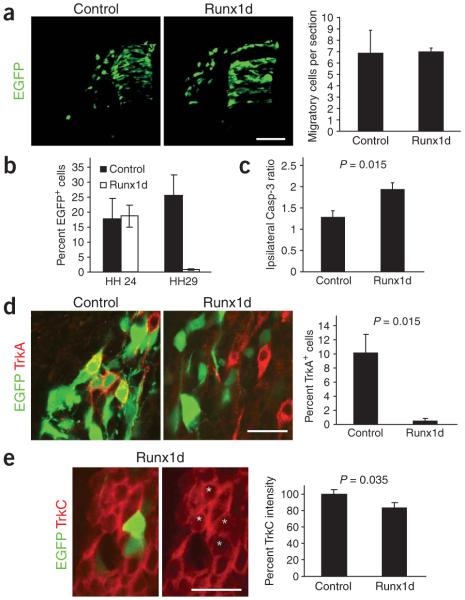
Runx1d-induced neuronal loss could be a consequence of a defect at any point of DRG development. We examined embryos 24 h after transfection (HH19) to determine whether Runx1d affected neural crest delamination and migration. No difference in the number of migratory NCCs was seen between control and Runx1d embryos (Fig. 4a) and no difference in the number of EGFP+ neurons in the DRG was seen at HH24 (72 h after transfection) (Fig. 4b). However, sections from HH24 embryos showed a significant increase of active caspase-3 positive cells as a consequence of forced Runx1d expression (Fig. 4c) and at HH29, there was an almost complete loss of neurons (Fig. 4b). We conclude that neurons overexpressing Runx1d are lost by apoptosis between HH24 and HH29, coinciding with the onset of the period of naturally occurring cell death.
Figure 4.
Runx1d-induced apoptosis is preceded by a loss of Trk expression. (a) Runx1d does not affect NCC migration. Control and Runx1d-overexpressing chick embryos at HH19 and quantitative measurements of the number of migratory NCCs per section with and without Runx1d. (b) Percentage of EGFP+ cells relative to the total number of DRG neurons in control and Runx1d-overexpressing embryos at stages HH24 and HH29. Note that the loss of neurons occurs between stages HH24 and HH29. (c) Loss of neurons is preceded by an increase in active caspase-3. The number of active caspase-3+ cells in Runx1d-overexpressing and contralateral control DRG was counted at HH24 (the ratio of ipsi- to contralateral is presented). (d) Death of neurons is preceded by a nearly complete loss of TrkA expression at stage HH24. Micrographs showing TrkA expression in control and Runx1d-overexpressing DRG neurons and quantitative measurements of the percentage of cells expressing TrkA in control and Runx1d-overexpressing embryos. Note that the Runx1d-overexpressing EGFP+ neurons lack TrkA expression. (e) Death of neurons is also preceded by a reduction of TrkC levels at HH24. Micrographs showing TrkC expression in control and Runx1d-overexpressing DRG neurons at stage HH24 and quantitative data on the mean intensity of TrkC expression in control and Runx1d-expressing DRG neurons. Note loss of TrkC expression levels in the two Runx1d-transfected cells (green stars) as compared to adjacent untransfected control cells (yellow stars). Scale bars, 50 μm (a) and 20 μm (d,e). ***P ≤ 0.001, one-tailed t-test. Error bars = s.e.m.
A loss of Trk expression precedes Runx1d-induced death
We examined whether Runx1d-induced apoptosis could involve a perturbation in neurotrophin-elicited neuronal survival by studying if Runx1d affects Trk expression just before apoptosis at stage HH24. TrkA expression in Runx1d-overexpressing neurons was completely abolished in nearly all neurons (Fig. 4d). There was no detectable difference in the number of cells coexpressing EGFP and TrkC between control and Runx1d-overexpressing embryos. However, quantitative measurements of expression levels between Runx1d-overexpressing cells and untransfected adjacent cells showed a small but significant downregulation of TrkC in Runx1d-overexpressing neurons (P = 0.035; Fig. 4e). Taken together, these results indicate that Runx transcription factors are necessary for TrkA expression and when blocked, sensory neuron death is preceded by a loss of TrkA expression.
Runx1b overexpression induces loss of NCC multipotency
It has been previously shown that migratory NCCs express the high-mobility group transcription factor Sox10. Moreover, despite its late role in Schwann cell formation29, in vitro gain-of-function studies have shown that Sox10 contributes to the maintenance of NCSC multipotency3. When migratory NCCs arrive at the location of the future DRG and commit to a neuronal fate, Sox10 expression is lost. We examined expression of Sox10 in migratory NCCs over-expressing Runx1b 24 h after transfection. Runx1b led to a nearly fourfold decrease of Sox10-expressing migratory NCCs (Fig. 5). These data indicate that forced Runx1 expression suppresses the multipotency of NCCs.
Figure 5.
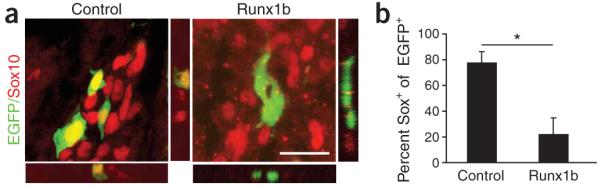
Runx1b-expressing cells lose Sox10 expression. (a) Images showing control and Runx1b overexpression in chick migratory NCCs at HH19 stained for the HMG transcription factor Sox10. (b) Quantitative data from measurements of colocalization of EGFP and Sox10 showing the loss of Sox10 expression in cells expressing Runx1b. Scale bar (a), 20 μm. *P ≤ 0.05, one-tailed t-test. Error bars = s.e.m.
Runx1 is sufficient for TrkA expression
We tested whether forced Runx1b expression is sufficient to induce TrkA expression in the migratory NCCs. Very few cells express TrkA in control animals at this stage (Fig. 6a). Runx1b overexpression resulted in precocious TrkA expression in transfected NCCs and ectopic expression in the neural tube. Runx1b did not induce expression of TrkB (data not shown) or TrkC, present in mechanoreceptive and proprioceptive neurons, respectively. Consistent with the result that Runx1 is not neurogenic in bNCSCs, Runx1b did not lead to precocious expression of neuronal differentiation markers such as Isl1 and Tuj1. Thus, Runx1 specifically induces TrkA expression, but is not sufficient to initiate a pro-neural program.
Figure 6.
Runx1 selectively controls TrkA expression. (a) Micrographs show stage-HH19 control and Runx1b-overexpressing neural tube and migratory NCCs stained for TrkA and the sensory neuronal marker Isl1, as well as for TrkC and the early neuronal marker Tuj1 (βIII-tubulin). Note that Runx1b induced premature and ectopic expression of TrkA but not TrkC. Confocal Z-stacks showed that TrkA is turned on in the Runx1b-overexpressing (green) cells. (b) RuntB siRNA efficiently silences β-galactosidase translated from a LacZ construct tagged in the 3′ untranslated region with the siRNA-targeted RuntB sequence compared to control without siRNA. Micrographs show β-galactosidase (blue) and EGFP (green) expression in control and siRNA-treated chick embryos at stage HH24. (c) Percent of β-galactosidase-expressing PC12 cells 24 h after transfection with siRNA1, siRNA2 and a combination of both together with the LacZ construct tagged with the RuntB siRNA-targeted sequence. Note that siRNA induced a significant knockdown of β-galactosidase both in vitro and in vivo. (d) Expression of endogenous RuntB mRNA detected by In situ hybridization in control and siRNA-treated chick DRG at stage HH29. Note that siRNA significantly reduced endogenous RuntB mRNA expression at this stage (n = 4 animals/construct). (e,f) Reducing endogenous RuntB expression with siRNAs selectively downregulates TrkA but not TrkC expression in chick DRG at stage HH29 as shown in micrographs (e) and the corresponding quantification (f). Scale bars, 100 μm (b,d) and 20 μm (a,e). Two-tailed t-test **P ≤ 0.01. Error bars represent s.e.m.
The Ntrk1 promoter has previously been partly characterized30,31. These studies show that the regulatory elements for TrkA expression reside upstream of the transcriptional start site. A 457-bp minimal enhancer DNA fragment upstream of Ntrk1 is sufficient to drive expression in neural crest-derived sensory and sympathetic neurons at early embryonic stages in transgenic animals31. The ectopic expression of TrkA induced by Runx1b in vivo suggested that Runx1 acts on the Ntrk1 promoter. A putative Runx binding site was found to be conserved in the Ntrk1 minimal enhancer of several species (TG(T/C)GGT-3′; Supplementary Fig. 5 online). PC12 cells were transfected with two luciferase fusion constructs, one including the full Ntrk1 promoter (1.5 kb) and the other including the Ntrk1 minimal enhancer, using the POU transcription factor Brn3a as positive control30. Runx1b activated both the full Ntrk1 promoter and minimal enhancer, similarly to Brn3a. Although we have not shown a direct binding of Runx1 to the Ntrk1 promoter, our results are consistent with a direct binding and conclusively show that Runx1 can activate TrkA expression in vivo as well as the Ntrk1 promoter in vitro.
Runx1 is specifically required for nociceptive neurons
The incompatibility in development of all classes of sensory neurons by Runx1d overexpression could result from its ability to compete with and block the activities of several Runx proteins. To examine if Runx1 is specifically required for the survival of the TrkA+ subpopulation of neurons, we knocked down RuntB expression in chick embryos using small interfering RNAs (siRNAs). Two different siRNAs against RuntB attenuated expression in both in vitro and in vivo assays. In these assays, β-galactosidase translated from a reporter construct in which LacZ was fused in the 3′-untranslated region to the siRNA targeted RuntB cDNA sequence (Fig. 6b,c). The siRNA also efficiently reduced endogenous RuntB mRNA expression at HH29 (Fig. 6d). Reducing RuntB levels led to a selective loss of TrkA-expressing neurons at HH29 (Fig. 6e,f). Consistent with the fact that the TrkC subpopulation of sensory neurons does not express RuntB, neither TrkC expression levels (data not shown) nor cell number was affected by RuntB siRNA (Fig. 6f). This shows that Runx1 is necessary for development of TrkA+ nociceptive sensory neurons.
DISCUSSION
We found that Runx protein activity is necessary for development of all subclasses of sensory neurons and that Runx1 selectively directs establishment of the TrkA+ nociceptive subclass by promoting cell survival, TrkA expression and neurite outgrowth. Diversification of the sensory neuron lineage has been known for decades to be closely linked to the expression of specific Trk receptors in functional subclasses of sensory neurons. Recent data from mice in which Ntrk3 replaces Ntrk1 in a Bax-null background to prevent normal cell death has indicated that Trk receptor signaling has a direct role in subclass specification14.
Runx factors control lineage commitment and genetic interruption of these pathways prevents normal development. Runx1 plays an important role in hematopoietic development and Runx1-/- mice completely lack fetal liver hematopoiesis and die around E11.5. The failure is caused by a loss of competence to generate myeloid and erythroid progenitors in this lineage27,28. Runx2-/- mice show arrested osteoblast differentiation and thus are totally devoid of bone due to the requirement of Runx2 activity for turning mesenchymal cells into osteoblasts32,33. The Runx2 transcriptional programs are involved in osteoblast-specific gene expression and induction of osteocalcin expression32-34. On the basis of their expression patterns, potential roles of Runx proteins in the nervous system have been suggested. Recently, Runx1 was shown to be important for development of cholinergic branchiovisceral motor neurons and olfactory and cortical neural precursors35,36. Runx3 is also important for development of sensory neurons16,17. Since the Runt domain of all Runx proteins binds the same DNA consensus sequence and the affinity of the short Runx1d protein is markedly higher than that of full-length isoforms, Runx1d is able to compete with and block the activities of all Runx proteins. The finding that blocking Runx activities did not affect migration of NCCs but induced a loss of neurons in all subclasses at the time of programmed cell death shows that Runx activities are required in the sensory and neuronally committed cells in the condensed DRG. The role of Runx1 for survival and development of nociceptors involved expression of TrkA, without which the neurons died during the period of programmed cell death due to deprivation of survival signaling37. However, Runx1 also affected survival in a cell-autonomous mechanism independent of TrkA, as seen in bNCSC cultures. This potential dual role of Runx1 combined with its effects on axonal growth makes it a candidate for a molecular link between the early neurons that survive cell autonomously and their acquisition of trophic factor dependency by TrkA expression and signaling when their axons arrive in the target field7.
Selective role of Runx1 and Runx3 for sensory subclasses
Previous reports on the activity of Runx proteins in the nervous system have suggested that they support different sensory subpopulations, but these studies did not investigate a direct role in specification of sensory neurons. Runx3 is required for sensory progenitors to generate TrkC proprioceptive neurons in mice, and in its absence, TrkC expression is initiated but fails to be maintained16,17. These two reports on Runx3-/- mice are somewhat conflicting. In one study17, no differences were observed in the number of TrkC+ or ER81+ DRG neurons, but these neurons failed to project to their central and peripheral targets37. In the other16, a decrease in the number of proprioceptive neurons (Parvalbumin+, ER81+ and TrkC+ neurons) was found and attributed to cell death resulting from a failure of these neurons to reach their targets. In Runx1-/- mice, a decrease in total number of neurons was observed in the trigeminal ganglia36. Our results showed a requirement of Runx protein activity for development of all classes of sensory neurons. Moreover, since members of the runt family are expressed in a non-redundant manner in DRG neurons (for example, Runx3 in TrkC+ neurons18 and Runx1 in TrkA+ neurons; Fig. 1 and Supplementary Fig. 1), we assume that blocking Runt activities with Runx1d reflects a specific block of Runx3 and Runx1 activities independently in the different sensory subclasses of neurons. Our Runx1 siRNA experiment, which selectively affected the TrkA population of sensory neurons, confirms this conclusion. Thus we conclude that Runx1 is required for specification of nociceptive TrkA-expressing neurons and Runx3 for development of proprioceptive TrkC-expressing neurons.
Transcriptional control of TrkA expression in sensory neurons
Brn3a, a POU domain transcription factor, is essential for sensory neuron development. Brn3a is expressed in cells of the developing trigeminal, geniculate, DRG and vestibulocochlear ganglia38,39. It is present in proliferating, neuronally committed sensory progenitors as well as in postmitotic neurons of the DRG40. Mice carrying a null mutation in Brn3a die shortly after birth and exhibit a loss of the majority of the trigeminal ganglion neurons41,42. In Brn3a-/- mice, there is no initiation of TrkC expression, and TrkA and TrkB expression is progressively lost in the trigeminal ganglion39,41,42. Brn3a therefore does not seem to direct sensory neurons to any particular functional subclass. Promoter studies show a direct interaction of Brn3a with the Ntrk1 promoter30. In the trigeminal ganglion, neuronal loss almost certainly results from the essential role of Brn3a in the initiation of TrkC expression and the maintenance of TrkA/TrkB receptor expression.
We found an increasing number of TrkA+ neurons between HH24 and HH29 in control animals, consistent with the birth of TrkA+ neurons (cell cycle exit and TrkA expression) until E6 (ref. 43). However, initiation of TrkA expression, which is unperturbed in the Brn3a-/- mice39,41, failed in the absence of Runx. This was evident from the nearly complete absence of TrkA in Runx1d-transfected cells at HH24. This finding agrees with the coincident onset of TrkA expression with Runx1. Furthermore, Runx1 was also by itself sufficient to induce TrkA expression both in vitro and in vivo. It is interesting to note that both Runx3 and Runx1 5′-upstream regions contain Brn3a binding sites18, and in Brn3a-/- mice, Runx1 mRNA levels are markedly decreased in trigeminal ganglia44, suggesting a reinforcement of Runx1 expression by Brn3a. Although it is not known whether Brn3a is sufficient to initiate TrkA expression, it is likely that Brn3a and Runx1 collaborate to maintain TrkA expression in vivo.
Genetic control of sensory neuron development
We present a schematic illustration of genetic interactions between Runx1 and other transcriptional regulators during DRG sensory neuron development (Supplementary Fig. 6 online). Expression of the high mobility group gene Sox10 is associated with the neural crest already at the time of delamination from the neural tube. Sox10 appears necessary for retaining NCC multipotency, as Sox10 overexpression maintains the neurogenic and gliogenic potentials of NCSCs, prevents cell cycle exit and inhibits differentiation3. We found that Runx1 expression is incompatible with preservation of Sox10 expression in the migratory NCCs. Presumably, Runx1 expression is therefore incompatible with multipotency. The Sox10+ premigratory and migratory NCCs contribute cells to both the sympathetic and sensory lineages. Some of them also express Ngn2 and are biased to adopt a sensory fate but remain competent to generate both neurons and glia45. Concurrent with the convergence of migratory NCCs into the DRG at E9.5 in the cervical enlargement of mice, Sox10 is lost in progenitor cells where Brn3a expression has commenced (data not shown). The onset of Brn3a expression is associated with a commitment to sensory and neuronal fates in proliferating progenitors40, which we found was followed 12-24 h later by a subclass-restricted expression of Runx1 in nociceptive committed neuronal progenitors of the DRG. Our results suggest that the activity of Runx1 for sensory neuron development depends on Ngn2 transcriptional activity. The nature of the instructive subtype determinant signals associated with the onset of Runx1 and Runx3 expression in subclasses remains unknown. Fibroblast growth factor (FGF) and retinoic acid are possible candidates. Basic FGF (bFGF) has been shown to induce Runx1 mRNA expression in the olfactory neuroblastoma cell line JFEN46 and notably, this induction was associated with an induction of TrkA. Furthermore, retinoic acid rapidly increases expression of all Runx factors in various cell lines as well as in vivo during bone development47.
METHODS
This work has been approved by the Swedish research animal committee in Stockholm (approval 404/03).
In situ hybridization
Embryos were fixed in 4% paraformaldehyde/PBS and sectioned at 14-μm thickness. Plasmids containing specific probes for mouse Ngn1, Ngn2, Runx1 and chick Ngn1, Ngn2 and RuntB were used to synthesize digoxigenin-labeled antisense riboprobes according to the supplier’s instructions (Roche). In situ hybridization was conducted as previously described21.
Cell culture
bNCSC sphere cultures were prepared by mechano-enzymatic dissociation as previously described21. Briefly, DRG including the boundary cap from E11 mice were dissected and dissociated in collagenase/dispase (1 mg/ml, Roche) and DNAse (0.55 mg/ml, Sigma) at room temperature (22-24 °C) for 30 min. Cells were plated in propagation medium consisting of N2 medium, B27 supplement (Gibco), bFGF and epidermal growth factor (20 ng/ml, RnD Systems).
in vitro electroporation
bNCSC cultures were collected and washed in PBS. cDNAs for Runx1, Runx1d, Ngn2, EGFP and RFP were cloned in an expression vector driven by the CMV enhancer chick β-actin promoter, and various combinations of these cDNAs were used for electroporation. We added 5 μg of each construct to 100 μl of PBS containing 50 to 100 bNCSC neurospheres. A square wave pulse generator (BTX) was used to deliver two consecutive pulses of 200 V for 15 ms in a 4-mm cuvette. After electroporation, cells were added to N2 medium containing BDNF, NGF, NT-3 and GDNF (10 ng/ml, Promega), 2% B27 supplement (Gibco) and retinoic acid (100 nM, Sigma) and seeded on culture plates coated with poly-d-lysine (50 μg/ml) and laminin (20 μg/ml, Invitrogen).
Cell counting and Sholl analysis
Living EGFP expressing cells with neuronal morphology and neurites exceeding twice the soma diameter were defined as neurons, then counted and analyzed by the Sholl method23 for neurite length and branching.
In ovo electroporation
cDNAs encoding Runx1b, Runx1d and EGFP were subcloned into an expression vector driven by the chick actin promoter. Constructs were injected into the neural tube of stage HH13 chick embryos. Electroporation by five pulses of 40 V/cm was performed using a square wave electroporator (BTX). After 24, 72 or 96 h, embryos were fixed in 4% PFA/PBS at 4 °C overnight and sectioned at 14-μm thickness. For the siRNA experiment, two siRNAs (1.5 μg/μl each) were similarly electroporated in combination with the EGFP expression vector. The siRNA sequences were as follows: siRNA1, 5′-AAAAACCAAGUGGCGAGGUUCAACGAC-3′ and siRNA2, 5′AGAACUA CUCUGCAGAGCUCAGAAAUG-3′, corresponding to nucleotides 411-439 and 371-398 in the chick RuntB coding sequence, respectively. As a control experiment, we transfected a construct in which β-galactosidase was fused to nucleotides 360-998 of the chick RuntB coding sequence into PC12 cells or electroporated them in the neural tube with and without siRNAs.
Immunohistochemistry
Sections were washed in PBS, then blocked for 1 h in PBS, 0.2% BSA, 0.1% Triton X-100 and 4% donkey serum. Rabbit antibodies specific to TrkA48, TrkB49 and TrkC50, as well as mouse antibodies to Tuj1 (Promega) and NeuN (Chemicon), were used at 1:500. Mouse antibody to Isl1 39.4D (Developmental Studies Hybridoma Bank, diluted 1:50) and guinea pig antibody to Sox10 (diluted 1:1,000). The secondary antibodies were Cy3, Cy2 and Cy5 (Jackson Immunoresearch). Staining was documented by confocal microscopy (Zeiss LSM 510). Optical sections were 2 μm in 25× overview pictures and 1 μm in 40× stacks and close ups.
For active Caspase-3, an antibody specific to cleaved caspase-3 (Cell Signaling) was used as described above. Positive cells were counted from confocal microscopy or fluorescence scanner micrographs (TECAN). Variation in Casp-3+ cells between embryos was high because of small differences in age since programmed cell death just starts around the analyzed stage43. Casp+ cells were therefore first normalized within each embryo by comparing with the contralateral DRG.
Quantitative measurements of colocalization were performed on confocal microscopy stacks. The total number of EGFP+ cells was counted on stack projections. At least three animals were used per condition.
Sections with Runx1d-transfected cells in the DRG and immunostained for TrkC were documented by scanning 40× optical stacks with a confocal microscope and analyzed using ImageJ. The soma of Runx1d-transfected cells were encircled and the mean fluorescence intensity was recorded. For each Runx1d-expressing cell, the closest untransfected cell was chosen as the control.
ACKNOWLEDGMENTS
We thank C. Tello for technical assistance and A. Käller for secretarial assistance. This work was supported by the Swedish Medical Research Council, the Swedish Foundation for Strategic Research (grant from the Center of Excellence in Developmental Biology), the Hedlunds Foundation, AstraZeneca Mölndal and the US Public Health Service (grant NS 16033).
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Carr PA, Nagy JI. Emerging relationships between cytochemical properties and sensory modality transmission in primary sensory neurons. Brain Res. Bull. 1993;30:209–219. doi: 10.1016/0361-9230(93)90246-8. [DOI] [PubMed] [Google Scholar]
- 2.Cheung M, et al. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 4.Serbedzija GN, Fraser SE, Bronner-Fraser M. Pathways of trunk neural crest cell migration in the mouse embryo as revealed by vital dye labelling. Development. 1990;108:605–612. doi: 10.1242/dev.108.4.605. [DOI] [PubMed] [Google Scholar]
- 5.Kasemeier-Kulesa JC, Kulesa PM, Lefcort F. Imaging neural crest cell dynamics during formation of dorsal root ganglia and sympathetic ganglia. Development. 2005;132:235–245. doi: 10.1242/dev.01553. [DOI] [PubMed] [Google Scholar]
- 6.Lawson SN, Biscoe TJ. Development of mouse dorsal root ganglia: an autoradiographic and quantitative study. J. Neurocytol. 1979;8:265–274. doi: 10.1007/BF01236122. [DOI] [PubMed] [Google Scholar]
- 7.Vogel KS, Davies AM. The duration of neurotrophic factor independence in early sensory neurons is matched to the time course of target field innervation. Neuron. 1991;7:819–830. doi: 10.1016/0896-6273(91)90284-7. [DOI] [PubMed] [Google Scholar]
- 8.Ernfors P, Merlio JP, Persson H. Cells Expressing mRNA for neurotrophins and their receptors during embryonic rat development. Eur. J. Neurosci. 1992;4:1140–1158. doi: 10.1111/j.1460-9568.1992.tb00141.x. [DOI] [PubMed] [Google Scholar]
- 9.Crowley C, et al. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- 10.Smeyne RJ, et al. Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature. 1994;368:246–249. doi: 10.1038/368246a0. [DOI] [PubMed] [Google Scholar]
- 11.Ernfors P, Lee KF, Kucera J, Jaenisch R. Lack of neurotrophin-3 leads to deficiencies in the peripheral nervous system and loss of limb proprioceptive afferents. Cell. 1994;77:503–512. doi: 10.1016/0092-8674(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 12.Patel TD, Jackman A, Rice FL, Kucera J, Snider WD. Development of sensory neurons in the absence of NGF/TrkA signaling in vivo. Neuron. 2000;25:345–357. doi: 10.1016/s0896-6273(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 13.Patel TD, et al. Peripheral NT3 signaling is required for ETS protein expression and central patterning of proprioceptive sensory afferents. Neuron. 2003;38:403–416. doi: 10.1016/s0896-6273(03)00261-7. [DOI] [PubMed] [Google Scholar]
- 14.Moqrich A, et al. Expressing TrkC from the TrkA locus causes a subset of dorsal root ganglia neurons to switch fate. Nat. Neurosci. 2004;7:812–818. doi: 10.1038/nn1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coffman JA. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol. Int. 2003;27:315–324. doi: 10.1016/s1065-6995(03)00018-0. [DOI] [PubMed] [Google Scholar]
- 16.Levanon D, et al. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue K, et al. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat. Neurosci. 2002;5:946–954. doi: 10.1038/nn925. [DOI] [PubMed] [Google Scholar]
- 18.Levanon D, et al. Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech. Dev. 2001;109:413–417. doi: 10.1016/s0925-4773(01)00537-8. [DOI] [PubMed] [Google Scholar]
- 19.Perez SE, Rebelo S, Anderson DJ. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development. 1999;126:1715–1728. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- 20.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J. Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- 21.Hjerling-Leffler J, et al. The boundary cap: a source of neural crest stem cells that generate multiple sensory neuron subtypes. Development. 2005;132:2623–2632. doi: 10.1242/dev.01852. [DOI] [PubMed] [Google Scholar]
- 22.Falk A, et al. Gene delivery to adult neural stem cells. Exp. Cell Res. 2002;279:34–39. doi: 10.1006/excr.2002.5569. [DOI] [PubMed] [Google Scholar]
- 23.Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- 24.Bae SC, et al. PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol. Cell. Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyers S, Lenny N, Hiebert SW. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol. Cell. Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka T, et al. An acute myeloid leukemia gene, AML1, regulates hemopoietic myeloid cell differentiation and transcriptional activation antagonistically by two alternative spliced forms. EMBO J. 1995b;14:341–350. doi: 10.1002/j.1460-2075.1995.tb07008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 29.Britsch S, et al. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma L, Lei L, Eng SR, Turner E, Parada LF. Brn3a regulation of TrkA/NGF receptor expression in developing sensory neurons. Development. 2003;130:3525–3534. doi: 10.1242/dev.00582. [DOI] [PubMed] [Google Scholar]
- 31.Ma L, Merenmies J, Parada LF. Molecular characterization of the TrkA/NGF receptor minimal enhancer reveals regulation by multiple cis elements to drive embryonic neuron expression. Development. 2000;127:3777–3788. doi: 10.1242/dev.127.17.3777. [DOI] [PubMed] [Google Scholar]
- 32.Komori T, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 33.Otto F, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 34.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 35.Theriault FM, et al. Role for Runx1 in the proliferation and neuronal differentiation of selected progenitor cells in the mammalian nervous system. J. Neurosci. 2005;25:2050–2061. doi: 10.1523/JNEUROSCI.5108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theriault FM, Roy P, Stifani S. AML1/Runx1 is important for the development of hindbrain cholinergic branchiovisceral motor neurons and selected cranial sensory neurons. Proc. Natl. Acad. Sci. USA. 2004;101:10343–10348. doi: 10.1073/pnas.0400768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levi-Montalcini R. The nerve growth factor 35 years later. Science. 1987;237:1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 38.Gerrero MR, et al. Brn-3.0: a POU-domain protein expressed in the sensory, immune, and endocrine systems that functions on elements distinct from known octamer motifs. Proc. Natl. Acad. Sci. USA. 1993;90:10841–10845. doi: 10.1073/pnas.90.22.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang EJ, et al. POU domain factor Brn-3a controls the differentiation and survival of trigeminal neurons by regulating Trk receptor expression. Development. 1999;126:2869–2882. doi: 10.1242/dev.126.13.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fedtsova NG, Turner EE. Brn-3.0 expression identifies early post-mitotic CNS neurons and sensory neural precursors. Mech. Dev. 1995;53:291–304. doi: 10.1016/0925-4773(95)00435-1. [DOI] [PubMed] [Google Scholar]
- 41.McEvilly RJ, et al. Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature. 1996;384:574–577. doi: 10.1038/384574a0. [DOI] [PubMed] [Google Scholar]
- 42.Xiang M, Gan L, Zhou L, Klein WH, Nathans J. Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc. Natl. Acad. Sci. USA. 1996;93:11950–11955. doi: 10.1073/pnas.93.21.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rifkin JT, Todd VJ, Anderson LW, Lefcort F. Dynamic expression of neurotrophin receptors during sensory neuron genesis and differentiation. Dev. Biol. 2000;227:465–480. doi: 10.1006/dbio.2000.9841. [DOI] [PubMed] [Google Scholar]
- 44.Eng SR, Lanier J, Fedtsova N, Turner EE. Coordinated regulation of gene expression by Brn3a in developing sensory ganglia. Development. 2004;131:3859–3870. doi: 10.1242/dev.01260. [DOI] [PubMed] [Google Scholar]
- 45.Zirlinger M, Lo L, McMahon J, McMahon AP, Anderson DJ. Transient expression of the bHLH factor neurogenin-2 marks a subpopulation of neural crest cells biased for a sensory but not a neuronal fate. Proc. Natl. Acad. Sci. USA. 2002;99:8084–8089. doi: 10.1073/pnas.122231199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nibu K, Li G, Kaga K, Rothstein JL. bFGF induces differentiation and death of olfactory neuroblastoma cells. Biochem. Biophys. Res. Commun. 2000;279:172–180. doi: 10.1006/bbrc.2000.3899. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka K, et al. Increased expression of AML1 during retinoic-acid-induced differentiation of U937 cells. Biochem. Biophys. Res. Commun. 1995a;211:1023–1030. doi: 10.1006/bbrc.1995.1913. [DOI] [PubMed] [Google Scholar]
- 48.Oakley RA, et al. Neurotrophin-3 promotes the differentiation of muscle spindle afferents in the absence of peripheral targets. J. Neurosci. 1997;17:4262–4274. doi: 10.1523/JNEUROSCI.17-11-04262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Bartheld CS, et al. Retrograde transport of neurotrophins from the eye to the brain in chick embryos: roles of the p75NTR and trkB receptors. J. Neurosci. 1996;16:2995–3008. doi: 10.1523/JNEUROSCI.16-09-02995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefcort F, Clary DO, Rusoff AC, Reichardt LF. Inhibition of the NT-3 receptor TrkC, early in chick embryogenesis, results in severe reductions in multiple neuronal subpopulations in the dorsal root ganglia. J. Neurosci. 1996;16:3704–3713. doi: 10.1523/JNEUROSCI.16-11-03704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]