Abstract
Although the catalytic subunit of the Schizosaccharomyces pombe telomerase holoenzyme was identified over ten years ago, the unusual heterogeneity of its telomeric DNA made it difficult to identify its RNA component. We used a new two-step immunoprecipitation and reverse transcription–PCR technique to identify the S. pombe telomerase RNA, which we call TER1. TER1 RNA was 1,213 nucleotides long, similar in size to the Saccharomyces cerevisiae telomerase RNA, TLC1. TER1 RNA associated in vivo with the two known subunits of the S. pombe telomerase holoenzyme, Est1p and Trt1p, and neither association was dependent on the other holoenzyme component. We present a model to explain how telomerase introduces heterogeneity into S. pombe telomeres. The technique used here to identify TER1 should be generally applicable to other model organisms.
The principal role performed by the telomerase ribonucleoprotein particle is the addition of telomeric repeats to the ends of eukaryotic chromosomes. The action of this specialized reverse transcriptase is necessary due to the inability of conventional DNA polymerase to replicate fully the ends of linear DNA molecules. The templating RNA and the catalytic subunit that comprise the core of the telomerase holoenzyme have been identified in a number of evolutionarily diverse organisms, including Tetrahymena thermophila, Saccharomyces cerevisiae and humans1,2.
In addition to providing the solution to the ‘end replication problem,’ telomeric DNA is coated by proteins that function in other aspects of telomere biology. These processes include transcriptional gene silencing, telomere length regulation and chromosome end protection (reviewed in ref. 3). Many of these features of telomere biology are similar between higher eukaryotes and the fission yeast Schizosaccharomyces pombe. For example, similar to human TRF1 and TRF2, S. pombe has two telomere binding proteins that contain Myb domains, Taz1p and Tbf1p, both of which regulate telomere length4,5. Notably, the discovery of the S. pombe telomere G-strand end-binding protein Pot1p and the S. pombe catalytic telomerase subunit Trt1p made it possible to identify their human orthologs by computational means6,7. Also, the Ku70 subunit of the telomere-binding Ku heterodimer was found by the reciprocal approach in which the S. pombe genome was queried with sequence from its human ortholog8.
Despite extensive studies of telomerase and telomere function in S. pombe, its telomerase RNA has not been identified. Unlike the catalytic subunit of telomerase and other telomere binding proteins, telomerase RNAs cannot be identified easily by computational approaches because these molecules are highly divergent among species, varying greatly in both size and sequence2. Even between closely related species, evolutionary covariation of base pairs maintains structure without primary sequence9–12. The difficulties to identify the S. pombe telomerase RNA gene are exacerbated by the heterogeneity of its telomeric DNA because only limited sequence information is provided by its derived consensus sequence13.
Although the sequences of previously identified telomerase RNAs are not often useful to identify the molecule in other species, there are evolutionarily conserved attributes of these RNAs that can be exploited. We used two of these features to enrich for and then identify the S. pombe telomerase RNA. The tight interaction between the catalytic subunit Trt1p and the templating RNA was used in an initial immunoprecipitation step, which was followed by a second immunoprecipitation using an antibody directed against the 2,2,7-trimethyl guanosine cap structure, which is conserved in S. cerevisiae14 and human15 telomerase RNAs. A reverse transcription–PCR (RT-PCR) reaction conducted with the enriched RNA and oligonucleotides specific for the putative templating sequence of the telomerase RNA identified a single previously uncharacterized intergenic region. Phenotypic analysis demonstrated that this locus encodes the sole S. pombe telomerase RNA, hereafter referred to as ter1+. Characterization of TER1 revealed both similarities and differences compared to the S. cerevisiae telomerase RNA, TLC1. TER1 RNA was large, as is TLC1 RNA and mutational analysis confirmed the presence of a template proximal helix that functioned as a template boundary element. Also, as in S. cerevisiae, TER1 RNA interacted in vivo with the telomerase subunit Est1p16,17, and this association was Trt1p independent18. However, the Pku80p subunit of the Ku heterodimer did not stably associate with TER1 RNA in vivo, whereas in S. cerevisiae this interaction recruits the catalytic subunit Est2p to the telomere in G1 phase19. Based on the sequence of the template region of TER1 RNA, we present a model to explain how heterogeneous sequence could be introduced into S. pombe telomeres.
RESULTS
Identification of a candidate S. pombe telomerase RNA locus
To identify the S. pombe telomerase RNA, we used two successive immunoprecipitations. The first immunoprecipitation (anti-Myc) was directed against a Myc epitope–tagged version of the telomerase catalytic subunit, Trt1p, and the second was directed against the 2,2,7-trimethylguanosine (TMG) cap structure found on a small subset of eukaryotic RNAs (Fig. 1a). The RNA isolated after the anti-TMG immunoprecipitation was used as template in directed RT-PCR (Supplementary Fig. 1 online).
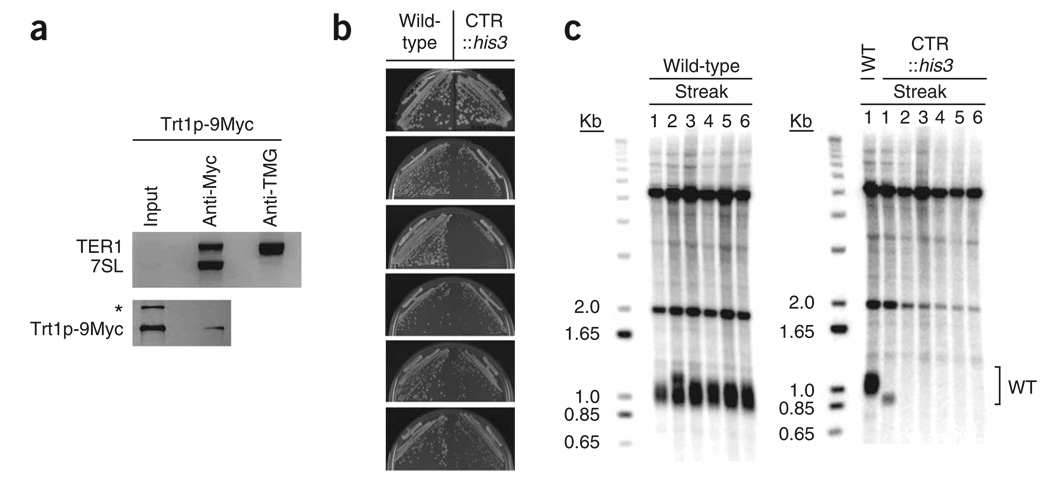
Figure 1.
Loss of viability and telomeric DNA in cells lacking the candidate gene for telomerase RNA. (a) Two successive immunoprecipitations greatly enrich candidate telomerase RNA (CTR). Top, multiplex RT-PCR analysis of aliquots from input (0.3% of cleared extract), anti-Myc immunoprecipitation (3% of immunoprecipitate) and anti-TMG immunoprecipitation (64% of immunoprecipitate) with oligonucleotides that amplify TER1 and 7SL RNA recovered from a Trt1p-9Myc overexpression strain. Eighty percent of precipitate recovered from the anti-Myc immunoprecipitation was used in the anti-TMG immunoprecipitation. Input and anti-Myc immunoprecipitation aliquots were amplified with 30 cycles and the anti-TMG immunoprecipitation aliquots were amplified with 40 cycles. Bottom, western blot analysis of input (1% of cleared extract) and anti-Myc immunoprecipitation (4% of immunoprecipitate) from a Trt1p-9Myc overexpression strain. *, cross-reacting nonspecific band. (b) Six successive restreaks of the parental WT haploid and an isogenic strain in which the CTR gene was replaced with his3+ shown from top to bottom. Streaks were grown on YES media at 30 °C for 3–4 days. (c) Telomere length analysis of successive streaks of parental WT strain (left) and CTR disrupted strain (right) after successive restreaks. Bracket, position of WT telomere signal. The bands at 2.0 and 5.0 Kb probably result from cross-reaction with the telomeric probes21.
The first immunoprecipitation was carried out in a trt1 deletion strain in which plasmid-borne Trt1p-9Myc was overexpressed, a method originally developed20,21 to generate an in vitro enzymatic assay for S. pombe telomerase. We reasoned that the anti-Myc immunoprecipitate must contain the telomerase RNA because it is capable of adding telomeric DNA to oligonucleotides. Furthermore, as the only Trt1p in this strain is epitope-tagged and overexpressed, the assay conditions are likely to capture most of the telomerase RNA in the cell, which should increase the ratio of telomerase RNA to other contaminating RNAs in the immunoprecipitate.
Only a small subset of the total population of RNA in the eukaryotic cell possesses a 5′ TMG cap, including spliceosomal small nuclear (sn) RNAs and small nucleolar RNAs, but not ribosomal RNAs22. More recently, the 5′ ends of S. cerevisiae14 and human15 telomerase RNAs were shown to be hypermethylated (although ciliate telomerase RNA is not23,24). As the evolutionary split between S. pombe and S. cerevisiae from metazoa occurred ~1.5 billion years ago and the progenitors of present day S. pombe diverged from S. cerevisiae ~1.1 billion years ago25, it seemed likely to us that the telomerase RNA in S. pombe would also harbor a TMG cap. We exploited the rare 5′ cap structure to further enrich for the telomerase RNA, using an anti-TMG antibody for a second immunoprecipitation from the deproteinized and DNase I treated anti-Myc immunoprecipitate. The 7SL RNA component of the signal recognition particle was used to demonstrate the large relative enrichment of TER1 obtained by employing the second anti-TMG immunoprecipitation step (Fig. 1a).
To amplify and clone the telomerase RNA from the anti-TMG immunoprecipitate, we modified a random PCR (rPCR) protocol26 that nonspecifically amplifies very low amounts of RNA (10−8 µg). We assumed that the templating RNA contains the reverse complement (UAACCG) of the minimal common sequence added to exogenous oligonucleotide primers in S. pombe in vitro telomerase assays20,27. Thus, to generate specificity for the S. pombe telomerase RNA, we added these six nucleotides to the 3′ end of one of the oligonucleotides (Supplementary Fig. 1a). In case the templating sequence of S. pombe telomerase RNA was immediately downstream of its 5′ end, as it is in mouse telomerase RNA12, the cDNA was also generated nonspecifically, with specificity achieved during the polymerization of the second strand (Supplementary Fig. 1b).
The two modified rPCR techniques yielded one previously unidentified locus (4/33 clones) (Supplementary Table 1 online). The four subcloned sequences encompassed a 521–base pair (bp) sequence on cosmid SPAC16A10 at coordinates 8748…8229. Regardless of how the first cDNA strand was primed, the main contaminant was ribosomal RNA (27/33 clones). The presence of rRNA clones was not due solely to the vast quantities of rRNA in the cell, as 11 of the rRNA isolates contained the specifying UAACCG sequence.
Disruption of the candidate locus causes senescence
To determine if the region identified is the S. pombe telomerase RNA, it was deleted by a one-step gene disruption that replaced 521 bp surrounding the putative templating region with his3+. Deletion of either trt1+6 or est1+21, the previously identified components of the S. pombe telomerase holoenzyme, causes an est (ever shorter telomere) phenotype, characterized by progressive loss of telomeric DNA accompanied by decreased cellular growth. Whereas a wild-type strain showed normal colony formation through six successive restreaks, the isogenic strain in which the candidate locus was deleted failed to produce robust colonies by the third restreak (Fig. 1b). The wild-type strain maintained telomere length during the six restreaks, whereas the null showed rapid loss of telomere signal (Fig. 1c). Two other independently derived deletions of the candidate locus lost telomeric signal by the third and fourth restreak (data not shown). In none of the individual strains did the subpopulation of survivors produce a telomeric signal, suggesting that, as in a trt1 deletion strain28, circularization of chromosomes is the dominant survival mode after deletion of this locus. Thus, the identified locus is required for the stable maintenance of telomeric DNA and is named ter1+ (telomerase RNA).
The ter1 locus templates the addition of telomeric DNA
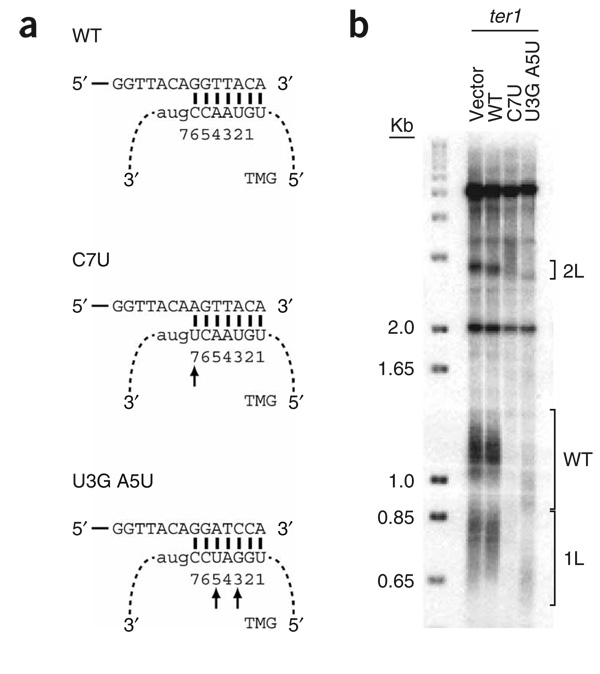
To demonstrate that the candidate locus provides the templating information for the addition of telomeric DNA, the putative templating sequence was altered. We generated two mutant versions of TER1, a single base substitution (C7U) and a double substitution (U3G A5U). The double substitution was designed to introduce BamHI sites into telomeric DNA (Fig. 2a). Wild-type TER1 and the two mutant RNAs were overexpressed from a plasmid in a ter1+ strain that had unique DNA sequence inserted adjacent to the telomeres of chromosomes 1L and 2L (ref. 29). The presence of this unique DNA facilitates cloning of telomeres (Methods). By ~30 cell divisions, the strain overexpressing the C7U allele had virtually no detectable telomeric DNA, whereas telomeres were short but visible at the comparable time in the U3G A5U strain (Fig. 2b). Unexpectedly, the C7U allele conferred a more severe phenotype, as it would be predicted to have a lesser effect on the binding of the single-stranded S. pombe telomere binding protein, Pot1p, than the U3G A5U double substitution30. The telomeres of the strain expressing the U3G A5U allele were shorter than those in the control strains, but could be cloned and sequenced31. Representative 1L telomere sequences cloned from cells overexpressing wild-type TER1 were of uniform length (234 ± 30 bp) and did not contain any BamHI sites (Supplementary Fig. 2a online). The absence of BamHI sites in wild-type telomeric repeat DNA also extends to the total collection of ~54 kb of published native S. pombe telomere sequences32. In contrast, the cloned telomeres from the U3G A5U double substitution mutant were uniformly shorter (146 ± 54 bp), and individual telomeres contained up to seven BamHI sites (Supplementary Fig. 2b).
Figure 2.
Expression of a ter1 allele with a mutated template promotes the incorporation of altered repeats into native telomeres. (a) Schematic of wild-type (WT) template, single substitution C7U and double substitution U3G A5U templating mutants’ interaction with telomere sequence and extension. Solid lines denote telomeric DNA and dashed lines indicate TER1 RNA. Numbers beneath capitalized nucleotides indicate positions of templating residues in TER1 RNA. Arrows indicate mutated nucleotides. (b) Telomere length analysis of vector alone or vector carrying either WT or template mutant alleles C7U or U3G A5U introduced into a ter1+ strain with his3+ and ura4+ inserted adjacent to the 1L and 2L telomeres, respectively29. Positions of the 1L, 2L, and WT length telomeres are indicated by brackets.
TER1 is a large RNA
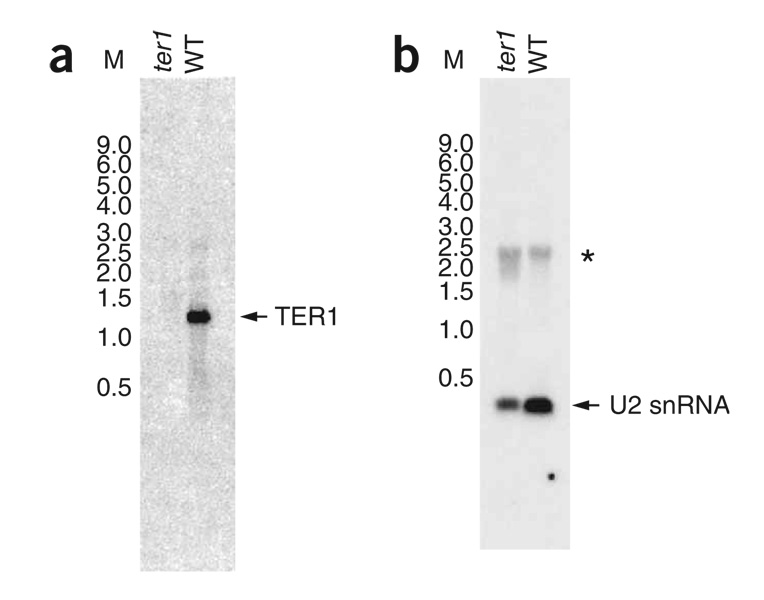
To determine the size of the ter1+ gene product, we performed a northern blot on total RNA from a ter1+ and a ter1 deletion strain. Based on this analysis, ter1+ encodes a 1,200- to 1,300-nucleotide (nt) RNA that is absent from the ter1 deletion strain (Fig. 3a). Reprobing the blot for the U2 snRNA demonstrated equivalent loading of total RNA and accurately sized RNA standards (Fig. 3b). To identify the 5′ end of the RNA, run-off cDNA specific for TER1 RNA was generated, tailed with dCTP and then cloned using a poly(G) oligonucleotide and a specific internal ter1 oligonucleotide. The identity of the 3′ end was established by application of a protocol devised to identify the 3′ end of the human telomerase RNA33. The most distal 5′ and 3′ ends determined in this way were deemed to be the ends of the TER1 RNA and indicate that the size of TER1 is 1,213 nt (Supplementary Fig. 3 online).
Figure 3.
TER1 RNA is ~1,200 nt. (a) Total RNA from a ter1 haploid deletion strain and an isogenic wild-type (WT) strain were probed with TER1 sequence. TER1 RNA, arrow. (b) Same blot as in a, stripped and reprobed with U2 snRNA sequence. U2 snRNA is indicated by an arrow. *, nonspecific hybridization signal that comigrates with ribosomal RNA (data not shown). Molecular weight RNA Millennium Size Markers-Formamide (Ambion) (M) is in kilobases.
TER1 secondary structure prediction compared to TLC1
A combination of phylogenetic comparisons, mutational analysis and biochemical probing has demonstrated the robustness of a secondary structure model for the 1,158-nt TLC1 RNA that contains three long quasihelical arms9,10. Each of these predicted arms is the binding site for either Est1p17, the Sm ring14 or the Ku heterodimer19,34, while the central region contains the templating sequence and interacts with the catalytic subunit, Est2p12,35,36. The three arms of TLC1 are hypothesized to function as a flexible scaffold that facilitates the interaction of the RNA with its protein partners9.
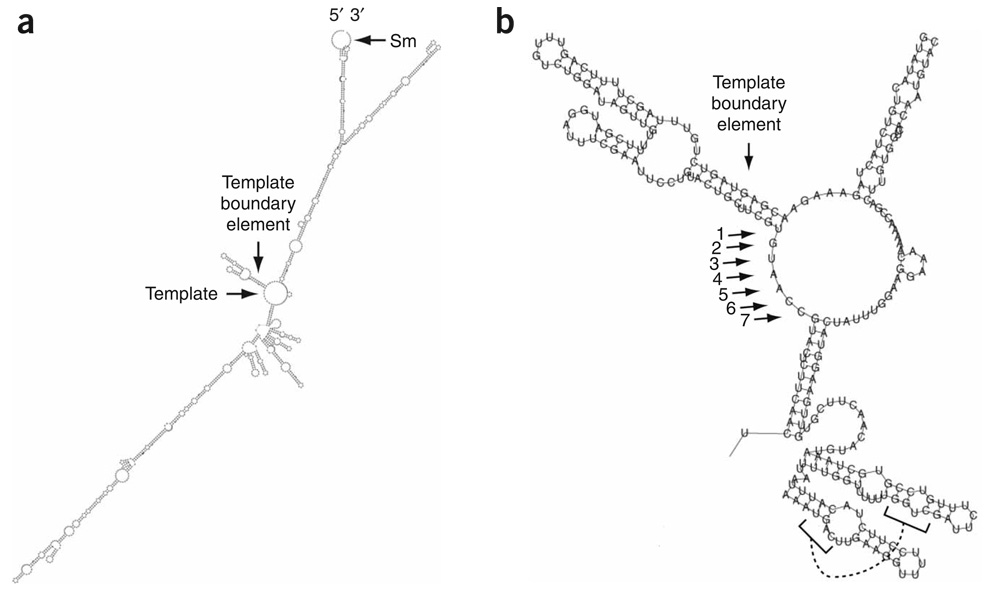
The Vienna RNA Secondary Structure Package37 was used to predict a secondary structure for the 1,213-nt TER1 RNA (Fig. 4a). The predicted TER1 RNA secondary structure had two distinct long arms that extend from the central core, not three as in TLC1. However, similarities between the TER1 and TLC1 predicted structures, as well as for other telomerase RNAs1, were readily apparent and increase our confidence that the TER1 predicted secondary structure is biologically relevant. First, as in TLC1, the templating region of TER1 was located at the center of the RNA. Second, the template region was predicted to be single-stranded as seen for all telomerase RNAs whose structures have been analyzed1,12. Finally, as in ciliates, mammals and yeasts, the templating sequence was immediately adjacent to a double-stranded region called the template boundary element1,12.
Figure 4.
Predicted secondary structure for TER1 RNA. (a) Output from the Vienna RNA Secondary Structure Prediction RNAfold program. The 5′ and 3′ ends of the RNA and template sequence, as well as the predicted template boundary element and Sm site, are indicated. (b) Enlarged view of templating region and predicted template boundary element. Arrows mark template nucleotide positions. A putative pseudoknot is designated by connected brackets.
The TER1 template boundary was predicted to be a 10-bp helix with two internal unpaired nucleotides (Fig. 4b), whereas the S. cerevisiae template boundary helix is 7 bp38. The predicted 5′ placement of the boundary element was supported by the consensus sequence for S. pombe telomeric DNA. The core consensus, GGTTACA, was generated from an alignment of the sequences of cloned S. pombe telomeres. Whereas the first six nucleotides are found in 132 of 136 sequenced telomeric repeats, the terminal adenosine is observed only 108 times13. The slightly lower representation of the last nucleotide is consistent with the position of the templating uracil at the start of the putative templating boundary element. Moreover, at a much lower frequency (17/136), a cytosine follows the seven-nucleotide consensus sequence13, which was consistent with the even more inaccessible position of the cognate guanosine two base pairs into the helix of the putative template boundary element (Fig. 4b). These data suggest that the base of the putative template boundary element breathes in vivo, allowing it to serve as a template in a subset of polymerization reactions. The impaired ability of the predicted S. pombe template boundary element to prevent DNA polymerization into the boundary element provides an explanation for heterogeneity at the 3′ end of the S. pombe telomeric repeat consensus sequence. In contrast, in S. cerevisiae TLC1, the template boundary element is separated from the template by a nucleotide; furthermore, in a wild-type strain background, polymerization templated by sequences in the template boundary element is not detected38.
The Kinefold pseudoknot prediction program39 indicated the potential for pseudoknot formation between nt 792–796 and 837–833. In both S. cerevisiae and S. pombe this region is separated from the template by a long quasihelical region, which is the Est1p binding arm in TLC1. Also similar to TLC1 (ref. 10), this region can be folded into an alternate two-hairpin structure (Fig. 4b; reviewed in ref. 2). Finally, a match to the U4 snRNA Sm site40 is located at the 3′ end of the TER1 RNA. In the TER1 secondary structure prediction this sequence is not base paired (Fig. 4a), as is also the case for the Sm site in the mature form of the TLC1 predicted structure9,10. Experimental analysis is necessary to validate these structures.
Mutational analysis provides evidence for a template boundary element
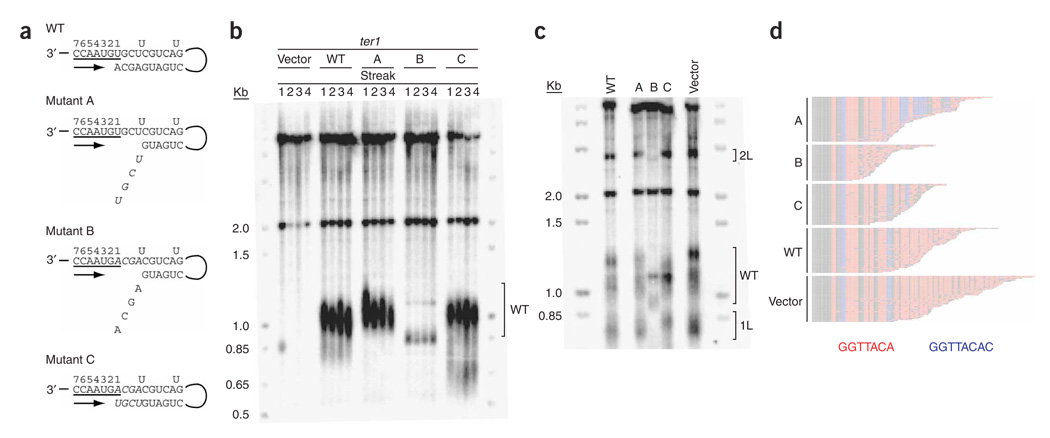
As has been done previously in Kluyveromyces lactis and S. cerevisiae38,41, we used a mutational approach to test the proposed template boundary element in the predicted secondary structure (Fig. 4). If the predicted template-proximal helix functions as a boundary element, it should limit the replication of telomeric repeats to GGTTACAc, where the penultimate adenosine is found at a slightly lower rate than the previous nucleotides and the terminal cytosine is much more rarely incorporated. Expression of a mutant that weakens the helix without changing the template is expected to increase the incorporation of the rare terminal cytosine into telomeric DNA. A set of three mutations in the first 4 bp of the helix was made to weaken (mutations A and B) and then restore (mutation C) the putative helix by compensatory base pairing (Fig. 5a). In mutation A, the four nucleotides at the 5′ end of the helix in the nontemplating strand were each changed to their complementary nucleotide. In mutation B, the four nucleotides at the 3′ end of the helix in the templating strand were changed to their complement. Mutations A and B were combined in mutation C thus restoring the putative helix.
Figure 5.
Disruption of the TER1 template-proximal helix causes telomeric shortening and increased incorporation of the infrequently added terminal cytosine, which can be rescued by a compensatory helix restoration mutant. (a) Schematic of mutations designed to test the predicted template-proximal helix. Nucleotides that encode the core S. pombe telomeric repeat sequence GGTTACA are underlined and nucleotides that are predicted to form a helix, except for two internal unpaired uracils (Fig. 4b), are also shown. Arrow, direction of DNA polymerization. Mutant A changed the first four nucleotides of the helix on the nontemplating strand to the complementary nucleotides (italics). Mutant B replaced the terminal four nucleotides of the helix on the templating strand to the complementary nucleotides (italics). Mutant C combined mutations A and B (italics). (b) Telomere length analysis of four successive restreaks of a ter1 deletion strain that expressed vector alone or vector carrying wild-type (WT) Ter1, Mutant A, Mutant B or Mutant C. Bracket, WT telomere length. (c) Vector individually carrying WT Ter1 or Mutant A, Mutant B, Mutant C or the vector alone were introduced into a ter1+ strain with his3+ and ura4+ inserted adjacent to 1L and 2L telomeres, respectively29. Brackets, positions of 1L, 2L and WT length telomeres. (d) 1L telomeric sequences from ter1+ cells overexpressing WT ter1+ or the A, B or C mutations from a plasmid. Analysis included 46 A telomeres, 37 B telomeres, 42 C telomeres, 46 WT telomeres and 47 vector telomeres. Core telomeric consensus sequence GGTTACA, red; extended GGTTACAC repeats, blue.
To determine their effects on telomere length, each mutant RNA was expressed from a plasmid in a ter1 deletion strain (Fig. 5b). Whereas mutation A did not cause telomere shortening even after four restreaks, mutation B caused an immediate and severe decrease in telomere length (~80 bases; Fig. 5b). The greater severity of mutation B compared to mutation A is probably due to a synergistic effect of weakening the helix and changing the sequence of the template. The shorter telomeres in mutant B may indicate reduced binding of telomere structural proteins, which results in enhanced degradation of telomeric DNA. Cells expressing mutation C, which restored the helix by compensatory base pairing (Fig. 5a), had wild-type length telomeres (Fig. 5b).
Next we tested mutations A, B and C for their effects on the sequence of telomeric DNA. For this analysis, each of the mutants, as well as wild-type ter1+ (or vector alone) was expressed from a plasmid in a ter1+ strain that has unique DNA inserted adjacent to the 1L and 2L telomeres to facilitate telomere cloning29. As in the ter1 deletion strain (Fig. 5b), expression of mutant B resulted in short telomeres, whereas mutants A and C had telomeres of about wild-type length (Fig. 5c). If the template adjacent helix is a boundary element, weakening this helix without changing the sequence of the template strand, as in mutation A, should result in increased incorporation of GGTTACAc repeats in cloned 1L telomeres. Reestablishing the helix by compensatory mutations, as in mutation C, should restore a wild-type level of this variant repeat. Indeed, in newly synthesized telomeric DNA (Fig. 5d), the percentages of GGTTACAc relative to GGTTACA were 23% (mutation A), 3.2% (mutation B), 4.3% (mutation C), 1.6% (wild type) and 2.6% (vector alone). The increased incorporation of the GGTTACAc repeat in mutant A telomeres and its restoration to near wild-type levels in mutant C strongly suggest that the proposed proximal helix is a template boundary element.
We also attempted phylogenetic comparisons with the telomerase RNAs from other fission yeast species to provide support for the proposed secondary structure, but the sequences obtained from three species were not suitable for such analysis. The ter1+ locus from Schizosaccharomyces malidevorans was identical in sequence to S. pombe. The Schizosaccharomyces kambucha ter1+ RNA was predicted to have an almost identical secondary structure to the S. pombe ter1+ RNA, but the two RNAs differed at only ten sites. In contrast, the putative ter1+ locus of the more distantly related Schizosaccharomyces japonicus was highly divergent (44% identical to S. pombe).
TER1 associates with telomerase holoenzyme components
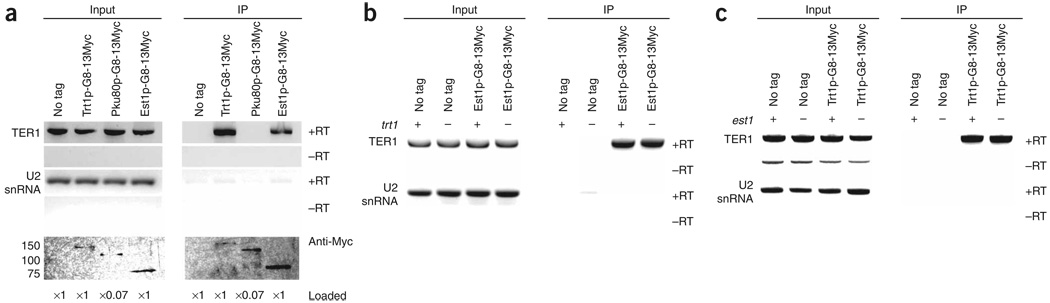
To identify proteins that interact with the TER1 RNA, we immunoprecipitated two components of the telomerase holoenzyme (Trt1p and Est1p) and the telomere binding protein Pku80p. Est1p was first identified in S. cerevisiae42, but is also present in S. pombe21 and humans43,44. In both S. cerevisiae and S. pombe, Est1p is essential for telomere maintenance by telomerase21,42, and in S. cerevisiae, this function requires a specific interaction between Est1p and TLC1 RNA17. To determine if this interaction is conserved in S. pombe, we used a strain expressing Est1-G8-13Myc from its endogenous locus under its own promoter. A whole cell extract from this strain was immunoprecipitated with antibody to Myc and the presence of TER1 RNA determined by RT-PCR (Fig. 6a). TER1 RNA was readily detected in the immunoprecipitate. This interaction was specific, as the abundant U2 snRNA was present in the input fraction but not in the anti-Myc immunoprecipitate. As in S. cerevisiae18,45, the Est1p-TER1 RNA interaction was not dependent on the catalytic subunit of telomerase, as this interaction was not reduced in a trt1 deletion strain (Fig. 6b). Likewise, TER1 RNA was readily detected in an anti-Myc immunoprecipitate from a strain expressing Trt1-G8-13Myc. Again, U2 snRNA was not detected in the immunoprecipitate. The interaction between telomerase RNA and the catalytic subunit was preserved in an est1 deletion strain (Fig. 6c).
Figure 6.
TER1 RNA is associated with the holoenzyme components Trt1p and Est1p in vivo, but not with Pku80p. (a) Coimmunoprecipitation of TER1 RNA with Trt1p-G8-13Myc, Pku80p-G8-13Myc or Est1p-G8-13Myc. RT-PCR signal for the TER1 RNA and U2 snRNA detected after extraction are indicated (+RT) in both the input and immunoprecipitated (IP) material. Mock RT-PCR reactions without reverse transcriptase are indicated (−RT). Signal in these lanes is due to genomic DNA contamination. An anti-Myc western blot of input and immunoprecipitated protein is shown. The relative amounts of protein extract loaded for the inputs and immunoprecipitations from each strain are indicated. Molecular masses (left) are in kDa. (b) The in vivo association of Est1p with the TER1 RNA is not dependent on Trt1p. Est1p-G8-13Myc immunoprecipitations were performed as in a except that the trt1 locus is disrupted. “+” and “−”, presence or absence of Trt1p. Strains are spore products from the same tetrad. (c) The in vivo association of Trt1p with the TER1 RNA is not dependent on Est1p. Trt1p-G8-13Myc immunoprecipitations were performed as in b except that the est1 locus is disrupted. “+” and “−”, presence or absence of Est1p. The minor band is from contaminating genomic DNA.
Ku is a conserved heterodimer of the Ku70 and Ku80 subunits that has a role in double stranded break repair by nonhomologous end joining in diverse organisms, including humans, S. cerevisiae and S. pombe. However, in organisms including S. pombe8, Ku also functions in telomere maintenance (reviewed in ref. 46), and lack of S. pombe Ku causes short but stable telomeres8. S. cerevisiae Ku interacts with a 48-bp stem-loop region of TLC1 in vitro34 and in vivo19, and this interaction is essential for Est2p telomere association in G1 phase and for wild-type levels of Est1p and Est2p telomere association in late S/G2 phase19. However, immunoprecipitates in a strain expressing Pku80p-G8-13Myc did not contain detectable TER1 RNA (Fig. 6a), suggesting that the Ku80p-telomerase RNA interaction is not conserved in S. pombe.
DISCUSSION
Telomerase RNAs have been isolated from ciliates, yeasts and mammals, but they have not been identified in several model organisms, such as S. pombe, Caenorhabditis elegans, and Arabidopsis thaliana, even though the genomes of these organisms are sequenced. Because both the primary sequence and the size of telomerase RNAs vary widely, it is difficult to use computational approaches to identify telomerase RNA genes. These difficulties are compounded in an organism like S. pombe by the heterogeneous sequence of its telomeric DNA, which made it hard to predict the sequence of its template. Biochemical approaches are also difficult because in many organisms telomerase is present in very low amounts. For example, there are only ~30 telomerase RNA molecules per baker’s yeast cell47, compared to ~25 × 103 to 140 × 103 rRNA molecules48.
We developed a novel method to isolate S. pombe telomerase RNA that could be useful in the identification of telomerase RNA from other model organisms. This approach involved two sequential immunoprecipitations, the first against the epitope-tagged and over-expressed Trt1p catalytic subunit and the second against the TMG cap that we predicted would be at the 5′ end of S. pombe telomerase RNA. We then modified an RT-PCR strategy designed to amplify low abundance RNAs26 and increased its specificity for telomerase RNA by adding the 6 bases predicted from the sequence of S. pombe telomeric DNA to one of the primers (Supplementary Fig. 1). This approach was highly successful, as 4 of 33 clones contained portions of ter1+. The S. pombe ter1+ locus was independently identified by another research group, as reported in an accompanying paper49.
Definitive proof that ter1+ encodes telomerase RNA came from the incorporation of the predicted altered sequence into telomeric DNA upon overexpression of the U3G A5U ter1 template mutant (Supplementary Fig. 2b). ter1+ is the only functional S. pombe telomerase RNA gene, as its deletion caused an est phenotype (Fig. 1b,c). As in S. cerevisiae, TER1 RNA interacts with both Trt1p and Est1p (Fig. 6a). Both interactions are likely direct, as the Trt1p interaction was Est1p-independent (Fig. 6c), and the Est1p interaction was Trt1p-independent (Fig. 6b). A direct association between S. pombe Est1p and telomerase RNA is somewhat unexpected as S. pombe Est1p, like human Est1p, lacks an obvious RNA binding domain21,43,44. Of course, the Est1p-TER1 interaction could be mediated by an as yet undiscovered subunit of the S. pombe telomerase holoenzyme.
Pku80p did not stably associate with TER1 (Fig. 6a), in contrast to what is seen with baker’s yeast and human telomerase RNAs. Although S. pombe Ku might interact directly with the catalytic subunit to mediate telomerase-telomere interaction, it is also possible that S. pombe Ku does not affect telomerase. Indeed, it is already clear that the S. pombe and S. cerevisiae heterodimeric Ku complexes have different telomere functions, as depletion of S. cerevisiae but not S. pombe Ku confers a telomere defect that results in temperature sensitivity8,50, and S. cerevisiae but not S. pombe Ku is critical for telomeric silencing51,52. Consistent with these findings, overexpression of TER1 did not reduce telomeric silencing (data not shown), whereas overexpression of TLC1 does, presumably because it titrates Ku from telomere ends53. Thus, there are likely to be both similarities and differences in the way telomerase is regulated in these two model organisms.
Telomerase RNAs vary widely in size, from the 149-nt Tetrahymena RNA23 to the 450-nt human RNA54 to the large RNAs in budding yeasts, such as S. cerevisiae9,10,55 (~1,200 nt) and K. lactis56 (~1,300 nt). Many aspects of S. pombe telomere biology are more similar to humans than to S. cerevisiae (reviewed in ref. 57). Thus, we were surprised at the large size of S. pombe telomerase RNA (Fig. 3, Supplementary Fig. 3).
Given that there is a single telomerase RNA gene, how does it generate telomeric DNA of nonuniform sequence? The expanded consensus sequence for S. pombe telomeric DNA is ggggggGGTTACAc, with lower case letters denoting rare insertions13. Polymerization using the templating guanosine that is the second base in the template boundary element (Fig. 4b, −1 position) suggests a model for the rare terminal cytosine insertion. The compensatory base pair analysis (Fig. 5) provides evidence that the predicted template-proximal helix forms in vivo and functions as a boundary element to reduce inappropriate templating of nucleotides. Unlike K. lactis or S. cerevisiae, the first nucleotide (+1 U) of the TER1 template and a second infrequently used nucleotide (−1 G) are within the template boundary element.
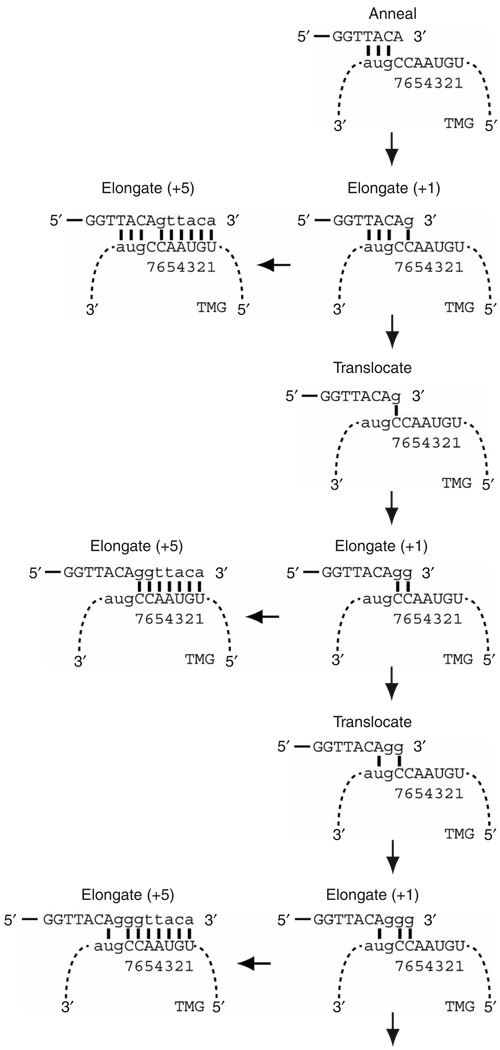
We propose that the leading variable length guanosine tracts are incorporated into telomeres by a +1 elongation/translocation mechanism in which an internal portion of the consensus GGTTACA telomeric repeat first anneals to TER1 (Fig. 7). After extending by a single guanosine, telomerase can either complete the addition of the repeat or translocate the template 5′ to 3′ on the telomere from the cytosine at position six to the cytosine at position seven. After another single-nucleotide extension, telomerase can either finish the repeat or translocate again. Successive rounds of single-nucleotide extension and translocation templated by the sequential guanosines could continue until there are constraints imposed by the unpaired added guanosines in the active site. In addition, or alternatively, if full elongation and translocation are equally likely events, then there is less than a 0.5% probability that six guanosines would be added between telomeric repeats. This low probability prediction is borne out by the observation that only 1 cloned telomeric repeat out of 136 has five and six preceding guanosines13. This explanation is plausible for each of the six positions in the guanosine tract that precedes the GGTTACA consensus (Supplementary Fig. 4 online).
Figure 7.
Model for interaction between the S. pombe telomeric DNA and TER1 RNA and the addition of telomeric sequence. The proposed model provides a mechanism for the introduction of variable length G-tracts before the consensus telomere repeat sequence13. Telomere, unbroken line; TER1 RNA, dashed line. Template nucleotide position is indicated beneath the RNA. After the initial annealing step between the telomere and the TER1 template, the catalytic subunit extends the telomere by a single guanosine residue. After the +1 elongation, telomerase can either copy the rest of the template or translocate the template along the telomere so that the added +1 guanosine templated by the cytosine at position six can now base pair with the cytosine at position seven. After translocation, the position six cytosine is now accessible to template the addition of a second guanosine. Successive rounds of +1 elongation and translocation can explain the introduction of multiple guanosines before the consensus sequence.
This model can also help explain the severe telomere loss phenotype of the C7U mutant (Fig. 2a,b). The change to uracil permits the terminal adenosine in the telomeric DNA repeat to anneal to the template. After the +1 addition of a guanosine there would be five consecutive base pairs between positions 6 and 10 in the template and the telomere, compared to four interrupted base pairs in the wild-type TER1 telomere interaction (Fig. 7). In addition to the higher binding energy, which may make a translocation event less likely, the base pair formed between the mutant uracil and the +1 added guanosine after translocation is a non-Watson-Crick wobble pair that is less stable58 than the wild-type interaction (Supplementary Fig. 5 online). These data suggest that overexpression of a mutant TER1 that prevents the addition of variable length guanosine tracts into the telomere has a severe impact on telomere replication or function.
METHODS
TER1 RNA isolation and cloning
The TER1 RNA was isolated by two successive immunoprecipitations followed by RT-PCR and subcloning. The first immunoprecipitation was performed using modified conditions21 from an in vitro S. pombe telomerase activity assay20. Immunoprecipitation was performed with 100 µl of prepared anti-c-Myc agarose conjugate (Sigma) and the equivalent of 100 ml of mid-log phase, strain CF830 S. pombe cells expressing plasmid-borne Trt1p-9Myc for 1 h at 4 °C in the presence of RNasin (Promega) and SUPERasein (Ambion). After washing, the beads were treated with proteinase K (Ambion) and extracted with phenol:chloroform:isoamyl alcohol, pH 6.6 (Ambion) and precipitated. The pellet was resuspended and treated with 100 units of amplification grade DNase I (Invitrogen). The RNA was precipitated again and resuspended in 500 µl of NET150 (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% v/v Igepal (Sigma)). The second immunoprecipitation was performed for one hour with 40 µl of prepared α-TMG mouse antibody agarose conjugate (Calbiochem) preblocked with 1 mg ml−1 BSA (Sigma), 100 µg ml−1 glycogen (Roche) at 4 °C. The beads were washed 5 times with NET150, resuspended in PK buffer (100 mM Tris, pH 7.5, 12.5 mM EDTA, 150 mM NaCl, 1% w/v SDS), extracted with acidic phenol:chloroform: isoamyl alcohol (Ambion) and precipitated.
The RNA precipitate from the successive immunoprecipitations was resuspended and divided for use in two separate RT-PCR strategies, which were used to clone and identify the ter1+ locus transcription product. Both strategies are modified versions of the rPCR technique26. The first strategy annealed an oligonucleotide with a universal sequence at the 5′ end followed by six random nucleotides that terminated in the reverse complement sequence of the inferred telomerase RNA templating sequence20,27. After reverse transcription (Applied Biosystems), the cDNA was annealed to a second oligonucleotide with a universal sequence at the 5′ end followed by six random nucleotides. The oligonucleotide was extended with Klenow (New England Biolabs). After purification (Qiagen), the double stranded cDNA was used in a PCR reaction (Takara) with oligonucleotides containing an EcoRI restriction site followed by the reverse complement of the universal sequence. The PCR product was subsequently subcloned and sequenced (Genewiz) (see Supplementary Fig. 1a for reaction outline). For the second RT-PCR strategy a nonspecific oligonucleotide was used to generate the cDNA. After purification, an oligonucleotide with specificity for the complement of the inferred templating region was used. The succeeding steps are the same as for the first RT-PCR strategy (Supplementary Fig. 1b).
RNA manipulation
The 5′ end of the TER1 RNA was identified by the addition of a poly(C) tract to a TER1 specific cDNA transcribed from a preparation of total cellular RNA by terminal deoxynucleotidyl transferase (Roche) followed by PCR amplification with a homopolymeric d(G18) oligonucleotide, subcloning and sequencing. The 3′ end of the TER1 RNA was determined by applying the method for determining the 3′ end of human telomerase RNA to a total RNA preparation from a strain harboring plasmidborne ter1+33. Northern blotting, stripping and reprobing were performed with random primed probes (GE Healthcare), which anneal between TER1 +134 and +376 nts and over the entire length of U2 snRNA, and Hybond-XL membrane as per the manufacturer’s instructions (Amersham Biosciences).
RNA secondary structure and pseudoknot prediction
The TER1 RNA secondary structure folding was predicted by the Vienna Secondary Structure Prediction RNAfold Web interface37. The default settings were used except that isolated base pairs were avoided (to allow single strand regions). A prediction using the same conditions for TLC1 revealed a secondary structure with three distinct arms (data not shown) that is similar to the reported secondary structure9. The Kinefold server39 was used with default settings to identify potential pseudoknot formation between nt 643 and 862 in the TER1 sequence. A prediction using the same settings for nt 724–813 of TLC1 accurately predicted 9 of 13 bp in the pseudoknot helix H4 (ref. 12) (data not shown).
Strain construction
The ter1+ locus was disrupted in h+ ade6-M216 his3-D1 leu1-32 ura4-D18 cells by PCR-mediated one-step gene replacement with his3+. Thirteen Myc epitopes were introduced at the est1+, trt1+ and pku80+ loci in a diploid strain (h+/− ade6-M210/ade6-M216 his3-D1/his3-D1 leu1-32/leu1-32 ura4-D18/ura4-D18) using the pFA6a-13Myc-kanMX6 tagging construct. Epitope tags were placed at the C terminus with an eight-glycine linker, which increases the functionality of epitope-tagged S. cerevisiae telomerase proteins59, and were introduced by addition of glycine codons to the internal downstream primers. Tagged est1+, trt1+ and pku80+ are expressed from their native promoters and are functional in recovered haploids, as determined by a nonsenescent phenotype and stable telomere maintenance at approximately −100 nt for tagged est1+ and wild-type telomere length for trt1+ and pku80+ (Supplementary Fig. 6 online). Open reading frames were sequenced and integrations were confirmed by Southern blot analysis. Gene disruptions of trt1 and est1 were made with his3+ in the est1-G8-13Myc::KanMX6 and trt1-G8-13Myc::KanMX6 diploid strains described above, respectively.
DNA manipulation
The ter1+/pSP1 plasmid was generated by PCR amplification of genomic DNA 275 bp upstream and 486 bp downstream of the ter1+ transcribed region. Template and template-proximal mutations were introduced by overlap extension PCR mutagenesis and sequenced (Genewiz). Genomic DNA extraction and telomere blots were performed as described60. Telomeric sequences from FY1862 (ref. 29) harboring ter1+, template and template-proximal mutation alleles were cloned by cytosine tailing with terminal deoxynucleotidyl transferase (Roche), PCR, cloning with a his3+ specific and poly(G18) oligonucleotides with restriction sites and sequence analysis (Genewiz)31.
Coimmunoprecipitation of TER1 RNA
Haploid strains expressing Est1p-G8-13Myc, Trt1p-G8-13Myc or Pku80p-G8-13Myc and an untagged isogenic control were grown in 100 ml of YES at 30 °C until mid-log phase (~1 × 107 cells ml−1). Cells were washed, resuspended in 1 ml of TMG100 (10 mM Tris-HCl, pH 8.0, 1 mMMgCl2, 10% v/v glycerol, 1 mM EDTA, 100 mM NaCl, 0.1 mM DTT) in the presence of Complete Mini EDTA-free protease inhibitors (Roche), 2 mM PMSF, RNasin (Promega) and SUPERasein (Ambion) and frozen in liquid nitrogen. The cell pellets were lysed using a Freezer Mill (SPEX CertiPrep) to > 95% lysis efficiency. After clearing the extract, protein concentration was determined (Coomassie plus, Pierce) and total protein normalized, typically to ~1 mg ml−1. We prepared and blocked 50 µl of anti-c-Myc agarose conjugate by washing 5 times in TMG100 with glycogen (100 µg ml−1), BSA (1 mg ml−1), herring sperm DNA (200 µg ml−1) and transfer RNA (100 µg ml−1) and added the agarose conjugate to the extract. The immunoprecipitation was incubated for 1 h at 4 °C in the presence of 0.5% v/v Tween-20. The beads were subsequently washed 5 times and treated with proteinase K. The immunoprecipitate was extracted with acidic phenol:chloroform:isoamyl alcohol and ethanol precipitated. Isolated RNA was treated with 20 units of amplification grade DNase I (Invitrogen) and ethanol precipitated.
To detect TER1 RNA in the input and immunoprecipitated RNA by RT-PCR, specific primers were designed to produce a 362 bp fragment. To ascertain the extent of nonspecific cellular RNA association, RT-PCR oligos designed to produce a 196 bp fragment from full length U2 snRNA were used. To demonstrate that the RT-PCR signal reflected RNA, mock RT-PCR reactions without reverse transcriptase were performed. cDNA was synthesized at 42 °C, and was followed by PCR conditions: 94 °C for 2 min, 50 °C for 40s, 72 °C for 30 s. 21 PCR cycles were carried out for the input and immunoprecipitated RNA. Input and immunoprecipitated samples for TER1 RT-PCR analysis were increased 60-fold relative to U2 snRNA. Reaction products were visualized by ethidium-stained 3% agarose gels (NuSieve). The same pattern of RT-PCR products was observed in experiments in which six-fold more starting product was used, thereby demonstrating the reproducibility of 21 PCR cycles.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
ACKNOWLEDGMENTS
We thank P. Baumann for sharing results before publication and J.B. Boulé, M. Mateyak, J. Phillips, S. Pinter, M. Sabourin, C. Tuzon and Y. Wu for critical reading of the manuscript. We thank O. Troyanskya and C. Huttenhower for computational assistance, R. Allshire (Wellcome Trust Centre for Cell Biology), P. Baumann (Stowers Institute for Medical Research), H. Lieberman (Columbia University) and M. Sipiczki (University of Debrecen) for strains, and K. Miller in J. Cooper’s lab, C. Tuzon and J. Bruzik for protocols. This work was supported by US National Institutes of Health grants GM43265 and R37 GM26938 and by a US National Research Service Award to C.J.W.
Footnotes
Published online at http://www.nature.com/nsmb/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Legassie JD, Jarstfer MB. The unmasking of telomerase. Structure. 2006;14:1603–1609. doi: 10.1016/j.str.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Vega LR, Mateyak MK, Zakian VA. Getting to the end: telomerase access in yeast and humans. Nat. Rev. Mol. Cell Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- 4.Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 5.Pitt CW, Valente LP, Rhodes D, Simonsson T. Identification and characterization of an essential telomeric repeat binding factor in Schizosaccharomyces pombe. J. Biol. Chem. 2007 doi: 10.1074/jbc.M708784200. published online 7 October 2007. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura TM, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 7.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 8.Baumann P, Cech TR. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell. 2000;11:3265–3275. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. USA. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dandjinou AT, et al. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 11.Chen JL, Greider CW. An emerging consensus for telomerase RNA structure. Proc. Natl. Acad. Sci. USA. 2004;101:14683–14684. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J, et al. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc. Natl. Acad. Sci. USA. 2004;101:14713–14718. doi: 10.1073/pnas.0405879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiraoka Y, Henderson E, Blackburn EH. Not so peculiar: fission yeast telomere repeats. Trends Biochem. Sci. 1998;23:126. doi: 10.1016/s0968-0004(98)01176-1. [DOI] [PubMed] [Google Scholar]
- 14.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 15.Jady BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin JJ, Zakian VA. An in vitro assay for Saccharomyces telomerase requires EST1. Cell. 1995;81:1127–1135. doi: 10.1016/s0092-8674(05)80017-0. [DOI] [PubMed] [Google Scholar]
- 17.Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J, Hidaka K, Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell. Biol. 2000;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher TS, Taggart AK, Zakian VA. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- 20.Haering CH, Nakamura TM, Baumann P, Cech TR. Analysis of telomerase catalytic subunit mutants in vivo and in vitro in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA. 2000;97:6367–6372. doi: 10.1073/pnas.130187397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beernink HT, Miller K, Deshpande A, Bucher P, Cooper JP. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr. Biol. 2003;13:575–580. doi: 10.1016/s0960-9822(03)00169-6. [DOI] [PubMed] [Google Scholar]
- 22.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 23.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 24.Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- 25.Hedges SB. The origin and evolution of model organisms. Nat. Rev. Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 26.Froussard P. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 1992;20:2900. doi: 10.1093/nar/20.11.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lue NF, Peng Y. Identification and characterization of a telomerase activity from Schizosaccharomyces pombe. Nucleic Acids Res. 1997;25:4331–4337. doi: 10.1093/nar/25.21.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura TM, Cooper JP, Cech TR. Two modes of survival of fission yeast without telomerase. Science. 1998;282:493–496. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- 29.Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- 30.Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11-389. J. Mol. Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 32.Sugawara NF. DNA Sequences at the Telomeres of the Fission Yeast S. pombe. Thesis. Harvard; 1989. [Google Scholar]
- 33.Zaug AJ, Linger J, Cech TR. Method for determining RNA 3′ ends and application to human telomerase RNA. Nucleic Acids Res. 1996;24:532–533. doi: 10.1093/nar/24.3.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livengood AJ, Zaug AJ, Cech TR. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chappell AS, Lundblad V. Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol. Cell. Biol. 2004;24:7720–7736. doi: 10.1128/MCB.24.17.7720-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seto AG, et al. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA. 2003;9:1323–1332. doi: 10.1261/rna.5570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xayaphoummine A, Bucher T, Isambert H. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Res. 2005;33:W605–W610. doi: 10.1093/nar/gki447. [online] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dandekar T, Tollervey D. Mutational analysis of Schizosaccharomyces pombe U4 snRNA by plasmid exchange. Yeast. 1992;8:647–653. doi: 10.1002/yea.320080808. [DOI] [PubMed] [Google Scholar]
- 41.Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- 42.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 43.Reichenbach P, et al. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 2003;13:568–574. doi: 10.1016/s0960-9822(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 44.Snow BE, et al. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 2003;13:698–704. doi: 10.1016/s0960-9822(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 45.Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- 46.Fisher TS, Zakian VA. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst.) 2005;4:1215–1226. doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 47.Mozdy AD, Cech TR. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rautio J, et al. Sandwich hybridisation assay for quantitative detection of yeast RNAs in crude cell lysates. Microb. Cell Fact. 2003:2–4. doi: 10.1186/1475-2859-2-4. [online] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leonardi J, Box JA, Bunch JT, Baumann P. Nat. Struct. Mol. Biol. 2007 doi: 10.1038/nsmb1343. advance online publication xxx. [DOI] [PubMed] [Google Scholar]
- 50.Feldmann H, Winnacker EL. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:12895–12900. [PubMed] [Google Scholar]
- 51.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manolis KG, et al. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 2001;20:210–221. doi: 10.1093/emboj/20.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson SE, et al. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 54.Feng J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 55.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 56.McEachern MJ, Blackburn EH. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 57.Kanoh J, Ishikawa F. Composition and conservation of the telomeric complex. Cell. Mol. Life Sci. 2003;60:2295–2302. doi: 10.1007/s00018-003-3245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugimoto N, Nakano M, Nakano S. Thermodynamics-structure relationship of single mismatches in RNA/DNA duplexes. Biochemistry. 2000;39:11270–11281. doi: 10.1021/bi000819p. [DOI] [PubMed] [Google Scholar]
- 59.Sabourin M, Tuzon CT, Fisher TS, Zakian VA. A flexible protein linker improves the function of epitope-tagged proteins in Saccharomyces cerevisiae. Yeast. 2007;24:39–45. doi: 10.1002/yea.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller KM, Cooper JP. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol. Cell. 2003;11:303–313. doi: 10.1016/s1097-2765(03)00041-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.