SUMMARY
The neuroligins are postsynaptic cell adhesion proteins whose associations with presynaptic neurexins participate in synaptogenesis. Mutations in the neuroligin and neurexin genes appear to be associated with autism and mental retardation. The crystal structure of a neuroligin reveals features not found in its catalytically active relatives, such as the fully hydrophobic interface forming the functional neuroligin dimer; the conformations of surface loops surrounding the vestigial active center; the location of determinants that are critical for folding and processing; and the absence of a macromolecular dipole and presence of an electronegative, hydrophilic surface for neurexin binding. The structure of a β-neurexin-neuroligin complex reveals the precise orientation of the bound neurexin and, despite a limited resolution, provides substantial information on the Ca2+-dependent interactions network involved in trans-synaptic neurexin-neuroligin association. These structures exemplify how an α/β-hydrolase fold varies in surface topography to confer adhesion properties and provide templates for analyzing abnormal processing or recognition events associated with autism.
INTRODUCTION
Synapse formation requires the assembly of highly ordered complexes containing ion channel, receptor, neurotransmitter storage, cell adhesion, and scaffolding proteins. Interactions between cell adhesion molecules contribute to the formation of complementary presynaptic and postsynaptic sites (Lardi-Studler and Fritschy, 2007). Various signaling events including neurotransmitter release, soluble factor secretion, and direct trans-synaptic interactions between cell adhesion molecules have roles in synapse formation and development (Waites et al., 2005). The neuroligins are a recently characterized family of postsynaptic cell adhesion proteins shown to be critical for synapse maturation and maintenance in vivo (Nguyen and Sudhof, 1997; Varoqueaux et al., 2006) and sufficient for triggering the initial formation of synaptic contacts in cultured neurons (Scheiffele et al., 2000). In vitro, the neuroligins bind in a Ca2+-dependent fashion to the lamininneurexin-sex hormone binding globulin (LNS) domains of α-neurexins (which encompass six LNS domains) and β-neurexins (one domain only), thus establishing an intricate, heterophilic trans-synaptic recognition code (Boucard et al., 2005; Comoletti et al., 2006; Graf et al., 2006; Nguyen and Sudhof, 1997). Since the neuroligins are expressed in a variety of tissues, they likely have recognition and adhesions functions extending to peripheral secretory tissues.
The five mammalian neuroligin genes (numbered NLGN1 to 4 and 4Y) encode membrane-anchored proteins with more than 70% sequence identity in their N-terminal, extracellular domains (Figure 1). This region is linked through a highly O-glycosylated region to a single transmembrane span and a C-terminal, small intracellular domain. The neuroligin extracellular domain exhibits 32%-36% sequence identity and presumably shares overall shape with the globular domain of acetylcholinesterase (AChE) (Ichtchenko et al., 1996) (Figure 1). This makes it a member of the α/β-hydrolase fold superfamily of proteins (Ollis et al., 1992; Cygler et al., 1993), which includes catalytically active cholinesterases, lipases, and carboxylesterases, along with the noncatalytic adhesion proteins neurotactin (de la Escalera et al., 1990), glutactin (Olson et al., 1990), and gliotactin (Auld et al., 1995) (see Figure S1 available online), and the thyroglobulin C-terminal domain (Schumacher et al., 1986).
Figure 1. Sequence Conservation within the Neuroligins and Comparison with Human Liver Carboxylesterase and mAChE.
The NL4 sequence displayed is that of the recombinant Gln44-Thr619 protein used for crystallization; the FLAG epitope is not shown. The positions for alternatively spliced inserts A and B are indicated by vertical arrowheads. The NL4 and mAChE numbering and their secondary structure elements are displayed above and below the alignment, respectively. The Cys-loop and loops L1 (α3 6,7-α4 6,7, Thr332-Ile338 in NL4), L2 (α1 7,8-α2 7,8, Asp381-Val390), L3 (β8-α1 8,9, Gln477-Ser487) and L4 (α1 8,9-α2 8,9, Gly503-Ser513) are indicated by gray bars under the alignment. The NL4 residues buried at the Nrxβ1 binding interface are shaded as gray boxes. The mAChE catalytic triad residues are indicated by asterisks; the two NL4 glycosylation sites by triangles; the residue pair involved in Ca2+ binding by filled circles; and the NL3/NL4 residues mutated in autism patients by filled squares. The LRE motif is indicated with a black bar above the sequences.
However, several characteristics of the neuroligin molecule extend beyond predictions based solely on AChE homology. For example, one finds a Gly to Ser substitution in the catalytic triad at the active center, sufficient for eliminating catalytic function; a distinct linkage for the third disulfide bond, perhaps defining a different modular organization of the molecule; divergent patterns of consensus sites for N-linked glycosylation, between neuroligins and their α/βhydrolase fold relatives as well as between the five neuroligins (Hoffman et al., 2004); and the possibility for insertion of two alternatively spliced sequences of 20 (insert A) and 9 (insert B) residues to generate several additional isoforms (Bolliger et al., 2001; Ichtchenko et al., 1996).
Mutations in the coding regions of the genes for neuroligin-3 and -4 (NL3 and NL4), including point mutations, truncations, and sequence deletions, were found to be associated with autism spectrum disorders (ASD) and mental retardation, indicating a strong genetic influence on developmental disorders (Blasi et al., 2006; Jamain et al., 2003; Laumonnier et al., 2004; Talebizadeh et al., 2006; Yan et al., 2005). These findings suggest that abnormal presentation of neuroligin at the postsynaptic surface, its altered association with neurexin, or both governs synaptic structural and cognitive disorders (Tabuchi et al., 2007).
Crystal structures of α- and β-neurexin LNS domains show a lectin, jelly-roll fold and reveal the positions for alternatively spliced surface loops on the hypervariable loop edge, located opposite the roll N and C termini (Rudenko et al., 1999; Sheckler et al., 2006). The presence or absence of these loops influences neurexin recognition for neuroligin. The expression of different neuroligin genes and presence of alternatively spliced insert B at the neuroligin surface also dictate affinity for neurexin (Boucard et al., 2005; Comoletti et al., 2006).
Neuroligin association with neurexin and its influence on cell adhesion only require extracellular sequences (Scheiffele et al., 2000). Hydrodynamic analyses established that the neuroligin extracellular domain, isolated through insertion of an early stop codon before the transmembrane span, forms homodimers in solution (Comoletti et al., 2003, 2006). Small-angle X-ray scattering (SAXS) combined with rigid-body modeling data led to experimentally based neuroligin models (Comoletti et al., 2007), using the crystal structure of mouse AChE (mAChE) (Bourne et al., 2003) and data on Cys connectivity and positions for oligosaccharide linkages (Hoffman et al., 2004) as constraints. These data are consistent with neuroligin dimerization through a four-helix bundle, as found for AChE (Bourne et al., 1995; Sussman et al., 1991), and an extended C-terminal linker emerging from the bundle.
A combination of X-ray and neutron scattering data recorded on neuroligin-1 (NL1) in complex with soluble β-neurexin-1 (Nrxβ1) determined that two Nrxβ1 subunits associate on cross-opposite sides of the long axis of the neuroligin dimer (Comoletti et al., 2007). Final positioning of the bound neurexins was achieved using rigid-body modeling and constraints derived from earlier binding and mutagenesis data. This resulted in an initial image of the complex wherein the Nrxβ1 hypervariable loop edge faces the neuroligin surface area proximal to splice site B. However, the low-resolution SAXS data only delineate the overall shape of the complex and the Nrxβ1 binding position, and do not reveal the precise orientation of the bound Nrxβ1 or the surface determinants involved.
A soluble form of NL4, which is naturally devoid of splice inserts and contains only two N-glycosylation sites (Bolliger et al., 2001)(Figure 1), proved to be amenable to obtaining well-diffracting crystals. Herein we present a 2.2Å resolution crystal structure of the extracellular domain of NL4 (Table 1), also a structure of a synaptic cell adhesion member of the α/β-hydrolase fold family of proteins. This structure permits a detailed analysis of those structural features that are unique to the neuroligins when compared with their catalytically active relatives, and that could not be predicted from homology-based models. The structure also defines positions for surface determinants known to be critical for folding and processing and for neurexin interaction. Moreover, a 3.9Å resolution structure of a complex between NL4 and a soluble Nrxβ1 form, also devoid of a splice insert, was also solved. This structure reveals the exact position and orientation of the bound neurexin relative to the neuroligin, along with a restricted, Ca2+-mediated, interfacial area consistent with a labile trans-synaptic complex. Hence, these structures, as compared with earlier models, provide an improved template for analysis of abnormal processing and/or recognition events that could be associated with ASD.
Table 1.
Data Collection and Refinement Statistics
| NL4: Low Resolution | NL4: High Resolution | Nrxβ1-NL4 Complex | |
|---|---|---|---|
| Data collection | |||
| Beamline at the ESRF | ID23 | ID14-EH1 | ID29 |
| Wavelength (Å) | 0.976 | 0.934 | 0.979 |
| Space group | C21 | P21212 | P21212 |
| Cell dimensions a, b, c (Å) α,β,γ (°) | 197.9,63.6,138.4 112.9 | 139.7,154.0,81.3 | 158.5,198.7,85.7 |
| Resolution range (Å)a | 15-3.2 (3.3-3.2) | 30-2.2 (2.25-2.20) | 30-3.9 (4.0-3.9) |
| Total observations | 79,446 | 587,014 | 121,593 |
| Unique reflections | 25,664 | 89,603 | 25,251 |
| Multiplicity | 3.2 (3.2) | 6.6 (6.6) | 4.8 (4.9) |
| Completeness (%) | 96.3 (98.8) | 99.9 (100) | 99.4 (99.9) |
| Rsym(%)b | 12.2 (41.5) | 6.5 (47.0) | 8.2 (55.9) |
| < I /σ(I) > | 8.9 (3.1) | 21.6 (4.5) | 16.1 (3.3) |
| Refinement | |||
| Resolution (Å) | 30-2.2 | 30-3.9 | |
| No. reflections | 85,129 | 25,214 | |
| Rwork/Rfree (%)c | 16.7/20.1 | 23.1/32.3 | |
| No. atoms | 9309 | 11,341 | |
| Protein | 8606 | 11,339 | |
| Ligand/sugar/ion | 38/56/17 | -/-/2 | |
| Water | 592 | - | |
| Average B-factors (Å2) | |||
| Protein | 38.8/41.5 | - | |
| Ligand/sugar/ion | 78.5/154.5/63.5 | - | |
| Water | 43.0 | - | |
| Rmsdd | |||
| Bond (Å) | 0.013 | 0.013 | |
| Angles (°) | 1.39 | 1.56 | |
| Chiral volume (A3) | 0.087 | 0.109 | |
| Ramachandran plot (%) | |||
| Most favored regions | 90.7 | 71.6 | |
| Additionally allowed regions | 9.3 | 26.8 | |
| PDB accession code | (not deposited) | 3BE8 | 2VH8 |
Values in parentheses are those for the last shell.
Rsym = Σhkl Σi|Ii(hkl) - < Ihkl > |Σhkl Σi Ii(hkl), where I is an individual reflection measurement and < I > is the mean intensity for symmetry-related reflections.
Rcryst = Σhkl||Fo| - |Fc||/Σhkl| |Fo|, where Fo and Fc are observed and calculated structure factors, respectively. Rfree is calculated for 5% of randomly selected reflections excluded from refinement.
Root mean square deviation from ideal values.
RESULTS AND DISCUSSION
Sequence-based searches for NL4 catalytically active relatives revealed highest identity with AChE and human liver carboxylesterase (Figure 1), while structure-based searches also pointed to bovine bile salt-stimulated lipase and bacterial p-nitrobenzyl esterase as close homologs. Hence mAChE, as a mammalian synaptic protein with the capacity for forming noncovalent dimers (Bourne et al., 1995), is an appropriate relative for comparative description of the NL4 structure.
The Neuroligin Subunit
The extracellular domain of NL4, which adopts an ellipsoidal shape with overall dimensions of 45 × 60 × 65Å, consists of a twisted, 12-stranded central β sheet surrounded by 14 α helices, typical of the α/β-hydrolase fold (Cygler et al., 1993; Ollis et al., 1992)(Figure 2). Of the three intra-molecular disulfide bridges conserved in the neuroligins, the first two, Cys110-Cys146 and Cys306-Cys317 (Cys69-Cys96 and Cys257-Cys272 in mAChE) (Figure 1), are conserved in most members of the fold family. The third bridge, Cys476-Cys510 (as earlier defined by peptide mapping and mass spectrometry analyses), is unique to the neuroligins (Hoffman et al., 2004) (Figure S1). This bridge constrains the included surface loop Gly503-Ser513 (or L4, or α1 8,9-α2 8,9; cf. Figure 1) into a conformation not observed in AChE that directly stabilizes the tip of the neighboring loop Gln 477-Ser487 (L3, β8-α1 8,9) through a stacking interaction of Trp484 with Arg437, and with Val377, to a more limited extent. In individuals with ASD, residues Arg437 and Lys378 are found to be replaced by Cys in NL3 and Arg in NL4, respectively (Jamain et al., 2003; Yan et al., 2005).
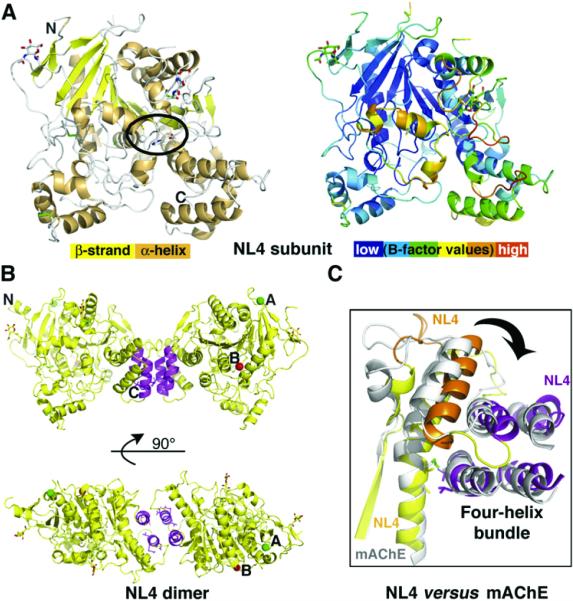
Figure 2. Overall View of the NL4 Structure.
(A) (Left) The NL4 subunit is displayed with yellow β strands, orange α helices, green disulfide bridges, and an encircled vestigial catalytic triad. The GlcNAc moieties are visible; the N and C termini are labeled. (Right) The NL4 subunit, oriented as in (A), is colored according to temperature factor (B-factor values). The flexibility of the Cys-loop (orange) is evident.
(B) Overall views of the NL4 dimer with the C termini pointing down (top) and with 90° rotation around the main axis (bottom). The four-helix bundle is displayed in magenta. The positions for splice sites A and B are indicated by green and red spheres, respectively.
(C) Close-up view of the superimposed four-helix bundle regions in the NL4 (yellow, orange, and magenta) and mAChE (gray) dimers, showing the twist of the helical region at the subunit interface.
In turn, the neuroligins lack the third bridge found in AChE (Cys409-Cys529 in mAChE), where it connects helix α4 7,8 to the C-terminal helix α10 (Figure 1). In fact, in NL4, absence of this bridge is compensated for by a set of strong hydrophobic interactions involving Ala451, Val454, Val587, and Leu591 (Cys409, Ala412, Cys529, and Asn533 in mAChE) in this region. Residue Cys259, which is free in all neuroligins, is deeply buried within helix α5,6, far from the protein surface. Ala substitution to this Cys in an NL1-soluble form resulted in improved intracellular processing and secretion yields (Comoletti et al., 2004).
GlcNAc moieties are linked to Asn102 and Asn511, which are conserved in all neuroligins. Residues Asp141 and Asn307, which correspond to potential Asn-linked glycosylation sites in neuroligin-2 (NL2) and NL1, respectively, are located at the NL4 surface, as are the positions for the spliced insert A (between Asp157 and Asp158) found in NL1, NL2, and NL3, and the spliced insert B (between Glu270 and Gly271) found in NL1.
The Neuroligin Dimer
Two NL4 subunits, related by a two-fold symmetry axis and linked through a tightly packed four-helix bundle made of helix α3 7,8 and the C-terminal helix α10 from each subunit (cf. Figure 1), assemble as a noncovalent antiparallel dimer (Figure 2). The same assembly was found for covalent and noncovalent AChE dimers (Bourne et al., 1995; Sussman et al., 1991). Hydrophobic interactions account for 100% of the ~875 Å2 interface area buried on each subunit, versus 77% and 870Å2 for mAChE, suggesting that the NL4 dimer is even more stable than the mAChE dimer. These values are consistent with analytical centrifugation data that showed that the neuroligin extracellular domain sediments as a dimer, even at low concentration (Comoletti et al., 2003), whereas diluted mAChE sediments as a monomer (Marchot et al., 1996). In vitro data on mutants within the four-helix bundle of NL1 (Lys578/Val579 and Glu584/Leu585 Ala mutations) (Dean et al., 2003) argue for the requirement of an oligomer for synaptogenic activity.
Compared with mAChE, in NL4 the displacement of the third disulfide bridge and its replacement with noncovalent, hydrophobic interactions, along with the distinct length and conformation of surface loop L2 (α1 7,8-α2 7,8, Asp381-Val390 in NL4) and shifted positions of loops α2 7,8-α3 7,8 (Tyr407-Gly412) and α3 7,8-α4 7,8 (Thr425-Pro433) in the four-helix bundle region (cf. Figure 1), result in a weaker connection between the core of the subunit and the C-terminal α3 7,8/α10 motif (Figure 2). In fact, this motif as a rigid body is shifted outward by ~14° from its position in mAChE; this motion is associated with a 15°-20° twist of one subunit relative to the second one around the dimer long axis.
A Leu-Arg-Glu (LRE) laminin binding motif conserved in all neuroligins is found in the two α3 7,8 helices in the four-helix bundle. While the Leu416 side chain establishes hydrophobic interactions within the bundle, the charged side chains of Arg417 and Glu418 point to the bulk solvent and are part of a highly polar, conserved network. LRE motifs are found at three distinctive positions in the extracellular domain of neurotactin (de la Escalera et al., 1990) (Figure S1). Whether these motifs in the neuroligins and neurotactin bind laminin or have another function is unknown.
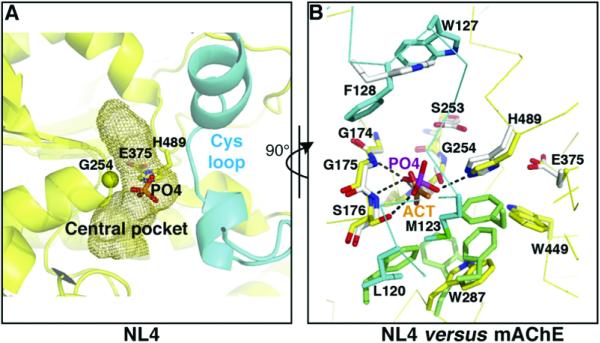
The Neuroligin Central Pocket
The central pocket, which in the cholinesterases contains the active center and oxyanion hole, lies centrosymmetric to the NL4 molecule (Figure 3 and Figure S2). The pocket in NL4, with its 200Å3 volume, is ~2-fold smaller than that in mAChE, mainly due to partial occupancy by the tip of the long Cys-loop (cf. below). However, and despite the Gly254 substitution to the active center Ser residue found in the catalytically active hydrolases (Ser203 in mAChE), the topology of the pocket and many of its lining residues are remarkably well conserved. Only small deviations are found upon superimposition of the vestigial triad residues Gly254, Glu375 (shared with AChE and only a few other relatives, instead of an Asp as found in most hydrolases), and His489; the oxyanion hole residues Gly175, Ser176 (Gly in mAChE), and Ala255 (Ile in NL3 but conserved in mAChE); Ser253 (Glu in mAChE); and Trp287 (conserved in mAChE) with their counterparts in mAChE. NL4 residue Ser253 replaces mAChE Glu202, whose mutation changes the catalytic constant (Gibney et al., 1990). A Gly254Ser mutation is not sufficient to confer AChE activity to NL1 (D.C., unpublished data).
Figure 3. The NL4 Central Pocket Compared with the mAChE Active Center.
(A) Close-up view of the tip of the Cys-loop (cyan) occluding access to the central pocket in NL4. The internal pocket, containing the phosphate, is displayed as a mesh surface. Residue Gly254 at the base of the nucleophilic elbow (β5-α5,6 turn) is shown as a yellow sphere.
(B) Close-up superimposition of the vestigial active center in NL4, rotated 90° from (A), and the active center in mAChE (rmsd, 0.3Å for 34 Cα atoms). The NL4 residues in the pocket are shown in yellow and those from the sealing Cys-loop in cyan. The mAChE residues in the pocket with Trp86 at the top are shown in white and those in the acyl pocket are shown in green. The NL4 Gly254 substitution to mAChE Ser203 is visible at the center. Bound phosphate (PO4) in NL4 mimics bound acetate (ACT) in mAChE. The phosphate oxygen atoms establish five hydrogen bonds with the main chain nitrogen atoms of Gly175, Ser176, and Ala255 (Gly121, Gly122, and Ala204 in mAChE) and the side chains of Ser176 and His489 (His447 in mAChE). The bond with Ala255 lies behind the figure plan and is not visible.
In the oxyanion hole, a tightly bound phosphate, positioned similarly to the carbonate/acetate found in mAChE structures (Bourne et al., 2003), forms five hydrogen bonds with NL4 residues (Figure 3). A chloride ion is also observed, 7Å from the phosphate. Hence the NL4 central pocket, while not serving a catalytic role, may confer additional flexibility in the NL4 molecule with its capacity for solvent and ion binding. Whether this feature proves advantageous to the neuroligin adhesion function remains to be documented.
Structural Variations within the α/β-Hydrolase Fold
NL4 and mAChE diverge primarily in the conformation and size of five surface loops located on the same face of the molecule: the Cys-loop (Cys110-Cys146); and loops L1 (α3 6,7-α4 6,7, Thr332-Ile338 in NL4), L2 (α1 7,8-α2 7,8, Asp381-Val390), L3 (β8-α1 8,9, Gln477-Ser487), and L4 (α1 8,9-α2 8,9, Gly503-Ser513) (cf. Figure 1). The Cys-loop (Figure 4, cyan) is homologous to the long Ω-loop (Cys69-Cys96) that in AChE forms one side of the rim of the active site gorge and appears to restrict substrate accessibility (Bourne et al., 1995; Sussman et al., 1991). In NL4 this loop, longer by nine residues and of a distinct sequence, folds as two short α helices almost perpendicular to each other and is followed by a short helical turn, in place of the single short α helix in mAChE. The tip of the loop curves oppositely to that in mAChE and bends, with a displacement of up ot 17Å inward, toward the center of the subunit, thereby occluding access to the central pocket. Several stabilizing polar and nonpolar interactions are found between residues of the loop edge and residues surrounding the central pocket.
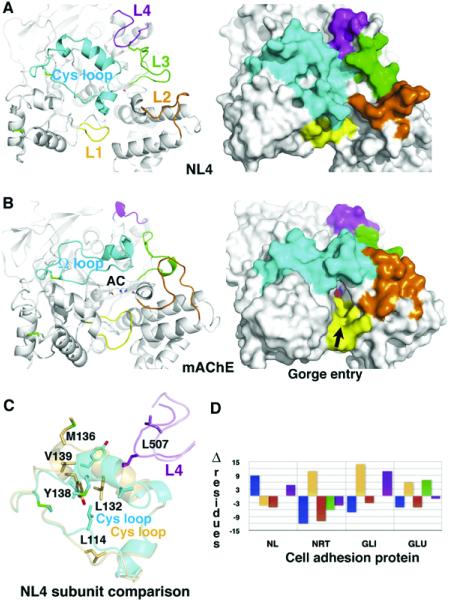
Figure 4. Comparison of the Surface Loops in NL4 and mAChE.
(A) The Cys-loop (Cys110-Cys146; cf. Figure 1) in NL4 and (B) the U-loop (Cys69-Cys96) in mAChE are displayed in cyan, loops L1 (Thr332-Ile338 in NL4) in yellow, loops L2 (Asp381-Val390) in orange, loops L3 (Gln477-Ser487) in green, and loops L4 (Gly503-Ser513) in magenta. The Cys-loop conformation is stabilized by several polar and nonpolar interactions between Leu120, His121, Asp122, and Trp127 at the loop edge, and Ser176, Glu179, Lys378, Gly490, and Leu507, which surround the central pocket. Label AC on mAChE denotes the active center. Molecular surfaces of NL4 ([A], right) and mAChE ([B], right) show the occluded surface in NL4 and the accessible gorge entry, marked by the arrow, in mAChE. (C) Superimposition of the two Cys-loops (cyan and orange) and interacting loops L4 (magenta and violet) from the two NL4 subunits in the dimer. Residues that undergo large displacements are displayed. (D) Graph of differences in the loop sizes (in numbers of residues) of NL4, neurotactin (NRT), gliotactin (GLI), and glutactin (GLU) compared with mAChE (color codes as in [A]).
The conformation of the Cys-loop in NL4 requires large compensative conformational rearrangements from four other surface loops located on the same face of the NL4 molecule that delineate the gorge rim in mAChE. Loop L1 (Figure 4, yellow) has a shorter edge segment than in mAChE and is ~4Å removed from the Cys-loop, precluding loop overlap. Loop L2 (Figure 4, orange) is shorter than in mAChE and moves away from the Cys-loop tip. Together with an ~3.5Å outward shift of helix α1 7,8, this positioning yields ~9Å separation between these loops. Loop L4 (Figure 4, magenta) is longer than in mAChE and adopts a conformation that appears to stabilize the Cys-loop conformation. It stretches toward the side of the Cys-loop, providing the Phe508 side chain for stacking interaction with Trp127 in the Cys-loop edge segment. The particular conformation of loop α1 8,9-α2 8,9 in NL4 requires conformational adaptation of the surrounding loop L3 (Figure 4, green), while both loops are stabilized by the neuroligin-specific Cys476-Cys510 bridge.
The Cys-loop in NL4 is also a homolog of the lid or flap in the lipases in the α/β-hydrolase fold family (Cygler et al., 1993; Grochulski et al., 1994). Structures of the apo and complexed forms of the Candida rugosa lipase show a closed flap occluding the active site cavity in the absence of substrate and an open loop folded backward to accommodate the bound substrate, respectively (Grochulski et al., 1994). In human pancreatic lipase, the open conformation is stabilized by the bound colipase partner (van Tilbeurgh et al., 1992). In p-nitrobenzyl esterase, the corresponding loop adopts a closed conformation similar to NL4, but it is 15 residues shorter and turned about 40° away from the active site entry, thus leaving the gorge unoccupied (Spiller et al., 1999). In AChE, the Ω-loop is believed to undergo rapid conformational fluctuations, allowing diffusion-limited substrate entry (Shi et al., 2001). In NL4, substantial mobility in the Cys-loop is illustrated by its greater B-factor values as compared with other regions (Figure 2 and Figure 4C), along with presence of the bound phosphate in the central pocket (Figure 3) and the existence of distinct conformations in the two dimer subunits (cf. Experimental Procedures).
The global change within the Cys-loop of NL4, compared with its enzyme relatives, requires large conformational and positional adaptation of the surrounding loops associated with deletions or insertions to construct a novel surface topography, which is well conserved within the neuroligins. Similarly, sequence alignment among the adhesion proteins, neuroligins, neurotactin, gliotactin, and glutactin reveals length compensation between the Cys-loop and its neighboring surface loops (Figure 4D and Figure S1). Hence, there is considerable variation in the loop sizes and conformations among members of the α/β-hydrolase fold family, where the adhesion proteins add an even greater degree of complexity.
Electrostatic Properties of Neuroligin
NL4 and mAChE show distinctive electrostatic features (Figure 5). NL4 displays an annular motif of electronegative surface potential covering a large area on the Cys-loop face of the molecule. A similar motif is found in AChE, where it surrounds the active site gorge entry and is thought to attract the positively charged ACh substrate (Ripoll et al., 1993), and in other adhesion members of the α/β-hydrolase fold family (Botti et al., 1998). This face in mAChE also encompasses the binding site for the peptidic ligand fasciculin (Bourne et al., 1995).
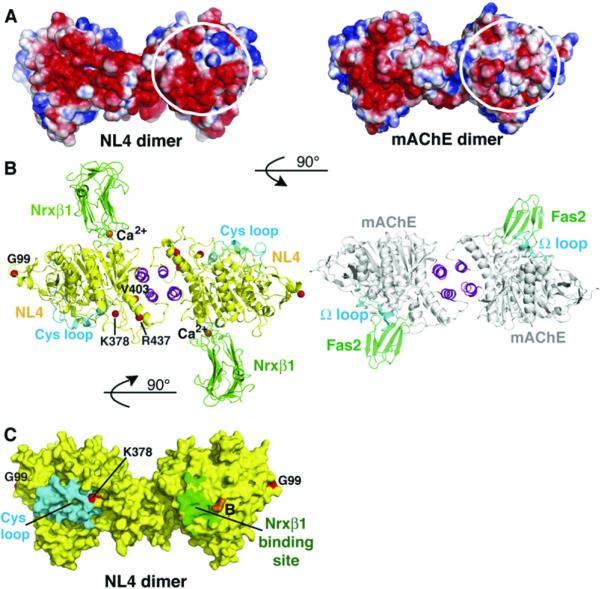
Figure 5. Electrostatic Potentials of the NL4 and mAChE Dimers and Comparison of the Dimeric Nrxβ1-NL4 and Fasciculin-mAChE Complexes.
(A) The NL4 (left) and mAChE (right) dimers are oriented with their C termini pointing down (same as in Figure 2B, top). In each dimer, the two-fold symmetry axis places opposite faces of each subunit side by side. The encircled surface area is negatively charged in NL4, but not mAChE. This results in a continuum of electronegative surface potentials from one subunit face to the other face in the NL4 dimer, in contrast to the restricted localization of this potential to a single subunit face in the mAChE dimer. Electrostatic surface potentials are contoured at -3/+3 kT/e, where red describes a negative and blue a positive potential.
(B) (Left) The Nrxβ1-NL4 complex with a yellow NL4, magenta four-helix bundle, cyan Cys-loops, and green Nrxβ1 molecules. The bound Nrxβ1 is oriented such that its curved β sheet is positioned in the prolongation of the curved long helix α4 7,8, which crosses the NL4 subunit. Red spheres denote the Gly99, Lys378, Val403, and Arg437 positions for mutations in NL3/NL4 of autism patients. (Right) The fasciculinmAChE complex with a gray mAChE, magenta bundle, cyan U-loops, and green fasciculins. Nrxβ1 (~25 kDa) and fasciculin (~7 kDa) bind on opposite faces of the NL4/mAChE dimer, through loops that connect b strands oriented perpendicular to the binding interfaces. The two complexes are oriented 90° compared with (A).
(C) The NL4 dimer, oriented as in (A), is displayed with a yellow molecular surface, a cyan Cys-loop, and a green area buried by bound Nrxβ1. The position for splice site B in NL1 is displayed in orange, and the Gly99 and Lys378 positions for mutations in autism patients are displayed in red. The other mutations are not visible with this orientation.
In contrast to mAChE, NL4 uniquely displays a second region of electronegative surface potential covering half the surface area on the face opposite the Cys-loop (Figure 5). This second motif covers a region more ordered than the Cys-loop region that is conserved in the other neuroligins, but not in neurotactin, gliotactin, or glutactin (Figure S1), as found upon analysis of primary theoretical models. In the symmetric NL4 dimer, the two electronegative motifs on each subunit merge into a continuous belt surrounding the two subunits, whereas in the mAChE dimer, the motif is restricted to one subunit. The second electronegative surface in the neuroligin corresponds to the Nrxβ1 binding face, as defined by the SAXS findings (Comoletti et al., 2007). As a result of the surface charge distribution, the overall net charge and dipole moment of the NL4 subunit (-25e and ~600 Debyes) are significantly larger and slightly lower, respectively, than those of mAChE (-9e and ~900 Debyes). The dipole vectors in the two molecules diverge by ~30°; the negative pole in each molecule is located close to the central pocket and the positive pole is on the side opposite the NL4 Cys-loop and mAChE Ω-loop regions. Hence, selectivity of the binding surface does not appear to depend on electrostatics. Rather, the balance of surface potential may be related to the adhesive function of neuroligin.
The Neurexin-Neuroligin Complex
To delineate the structural determinants of neurexin specificity for neuroligins, we have determined a crystal structure of the LNS domain of Nrxβ1 bound to NL4. Despite the overall limited resolution, the electron density map unambiguously reveals the position and orientation of the bound Nrxβ1 and those of the interacting side chains at the complex interface (Figure 5 and Figure S2). Nrxβ1 is bound through its hypervariable loop edge to the electronegative surface of the NL4 molecule opposite the Cys-loop region, where it protrudes some 40Å with its β strands oriented perpendicular to the long axis of the NL4 dimer. This makes the Nrxβ1-NL4 complex a mirror image of the mAChE complex with the β-strand three-fingered peptide fasciculin (Bourne et al., 1995)(Figure 5), exemplifying the variability in ligand selectivity and interaction capability of the α/β-hydrolase fold.
The concave side of the Nrxβ1 molecule faces the dimer four-helix bundle, which results in bound Nrxβ1 being rotated a quarter of a turn from its orientation in the SAXS model (Comoletti et al., 2007). The binding interface is very flat, and it encompasses a buried surface area of ~560Å2 on each partner in the complex (1.4Å probe radius) (Figure 6). This area, which is consistent with a low-affinity complex, corresponds to only 2.5% and 7% of the molecular surfaces of NL4 and Nrxβ1, respectively. Compared with the unbound NL4 and Nrxβ1 individual structures, no major conformational changes are observed at the interacting surfaces in the two partners. However, at the opposite side of the NL4 molecule, the Cys-loop appears to exhibit an 3Å outward displacement. Whether this rigid-body motion is related to Nrxβ1 binding is unknown.
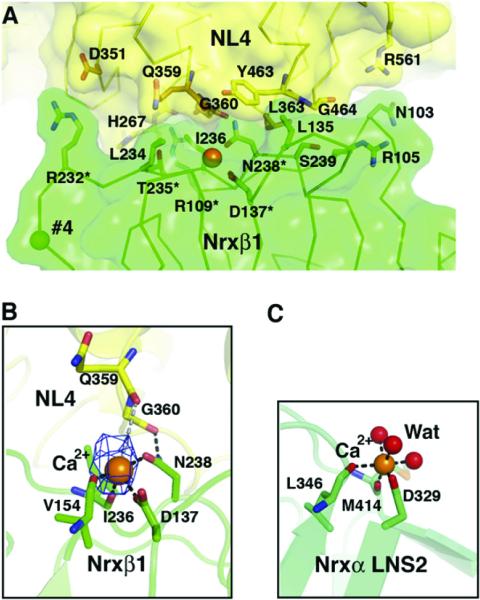
Figure 6. Close-up View of the Ca2+-Mediated Nrxβ1-NL4 Complex Interface.
(A) The Nrxβ1 molecular surface and key interacting side chains are displayed in green. The NL4 molecular surface is in yellow. The NL4 interacting side chains are in orange for the Ca2+-binding Gln359-Asn364 residues, and in yellow for the other residues. The Ca2+ ion is shown as an orange sphere central to the interface, and the position of splice site 4 in Nrxβ1 is shown as a green sphere. The asterisks denote Nrxβ1 residues whose mutation impairs binding to neuroligin (Graf et al., 2006).
(B) Close-up view of the Ca2+ coordination at the complex interface (same orientation as in [A]). The omit difference electron density map (cyan) for Ca2+ is contoured at 3.5 s. Bonds with Nrxβ1 residues and likely bonds with NL4 residues are displayed as black and gray dotted lines, respectively.
(C) Ca2+ coordination in the Nrxα LNS-2 domain structure (PDB accession code 2H0B; Sheckler et al., 2006) (same orientation as that of Nrxβ1 in [B]). The three coordinating water molecules are displayed as red spheres.
The presence of bound Ca2+ is an evident feature at the center of the complex interface (Figure 6). Here, Ca2+ is coordinated by the carboxylate side chain of Asp137, the amide of Asn238, and the carbonyl oxygens of Val154 and Ile236 from Nrxβ1 (same numbering as in Ushkaryov et al., 1994), which is consistent with the coordination observed in the Nrxα1 LNS-2 domain structure (Sheckler et al., 2006). The orientation of the Asn238 side chain is constrained by an additional interaction with the backbone carbonyl of Gly360 from NL4. Additional candidates for the coordination sphere include the carbonyl oxygens of NL4 residues Gln359 and Gly360, probably through water-mediated interactions, and a second water molecule in apposition with the Nrxβ1 surface, suggesting that NL4 residues may act as indirect ligands for Ca2+ coordination. A contribution from NL4 would compensate for the low Ca2+ affinity of neurexin alone (~400 μM for the Nrxα1 LNS-2 domain) (Sheckler et al., 2006). Higher resolution will be required to fully describe the Ca2+ coordination geometry and the reasons for Ca2+ selectivity over other cations (Comoletti et al., 2003; Nguyen and Sudhof, 1997).
Direct interactions between the complex partners arise principally from the NL4 Gln359-Asn364 stretch within loop α4 6,7-β7 (cf. Figure 1), with a minor contribution from Tyr463-Gly464 in loop α1 7,8-β8, His268/Glu270 in loop α5,6-β6, and residues within the hypervariable loop edge of Nrxβ1 (Table 2). Two salt bridges (Nrxβ1 Arg109 with NL4 Glu361/Glu270; and Nrxβ1 Arg232 with NL4 Asp 351) and a polar interaction (Nrxβ1 Asn103 with NL4 Arg561) are arranged as a triangle at the perimeter of the Ca2+ binding site and may serve as boundary clamps for the complex. Overall, these structural features are consistent with a labile or moderate-affinity complex (KD = 132 nM for Nrxβ1 and NL4) (Comoletti et al., 2006) imparting flexibility in the interaction partners of the membrane-tethered adhesion proteins, and perhaps contributing to plasticity of the synapse. Prior mutagenesis data on the Nrxβ1 Ca2+-binding residues Asp137 and Asn238 and on residues Arg109, Arg232, and Thr235 at the complex interface, but not Ser107, Thr108, Thr156, Asp157, Asn184, and Gln233, show diminished binding or synaptogenic activity (Graf et al., 2006), consistent with the spatial arrangement of partners in the crystalline complex.
Table 2.
Nrxβ1 and NL4 Residues in Contact or within Contact Distance in the Complex
| Nrxβ1 | NL4 | |
|---|---|---|
| Hydrogen bonds | Pro106 Oa | Asn364 Na |
| Ser107 Oa | Asn364 ND2a | |
| Arg109 NH1/NH2 | Ser266 Ob | |
| Glu270 OE1b | ||
| Glu361 OE1/OE2 | ||
| Thr235 OG1a | Gln359 NE2a | |
| Van der Waals contactsc | Ser107 | Leu363 |
| Arg109 | His267 | |
| Glu270b | ||
| Leu135 | Tyr463 | |
| Leu234 | Gln359 | |
| Ile236 | Gly360 | |
| Glu361 | ||
| Asn238 | Leu363 | |
| Ser239 | Leu363 | |
| Gly464 | ||
| Putative salt bridges | Arg109 | Glu270 |
| Glu361 | ||
| Arg232 | Asp351 | |
| Putative hydrogen bonds | Asn103 OD1 | Arg561 NH1/2 |
| Arg105 NH1/2 | Gly464 O | |
| Arg109 NH1/2 | Ser266 O | |
| His267 O | ||
| Asn364 OD1a | ||
| Thr235 OG1a | Glu361 Na | |
| Asn238 ND2b | Gln359 Ob | |
| Gly360 Ob | ||
| Ser239 OGa | Gly464 Na |
In italics are those residues that participate to the Ca2+ binding site.
Contact observed only in the one or the other subunit.
Contact observed only in the one or the other subunit.
Within a 4 Å distance between atoms from each partner in the complex.
Most interacting residues are conserved among neuroligins (Figure 1 and Figure 6), arguing for the present structure to be a representative model structure for other neuroligins. The only exception refers to Gly464, which is replaced by Gln in NL2. Since the Gly464 Cα is 4.6Å away from the facing Nrxβ1 Ser239, the bulkier Gln side chain is likely responsible for the lower affinity of NL2 (KD = 8840 nM) (Comoletti et al., 2006). In fact, an Ala substitution to Gly464 in NL1 abolishes Nrxβ1 binding (D.C., unpublished data). In contrast to the Cys-loop region, the shape of the Nrxβ1 binding region is surprisingly well conserved between NL4 and AChE, with only a small displacement of surface loop α4 6,7-β7. However, the key NL4 interacting residues are neither conserved in AChE nor in neurotactin, glutactin and gliotactin (Figure S1).
NL4 lacks both splice inserts A and B that are typically found in other neuroligins (Figure 1). The positions for these inserts are located on the neuroligin face opposite the Cys-loop where they are separated by 25Å. The presence of the short insert B, which borders the Nrxβ1 binding site, and its posttranslational processing for N-linked glycosylation, appreciably affect the Nrxβ1 binding affinity (Boucard et al., 2005; Comoletti et al., 2006), consistent with the current Nrxβ1-NL4 complex structure. By contrast, presence of the long insert A, which can display two distinctive sequences (Ichtchenko et al., 1996) with drastically opposite overall net charges, may affect binding indirectly by influencing the binding surface topography and overall electrostatics. In turn, splice site 4 in Nrxβ1 is remote from the binding interface (15Å from the bound Ca2+) and the presence of the 30-residue insert is likely to have a minor influence on the complex formation, in agreement with biochemical data (Boucard et al., 2005; Comoletti et al., 2006; Graf et al., 2006).
Autism and Neurexin-Neuroligin Mutations
Recently, several point mutations located within the extra-cellular domain of neuroligins have been reported in human NL3 and NL4 and appear to be associated with ASD. Mapping these mutations onto the molecular surface of the NL4 dimer shows that all are remote from the bound Nrxβ1(Figure 5). In human NL3, a Cys substitution to Arg451 (NL4 Arg437) gives rise to endoplasmic reticulum retention of the protein and decreases affinity for Nrxβ1(Comoletti et al., 2004). The fact that a homologous mutation in butyrylcholinesterase (Yen et al., 2003) causes an identical phenotype reveals a common mechanism of intracellular retention for relatives in the α/β-hydrolase fold family (De Jaco et al., 2006). In vivo studies of this mutation using a knockin mouse result in enhanced GABA inhibitory activity (Tabuchi et al., 2007). Other missense mutations identified within the region encoding the extracellular domain of each NL4 gene in unrelated patients with autism disorder are Gly99Ser, Lys378Arg and Val403Met (the latter two residues are invariant in all neuroligins). Gly99 is located at the surface in a turn preceding the short β3 strand, and its mutation does not seem to affect folding or binding. In contrast, Val403 is located near the end of helix α2 7,8 (Asn493-Leu406) and participates in the tight parallel packing of this helix onto the four-helix bundle. Mutation of this residue by a bulkier side chain may affect correct folding of the C-terminal domain and prevent formation of the functional neuroligin dimer. Lys378 is located at the beginning of the short helix α1 7,8 (Leu377-Val380) and establishes van der Waals contacts with Asp122 within the Cys-loop. Arg437, located within the long helix α4 7,8, is tightly packed against Trp484 and hydrogen bonded to the backbone carbonyls of Asp388 and Trp484 via a solvent molecule. The group of charged residues Asp388, Asp486, Glu434, and Lys338, surrounding Arg437, form a dense network at the surface that could be important for processing events.
In summary, the NL4 structure and its complex with Nrxβ1 reveal several key features for these adhesion molecules. First, the neurexin interaction surface of the neuroligin dimer is on the side opposite that for substrate access in the catalytically active neuroligin relatives. This demonstrates distinctive functional modules for cell adhesion and substrate binding in the α/β-hydrolase fold core. Second, a central pocket containing a vestigial active center is present in NL4, but its occlusion by the Cys-loop would preclude efficient substrate entry. Third, the loops that delineate the entry to the active center in the hydrolases differ greatly in size and positional conformation in NL4, where larger loops compensate for smaller neighboring loops and vice versa. The precise functional role of these loops in neuroligin remains to be investigated. Fourth, the Cys-loop, although tethered to the molecular core, displays conformational flexibility. This feature, along with the presence of entrapped solvent and bound phosphate in the central pocket, argues for potential motion of the Cys-loop in solution. The looser connection between the molecular core and four-helix bundle provides additional flexibility to the neuroligin dimer. Molecular plasticity may be a hallmark for the adhesion function. Fifth, neuroligin displays negatively charged surfaces on both its neurexin binding and Cys-loop faces. In contrast to AChE, a large electrostatic dipole in the neuroligins, which function as surface adhesion molecules, is not required to draw a substrate into the vestigial active center. Rather, absence of a large, permanent dipole for the neuroligin, along with the moderate affinity of the complex, may permit fluctuating interactions, which is a desirable property for adhesion function. All in all, the α/β-hydrolase fold, with its capacity for turning a dense core with a buried active center tailored for trapping and hydrolyzing a substrate into a flexible adhesion molecule with surface recognition properties, gives rise to surprising diversity in both molecular form and function.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant NL4, encompassing residues Gln44 to Thr619 of full-length human NL4 (Bolliger et al., 2001) (Figure 1) and N-terminally flanked with a FLAG epitope, numbered (-12)DYKDDDDKLAAA(-1), was expressed at high yields (~2 mg/l) as a soluble exported protein from stable HEK293 cells (Comoletti et al., 2006). It was purified from the culture medium on immobilized anti-FLAG M2 antibody (Sigma-Aldrich) (Comoletti et al., 2003). Further purification was by gel-filtration FPLC on prepacked Superdex-200 (GE Healthcare) equilibrated and eluted with 10 mM Na-HEPES (pH 7.4), 50 mM NaCl, and 0.01% NaN3 (buffer A). Quantification of NL4 by UV spectrophotometry used an extinction coefficient of 102 680 M-1cm-1 at 280 nm, calculated from the protein sequence, and a molecular mass of 72,663 Da, determined by MALDI-TOF spectrometry. Electrophoretic analyses used a PhastSystem apparatus and related gels and buffers (GE Healthcare). Purified NL4 was concentrated by ultrafiltration, filtered on sterile 0.22 μm membranes, and stored on ice.
Soluble recombinant Nrxβ1, encompassing residues Gly81-Val288 of the LNS domain of rat β-neurexin-1 (Ushkaryov et al., 1994) and N-terminally flanked with a 6xHis tag followed by a TEV cleavage site, was obtained by subcloning our initial pGEX construct (Comoletti et al., 2004) into pDEST17 (Invitrogen). This Nrxβ1 does not comprise those N-and C-terminal stretches found to be disordered in the Nrxβ1 structure (Rudenko et al., 1999). Nrxβ1 was expressed at high yields (~70 mg/l) from Rosetta PlyS cells, and purified from the culture medium by nickel affinity chromatography on a HisTrap FF Crude cartridge (GE Healthcare). Removal of the 6xHis tag used a 6xHis-tagged TEV protease (InVitrogen) in a 1:25 TEV-to-Nrxβ1 weight ratio (overnight incubation, 20°C), and a second nickel affinity chromatography step. A molecular mass of 19,312 Da consistent with efficient tag removal was found by MALDI-TOF spectrometry. Final purification was by gel-filtration FPLC on Superdex 200 in buffer A. The Nrxβ1-NL4 complex was formed in solution using a 1.2 molar excess of Nrxβ1 over NL4 in 10 mM HEPES (pH 7.4), 25 mM NaCl, and 2 mM CaCl2 (overnight incubation, 4°C). The buffer composition and final complex concentration were adjusted by ultrafiltration.
Crystallization, Data Collection, and Processing
Initial crystallization conditions were found from the PEG (Quiagen) and SM1 (Nextalbiotech) screens using vapor diffusion in 96-well Greiner plates and the Cartesian PixSys 4200 (Genomic Solutions) robot. Optimized conditions for crystallization of NL4 used hanging drops of 1.2 μl and a protein-to-well solution ratio of 1:1, with 10% PEG-3000 in 100 mM Na-phosphate-citrate (pH 4.2), 300 mM NaCl, and 0 mM (20°C; 3.2Å resolution data set) or 10 mM (4°C; 2.2Å resolution data set) CaCl2 as the well solution. Crystallization of the complex at 20° used sitting drops and 8% PEG-20000 in 100 mM Mes (pH 6.5) as the well solution. Crystals were briefly transferred to the mother liquor supplemented with 22%-25% glycerol and flash-cooled in liquid nitrogen. Space groups and resolution limits are reported in Table 1. Data were processed, merged, and scaled with XDS (Kabsch, 1993).
Structure Solution and Refinement
An initial 3.2Å resolution NL4 structure was solved by molecular replacement using, as a search model, one mAChE subunit (PDB accession code 1J06) (Bourne et al., 2003) and MOLREP from the CCP4 Program Suite (CCP4, 1994). The resolution achieved was sufficient to unambiguously position a four-helix bundled dimer of NL4 molecules within the asymmetric unit and build most of the backbone and side chains into the electron density using COOT (Emsley and Cowtan, 2004). This structure without further refinement was used as a template to solve the 2.2Å structure. The molecular replacement solution again positioned an NL4 dimer within the asymmetric unit. However, after ~90% of the protein model was built automatically with ARP/wARP (Perrakis et al., 1999), the region surrounding the Cys-loop (Cys110-Cys146) (Figure 1) appeared to be highly disordered. The structure was manually corrected using COOT, and structure refinement including TLS refinement was performed with Refmac in CCP4. TLS groups were generated using the TLS Motion Determination server (Painter and Merritt, 2006) and were manually adjusted to five groups per subunit corresponding to residues Ala(-1)-Cys110/Gln142-Tyr291/Phe340-Gln373/Trp449-Phe472/Lys561-Tyr582, Pro111-Asp141, Gln292-Ala339, Gly374-Gln448/Arg583-His598, and Tyr473-Ser560. Data collection and refinement statistics are summarized in Table 1.
The final NL4 structure comprises FLAG residues Asp(-8)-Ala(-1) and NL4 residues Gln44-Glu156, Ser164-Val540, and Glu556-Leu597 in subunit A of the dimer, and residues Ala(-2)-Ala(-1), Gln44-Ile159, Ser164-Pro541, and Val557-Leu597 in subunit B, out of the total 588 residues per subunit of the crystallized protein (Figure 1). GlcNAc moieties linked to Asn102 and Asn511; a phosphate (arising from the crystallization liquor) trapped in the central pocket; a citrate and a glycerol molecule (from the crystallization liquor and flash-cooling solution, respectively) trapped in solvent-accessible cavities at the protein surface; and three chloride and one sodium (from the crystallization liquor) are clearly identified. Despite the presence of CaCl2 in the crystallization conditions, no bound Ca2+ is observed, consistent with the ion being mainly coordinated by neurexin. The average root mean square deviation (rmsd) between subunits A and B is 0.5Å for 518 Cα atoms. Missing electron densities correspond to surface loops Asp157-Asn163, which in NL1 contains splice site A, and Pro541/Gln542-Val557/Ala558 and the C-terminal,~20-residue peptide that prolongs helix α10. The structure stereo-chemistry was analyzed with PROCHECK (Laskowski et al., 1993); no residues were found in the disallowed regions of the Ramachandran plot.
The structure of the Nrxβ1-NL4 complex was solved by molecular replacement using MOLREP and, as templates, the NL4 dimer structure and the Nrxβ1 structure (1C4R) (Rudenko et al., 1999). The model was refined with Refmac, including medium NCS restrains for the two NL4 molecules (excluding the Cys-loop) and the two Nrxβ1 molecules, and TLS refinement. The resulting electron density maps were used, when well-defined, to correct the backbone traces along the NL4 and Nrxβ1 molecules and position side chains and Ca2+ at the complex interface using COOT. An X-ray fluorescence emission spectrum confirmed the presence of Ca2+ in the frozen Nrxβ1-NL4 crystal. TLS groups were the same as for NL4, with an additional group for the entire Nrxβ1 molecule. NL4 residues Asp158-Asn163 in one subunit were built manually using COOT. Due to the limited resolution achieved, the remainder of the model was not refined further. The primary Nrxβ1-NL4 complex structure comprises NL4 residues Asp(-8)/Leu(-4)-Leu597 and Nrxβ1 residues His82-Val288.
Structure Analysis and Comparison with Other Structures
The volume of the NL4 central pocket was calculated using the CASTp server (Liang et al., 1998). Primary theoretical models of the other neuroligins and of neurotactin, gliotactin, and glutactin were designed using the mAChE structure as a template and the SWISS-MODEL server (Kopp and Schwede, 2006), and used without correction. The electro-static surface potentials were calculated using APBS (Baker et al., 2001) with the PyMOL APBS tools. Dipole moments were calculated from the Protein Dipole Moments Server (Felder et al., 2007). The volume of the internal pocket was calculated with VOIDOO (Kleywegt and Jones, 1994).
Sequence-and structure-based searches for NL4's closest catalytically active relatives used PSI-BLAST and SSM (Krissinel and Henrick, 2004), respectively. Highest structural hits were for mAChE, human liver caboxylesterase (2H7C), bovine bile salt-stimulated lipase (2BCE) and bacterial p-nitrobenzyl esterase (1C7J). Superimposition of the NL4 structure with each of the four enzymes, performed with LSQMAN (Kleywegt and Jones, 1997), resulted in an average rmsd in the 1.1-1.4Å range for at least 380 Cα atoms.
Figure 1 and Figure S1 were generated with ESPript (Gouet et al., 1999) and MUSCLE (Edgar, 2004), and Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, and Figure S2 were generated with PyMOL (DeLano, 2002).
ACKNOWLEDGMENTS
We are grateful to Jennifer Wilson (UCSD), Sandrine Conrod (CNRS FRE-2738), and the beamline staff of the European Synchrotron Research Facility (ESRF, Grenoble, France), for assistance in cell culture and protein expression, protein purification and crystallization, and data collection, respectively. This work was supported by the SPINE2-Complexes Consortium (Y.B., P.M., and a postdoctoral grant to P.L.); the CNRS DREI-SDV (P.M. and Y.B.); the CNRS and Fondation pour la Recherche Médicale (postdoctoral grants to I.F.); the Cure Autism Now Pilot Study (D.C.); and NIH grants R37-GM 18360 and UO-1 ES 10337 (P.T.) and GM07752 (M.M.).
Footnotes
Accession Numbers Atomic coordinates and structure factors have been deposited with the Protein Data Bank; accession codes are 3BE8 (NL4) and 2VH8 (Nrxb1-NL4 complex).
Supplemental Data The Supplemental Data for this article can be found online at http://www.neuron.org/cgi/content/full/56/6/979/DC1/.
References
- Auld VJ, Fetter RD, Broadie K, Goodman CS. Gliotactin, a novel transmembrane protein on peripheral glia, is required to form the blood-nerve barrier in Drosophila. Cell. 1995;81:757–767. doi: 10.1016/0092-8674(95)90537-5. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi F, Bacchelli E, Pesaresi G, Carone S, Bailey AJ, Maestrini E. Absence of coding mutations in the X-linked genes neuroligin 3 and neuroligin 4 in individuals with autism from the IMGSAC collection. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2006;141:220–221. doi: 10.1002/ajmg.b.30287. [DOI] [PubMed] [Google Scholar]
- Bolliger MF, Frei K, Winterhalter KH, Gloor SM. Identification of a novel neuroligin in humans which binds to PSD-95 and has a widespread expression. Biochem. J. 2001;356:581–588. doi: 10.1042/0264-6021:3560581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botti SA, Felder CE, Sussman JL, Silman I. Electro-tactins: a class of adhesion proteins with conserved electrostatic and structural motifs. Protein Eng. 1998;11:415–420. doi: 10.1093/protein/11.6.415. [DOI] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Bourne Y, Taylor P, Marchot P. Acetylcholinesterase inhibition by fasciculin: crystal structure of the complex. Cell. 1995;83:503–512. doi: 10.1016/0092-8674(95)90128-0. [DOI] [PubMed] [Google Scholar]
- Bourne Y, Taylor P, Radic Z, Marchot P. Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site. EMBO J. 2003;22:1–12. doi: 10.1093/emboj/cdg005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project, Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Flynn R, Jennings LL, Chubykin A, Matsumura T, Hasegawa H, Sudhof TC, Taylor P. Characterization of the interaction of a recombinant soluble neuroligin-1 with neurexin-1beta. J. Biol. Chem. 2003;278:50497–50505. doi: 10.1074/jbc.M306803200. [DOI] [PubMed] [Google Scholar]
- Comoletti D, De Jaco A, Jennings LL, Flynn RE, Gaietta G, Tsigelny I, Ellisman MH, Taylor P. The Arg451Cysneuroligin-3 mutation associated with autism reveals a defect in protein processing. J. Neurosci. 2004;24:4889–4893. doi: 10.1523/JNEUROSCI.0468-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoletti D, Flynn RE, Boucard AA, Demeler B, Schirf V, Shi J, Jennings LL, Newlin HR, Sudhof TC, Taylor P. Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for beta-neurexins. Biochemistry. 2006;45:12816–12827. doi: 10.1021/bi0614131. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Grishaev A, Whitten AE, Tsigelny I, Taylor P, Trewhella J. Synaptic arrangement of the neuroligin/beta-neurexin complex revealed by X-ray and neutron scattering. Structure. 2007;15:693–705. doi: 10.1016/j.str.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cygler M, Schrag JD, Sussman JL, Harel M, Silman I, Gentry MK, Doctor BP. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases, and related proteins. Protein Sci. 1993;2:366–382. doi: 10.1002/pro.5560020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jaco A, Comoletti D, Kovarik Z, Gaietta G, Radic Z, Lockridge O, Ellisman MH, Taylor P. A mutation linked with autism reveals a common mechanism of endoplasmic reticulum retention for the alpha,beta-hydrolase fold protein family. J. Biol. Chem. 2006;281:9667–9676. doi: 10.1074/jbc.M510262200. [DOI] [PubMed] [Google Scholar]
- de la Escalera S, Bockamp EO, Moya F, Piovant M, Jimenez F. Characterization and gene cloning of neurotactin, a Drosophila transmembrane protein related to cholinesterases. EMBO J. 1990;9:3593–3601. doi: 10.1002/j.1460-2075.1990.tb07570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C, Scholl FG, Choih J, DeMaria S, Berger J, Isacoff E, Scheiffele P. Neurexin mediates the assembly of presynaptic terminals. Nat. Neurosci. 2003;6:708–716. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. The PyMOL molecular graphics system. 2002 http://www.pymol.org.
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Felder CE, Prilusky J, Silman I, Sussman JL. A server and database for dipole moments of proteins. Nucleic Acids Res. 2007;35:W512–521. doi: 10.1093/nar/gkm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney G, Camp S, Dionne M, MacPhee-Quigley K, Taylor P. Mutagenesis of essential functional residues in acetylcholinesterase. Proc. Natl. Acad. Sci. USA. 1990;87:7546–7550. doi: 10.1073/pnas.87.19.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Graf ER, Kang Y, Hauner AM, Craig AM. Structure function and splice site analysis of the synaptogenic activity of the neurexin-1 beta LNS domain. J. Neurosci. 2006;26:4256–4265. doi: 10.1523/JNEUROSCI.1253-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochulski P, Li Y, Schrag JD, Cygler M. Two conformational states of Candida rugosa lipase. Protein Sci. 1994;3:82–91. doi: 10.1002/pro.5560030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman RC, Jennings LL, Tsigelny I, Comoletti D, Flynn RE, Sudhof TC, Taylor P. Structural characterization of recombinant soluble rat neuroligin 1: mapping of secondary structure and glycosylation by mass spectrometry. Biochemistry. 2004;43:1496–1506. doi: 10.1021/bi035278t. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Nguyen T, Sudhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J. Biol. Chem. 1996;271:2676–2682. doi: 10.1074/jbc.271.5.2676. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 1997;277:525–545. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]
- Kopp J, Schwede T. The SWISS-MODEL Repository: new features and functionalities. Nucleic Acids Res. 2006;34:D315–D318. doi: 10.1093/nar/gkj056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Lardi-Studler B, Fritschy JM. Matching of pre- and post-synaptic specializations during synaptogenesis. Neuroscientist. 2007;13:115–126. doi: 10.1177/1073858406296803. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur M, Moss D, Thornton J. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am. J. Hum. Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S. Analytical shape computation of macromolecules: II. Inaccessible cavities in proteins. Proteins. 1998;33:18–29. [PubMed] [Google Scholar]
- Marchot P, Ravelli RB, Raves ML, Bourne Y, Vellom DC, Kanter J, Camp S, Sussman JL, Taylor P. Soluble monomeric acetylcholinesterase from mouse: expression, purification, and crystallization in complex with fasciculin. Protein Sci. 1996;5:672–679. doi: 10.1002/pro.5560050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Sudhof TC. Binding properties of neuroligin 1 and neurexin 1beta reveal function as heterophilic cell adhesion molecules. J. Biol. Chem. 1997;272:26032–26039. doi: 10.1074/jbc.272.41.26032. [DOI] [PubMed] [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- Olson PF, Fessler LI, Nelson RE, Sterne RE, Campbell AG, Fessler JH. Glutactin, a novel Drosophila basement membrane-related glycoprotein with sequence similarity to serine esterases. EMBO J. 1990;9:1219–1227. doi: 10.1002/j.1460-2075.1990.tb08229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Ripoll DR, Faerman CH, Axelsen PH, Silman I, Sussman JL. An electrostatic mechanism for substrate guidance down the aromatic gorge of acetylcholinesterase. Proc. Natl. Acad. Sci. USA. 1993;90:5128–5132. doi: 10.1073/pnas.90.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G, Nguyen T, Chelliah Y, Sudhof TC, Deisenhofer J. The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing. Cell. 1999;99:93–101. doi: 10.1016/s0092-8674(00)80065-3. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Schumacher M, Camp S, Maulet Y, Newton M, MacPhee-Quigley K, Taylor SS, Friedmann T, Taylor P. Primary structure of Torpedo californica acetylcholinesterase deduced from its cDNA sequence. Nature. 1986;319:407–409. doi: 10.1038/319407a0. [DOI] [PubMed] [Google Scholar]
- Sheckler LR, Henry L, Sugita S, Sudhof TC, Rudenko G. Crystal structure of the second LNS/LG domain from neurexin 1alpha: Ca2+ binding and the effects of alternative splicing. J. Biol. Chem. 2006;281:22896–22905. doi: 10.1074/jbc.M603464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Boyd AE, Radic Z, Taylor P. Reversibly bound and covalently attached ligands induce conformational changes in the omega loop, Cys69-Cys96, of mouse acetylcholinesterase. J. Biol. Chem. 2001;276:42196–42204. doi: 10.1074/jbc.M106896200. [DOI] [PubMed] [Google Scholar]
- Spiller B, Gershenson A, Arnold FH, Stevens RC. A structural view of evolutionary divergence. Proc. Natl. Acad. Sci. USA. 1999;96:12305–12310. doi: 10.1073/pnas.96.22.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. A Neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talebizadeh Z, Lam DY, Theodoro MF, Bittel DC, Lushington GH, Butler MG. Novel splice isoforms for NLGN3 and NLGN4 with possible implications in autism. J. Med. Genet. 2006;43:e21–e28. doi: 10.1136/jmg.2005.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushkaryov YA, Hata Y, Ichtchenko K, Moomaw C, Afendis S, Slaughter CA, Sudhof TC. Conserved domain structure of beta-neurexins. Unusual cleaved signal sequences in receptor-like neuronal cell-surface proteins. J. Biol. Chem. 1994;269:11987–11992. [PubMed] [Google Scholar]
- van Tilbeurgh H, Sarda L, Verger R, Cambillau C. Structure of the pancreatic lipase-procolipase complex. Nature. 1992;359:159–162. doi: 10.1038/359159a0. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Waites CL, Craig AM, Garner CC. Mechanisms of vertebrate synaptogenesis. Annu. Rev. Neurosci. 2005;28:251–274. doi: 10.1146/annurev.neuro.27.070203.144336. [DOI] [PubMed] [Google Scholar]
- Yan J, Oliveira G, Coutinho A, Yang C, Feng J, Katz C, Sram J, Bockholt A, Jones IR, Craddock N, et al. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol. Psychiatry. 2005;10:329–332. doi: 10.1038/sj.mp.4001629. [DOI] [PubMed] [Google Scholar]
- Yen T, Nightingale BN, Burns JC, Sullivan DR, Stewart PM. Butyrylcholinesterase (BCHE) genotyping for post-succinylcholine apnea in an Australian population. Clin. Chem. 2003;49:1297–1308. doi: 10.1373/49.8.1297. [DOI] [PubMed] [Google Scholar]