SUMMARY
Studies in mammals have indicated a connection between circadian clocks and feeding behavior, but the nature of the interaction, and its relationship to nutrient metabolism, are not understood. In Drosophila clock proteins are expressed in many metabolically important tissues, but have not been linked to metabolic processes. Here we demonstrate that Drosophila feeding behavior displays a 24-hour circadian rhythm which is regulated by clocks in digestive/metabolic tissues. Flies lacking clocks in these tissues, in particular in the fat body, also display increased food consumption, but have decreased levels of glycogen and a higher sensitivity to starvation. Interestingly, glycogen levels and starvation sensitivity are also affected by clocks in neuronal cells but the effects of neuronal clocks generally oppose those of the fat body. We propose that the input of neuronal clocks and clocks in metabolic tissues is coordinated to provide effective energy homeostasis.
INTRODUCTION
Circadian clocks are endogenous oscillators found in almost all organisms ranging from cyanobacteria to humans (reviewed by Hardin, 2005; reviewed by Williams and Sehgal, 2001). The molecular machinery of circadian clocks consists of transcription/translation based feedback loops that maintain cycles of gene expression. In Drosophila, the major loop involves a positive limb, which includes the transcriptional activators CLOCK (CLK) and CYCLE (CYC), and a negative limb composed of the transcriptional repressors PERIOD (PER) and TIMELESS (TIM). CLK and CYC activate the transcription of per and tim, and PER and TIM translocate to the nucleus at a specific time of day to inhibit the transcriptional activity of CLK and CYC. This feedback results in the rhythmic expression of per and tim as well as of many other CLK-CYC target genes (Hardin, 2005; Williams and Sehgal, 2001).
Although a relatively under-explored area, the circadian control of metabolism is now increasingly recognized as critical for organisms to adapt to their environment. Many of the circadian effects on metabolism were discovered in the course of molecular analysis of rhythms. Clock genes were found to be expressed in metabolic tissues (Bray and Young, 2007; Shibata, 2004), microarray screens to identify cycling genes revealed that many metabolic genes are expressed with a circadian rhythm (Akhtar et al., 2002; Kohsaka, and Bass, 2007; Storch et al., 2002) and clock gene knockouts have been associated with some metabolic deficits (Bray and Young, 2007; Zvonic et al., 2007). For example, inactivation of BMAL1 (mammalian equivalent of CYC) suppresses a diurnal variation in glucose and triglyceride (Rudic et al., 2004). Furthermore, clock proteins including BMAL1 and CLOCK appear to regulate lipid metabolism, gluconeogenesis, and adipogenesis (Rudic et al., 2004; Shimba et al., 2005). In addition, human and animal disease model studies suggest a relationship between circadian rhythms and metabolic dysfunction. Shift workers and nighttime eaters, two groups of people who usually have disrupted circadian rhythms, have an increased risk of developing metabolic syndrome (Colles et al., 2007; Bray and Young, 2007). Consistent with this idea, Clock mutant mice reportedly display symptoms of metabolic syndrome (Turek et al., 2005). Conversely, the circadian rhythms of leptin, adipokines and adiponectin expression are blunted in obese and diabetic animals (Ando et al., 2005), and some patients with metabolic syndrome display a circadian rhythm phenotype. For instance, diabetic patients exhibit an altered daily oscillation of insulin secretion and sensitivity (Boden et al., 1999). Despite these associations, the connection between circadian clocks and metabolism is poorly understood.
Circadian clocks exist in both the central nervous system and in peripheral tissues (Balsalobre, 2002; Giebultowicz, 2000). In mammals, the central pacemaker is in the suprachiasmatic nuclei (SCN) of the hypothalamus (Aton and Herzog, 2005; Liu et al., 2007) and peripheral clocks are located in multiple organs including the liver (Shibata, 2004), heart (Young, 2006), pineal gland (Fukada and Okano, 2002), and adipose tissue (Zvonic et al., 2006), many of which have important metabolic functions. A restricted feeding paradigm can entrain the phase of clock gene cycling in some of these peripheral organs without affecting the central clock in the SCN (Damiola et al., 2000; Hara et al., 2001; Stokkan et al., 2001), indicating that peripheral clocks respond preferentially to metabolic changes. Microarray analyses showed that circadian clock controlled genes (CCGs) vary among different tissues, suggesting that tissue-specific CCGs mediate the specific function of these tissues (Ptitsyn et al., 2006; Storch et al., 2002). The restricted feeding paradigm also results in a food-anticipatory activity rhythm which appears to be driven by a circadian pacemaker outside the SCN (Fuller et al., 2008). These results suggest that the mutual regulation of metabolism and circadian rhythms requires extra-SCN clocks.
Drosophila has proved to be an extremely powerful model for understanding the molecular basis of circadian rhythms. However, its utility for addressing the circadian control of metabolism has been limited although it is increasingly being used to study metabolism in general. As in mammals, the Drosophila central clock that regulates eclosion and locomotor activity rhythms is located in a small group of neurons in the lateral part of the brain (Hardin, 2005), and molecular clocks have been identified in various peripheral tissues (Giebultowicz, 2000). Here we sought to exploit the powerful genetics available in Drosophila to determine the metabolic function of peripheral clocks. Because feeding is an activity closely related to energy metabolism, we first addressed the relationship between peripheral clocks and Drosophila feeding behavior. We found that Drosophila feeding behavior is under circadian control, and that clocks in metabolic tissues regulate the phase of the feeding rhythm. We also report that the clock in the Drosophila fat body affects energy storage and thereby the response to starvation. Interestingly, neuronal and peripheral clocks have opposing effects on glucose metabolism, which may serve to maintain energy homeostasis.
RESULTS
Drosophila feeding behavior exhibits a clock-controlled rhythm
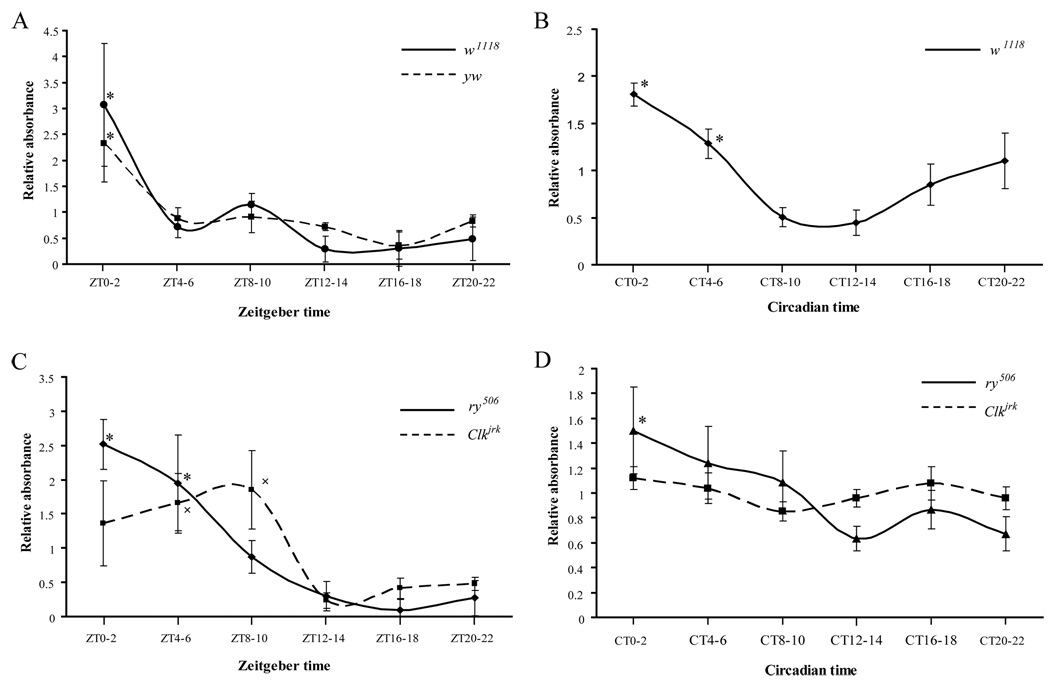
To investigate the relationship between peripheral clocks and metabolism, we first sought to determine if the circadian clock controls feeding behavior, an animal behavior which is closely related to energy metabolism. We used two assays to study Drosophila feeding behavior. In the first assay, we measured food consumption through the use of a food dye that can be measured spectrophotometrically. Groups of male w1118 flies (30 flies per group) entrained to 12:12 Light:Dark (LD) cycles for three days were fed food containing this dye for 2 hours at different times of day. After feeding, the flies were homogenized and the blue signal in the supernatant was measured in a spectrophotometer. The results showed that feeding behavior exhibits a strong rhythmic pattern in LD with a feeding peak in the early daytime and a trough at night (Fig 1A). Similar results were obtained with y w flies (Fig 1A).
Figure 1. Drosophila display a circadian rhythm of feeding.
(A) Feeding rhythms of w1118 and y w flies in the presence of LD cycles. Flies were fed dye-labeled food for 2 hours at each of 6 different time points over a 24-hour cycle and then collected and homogenized in PBS. Absorbance of the blue dye was measured at 625nm, and the amount per fly was calculated by dividing the OD625 value by the number of flies used in the experiment. For each time point, the relative food consumed was the ratio of the blue signal per fly to the average value across the 6 time points in the same experiment. The experiment was repeated at least 3 times. Asterisks denote a significant (p<0.05) difference in relative food consumption of w1118 or y w flies at the peak of the feeding curve (ZT0-2) compared to the trough (ZT8-10, ZT12-14, ZT16-18, and ZT20-22). (B) Feeding rhythm of w1118 on the third day of DD. Asterisks denote a significant (p<0.05) difference in relative food consumption of w1118 flies at the peak of the feeding curve (CT0-2 and CT4-6) compared to the trough (CT8-10 and CT12-14). (C and D) The feeding rhythm is controlled by the circadian clock as well as by light. Feeding curves for ry506 and Clkjrk in LD (C) and DD (D) conditions. In (C), asterisks denote a significant (p<0.05) difference in relative food consumption of ry506 flies at the peak of the feeding curve (ZT0-2 and ZT4-6) compared to the trough (ZT12-14, ZT16-18 and ZT20-22). The x denotes a significant difference in relative food consumption of Clkjrk at the peak of the feeding curve (ZT4-6 and ZT8-10) compared to the trough (ZT12-14, ZT16-18 and ZT20-22). In (D), the asterisk denotes a significant (p<0.05) difference in relative food consumption of ry506 flies at CT0-2 compared to the trough (CT12-14). Since the feeding rhythm was weak in ry506 flies, the experiment was repeated at least 9 times to ensure the presence of a clear rhythm. Statistical comparisons were made with one-way ANOVA. All data are presented as mean±SEM.
To determine if the feeding rhythm is under circadian control, we first measured the feeding behavior of flies maintained in constant darkness (DD). Following 3 days of entrainment to LD, flies were transferred to DD for 2 days before the feeding assay was performed. We found that w1118 flies maintained their feeding rhythm on the third day in DD, with a peak that started in the late night (CT20) and ended in the early daytime (CT4) (Fig 1B). y w flies displayed a similar rhythmic feeding pattern (data not shown). In contrast, a clock mutant, Clkjrk, consumed similar amounts of food during all 6 time periods on the third day in DD as compared to its control, ry506 (Fig 1D). However, Clkjrk flies displayed rhythmic feeding patterns in LD (Fig 1C). These data indicate that the feeding rhythm is under the control of an internal clock, but can also be driven by light. Indeed, a reversal of the light:dark cycle led to an immediate reversal of the feeding rhythm (data not shown).
The second assay we used was a revised CAFÉ assay, first reported by Ja W.W. (Ja et al., 2007). In this assay, flies were fed a 5% sucrose solution maintained in a capillary tube, and food consumption was measured by calculating the change of liquid volume in the tube. Since flies were kept in the same environment and fed the same food they had before the assay, the only perturbation was the introduction of the capillary tube; thus, experimental variance caused by exposure to new food, environment or handling was minimized. Also, the same group of flies was tested at all time points, thereby providing a longitudinal assay for feeding. Results of this assay corroborated those obtained in the above-described cross-sectional assay, with w1118 flies displaying a circadian rhythm of feeding in DD (Supplement Figure 1A).
A functional clock exists in the fat body
Given that several peripheral tissues expressing clock genes have functions related to nutrient sensing or to the regulation of energy homeostasis, we reasoned that clocks in these tissues may regulate feeding behavior. To test this idea, we first focused on a possible clock in the fat body..
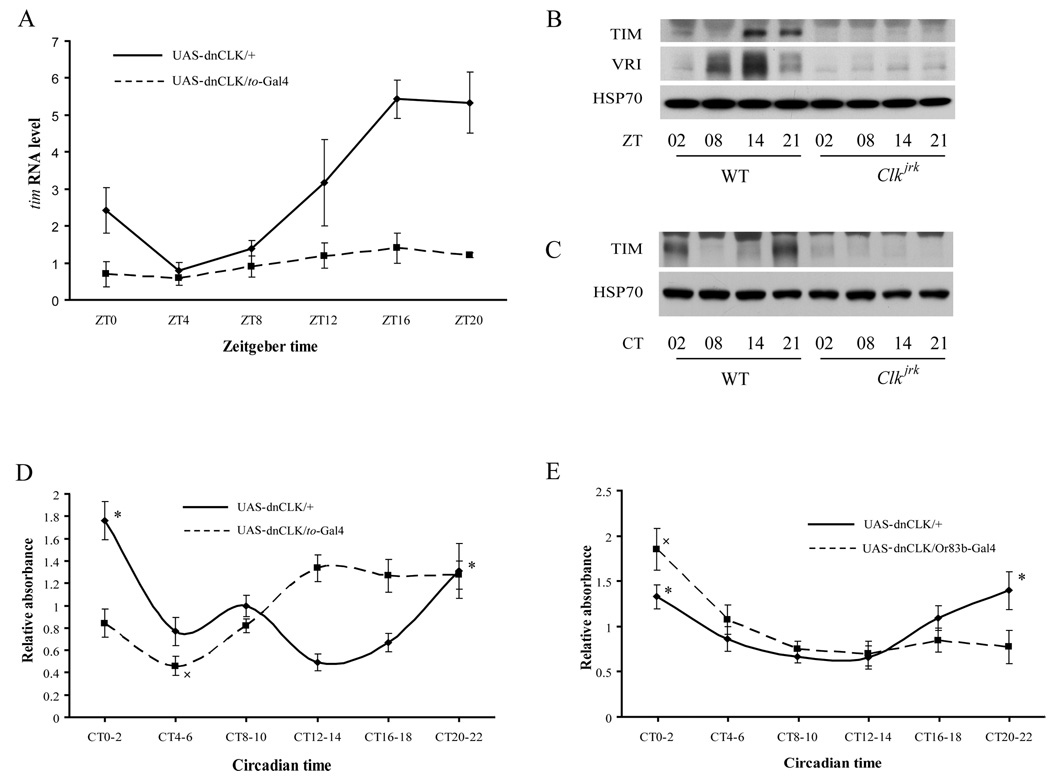
In Drosophila, the fat body plays an important role similar to that of the liver in mammalian energy metabolism (Scott et al., 2004). Clock protein expression in the fat body has been reported, but there is little information on cyclic regulation (Giebultowicz et al., 2001). To assess circadian control, we assayed expression of clock proteins in the fat body over the course of a 24 hour day. Western blots showed that the TIM clock protein cycles in the fat body of wild type flies, but not Clkjrk flies in both LD and DD (Fig 2B and 2C). Real-time quantitative-PCR (qPCR) results indicated that tim mRNA also cycles in the adult fat body (Fig 2A). To determine if the cycling is driven by a clock in the fat body, we expressed a dominant negative form of CLK (dnCLK) under the control of a driver that is expressed predominantly in the fat body, takeout-Gal4 (to-Gal4) (Dauwalder et al., 2002). Disruption of the fat body clock in this fashion dampened the cycling and decreased the overall levels of tim mRNA (Fig 2A). In addition, microarray analysis of the fat body revealed that dozens of genes, including important metabolic genes such as zw, cycle under the control of the fat body clock, (Supplemental Fig 2 and unpublished data). All these data support the existence of a functional clock in the fat body of adult flies.
Figure 2. Metabolic tissues contain clocks which regulate the feeding rhythm.
(A) Cycling of tim mRNA in the adult fat body. Flies were collected at different times in an LD cycle, and the abdominal fat body (with the attached epidermis) was used for the extraction of total RNA which was subjected to reverse transcription and qPCR analysis. In flies carrying UAS-dnCLK only, tim mRNA showed a strong rhythmic pattern, peaking at nighttime. However, the cycling of tim mRNA was dampened greatly, and the overall level of tim mRNA level was reduced, in flies carrying both UAS-dnCLK and to-Gal4. An average of three experiments is shown and statistical significance was determined by two-tailed Student’s t-test with unequal variance. (B) and (C) Cycling of TIM in the adult fat body of WT flies but not Clkjrk flies in both LD (B) and DD (C). (D) Feeding profiles of flies with clocks disrupted in metabolic tissues. Asterisks indicate significant differences (p<0.05) in relative food consumed by flies carrying UAS-dnCLK at the peak of the feeding curve (CT20-22 and CT0-2) as compared to the trough (CT12-14). The x indicates a significant difference (p<0.05) in relative food consumption of flies carrying UAS-dnCLK and to-Gal4 at the trough of the feeding curve (CT4-6) compared to the peak (CT12-14, CT16-18, CT20-22 and CT0-2). (E) Feeding profiles of flies with a disrupted olfactory clock. Feeding assays were performed on the third day in DD and data were evaluated as described in Figure 1. Asterisks indicate significant differences (p<0.05) in relative food consumption of control flies (carrying UAS-dnCLK alone) at the peak of the feeding curve (CT20-22 and CT0-2) as compared to the trough (CT8-10 and CT12-14). The cross indicates a significant difference (p<0.05) in relative food consumption of flies carrying UAS-dnCLK and Or83b-Gal4 at the peak of the feeding curve (CT0-2) as compared to the other 5 time periods. One-way ANOVA was used to compare relative feeding signals for flies of a given genotype at different time points. Two-way ANOVA was used to compare across strains.
Clocks in digestive/metabolic tissues, including the fat body, regulate the feeding rhythm
Next, we examined the feeding rhythm in flies expressing dnCLK under the control of the to-Gal4 driver. As noted above, the to-driver is expressed predominantly in the fat body although there is limited expression in some digestive tissues such as the hind gut (data not shown). Flies lacking clocks in these tissues displayed locomotor activity rhythms similar to those of control flies (Supplemental Fig 3). However, the phase of the feeding rhythm on the third day in DD was different from that of control flies- the trough was limited to a small period during the day (CT4-6), and the peak appeared in the early nighttime and lasted until the following day (Fig 2D). Similar results were obtained when the CAFÉ assay was used to measure the feeding rhythm (Supplement Fig 1B). This change in the feeding curve suggests that clocks in metabolic tissues, most likely in the fat body, regulate the phase of the feeding rhythm.
A peripheral clock located in olfactory receptor neurons of antenna is both necessary and sufficient for rhythms in olfactory responses (Tanoue et al., 2004). Thus, expression of dnCLK by a general driver for olfactory receptor cells, Or83b-Gal4, eliminates the olfactory rhythm (Tanoue et al., 2004). Given that the detection of food, mediated by sensory components such as olfactory receptors, is an important step in the initiation of feeding behavior, we asked if the clock in olfactory receptor neurons regulates the feeding rhythm. We found that flies lacking the olfactory clock displayed feeding rhythms on the third day in DD with a phase similar to that of control flies (Fig 2E), demonstrating that this clock does not participate in the regulation of feeding rhythms.
Clocks in digestive/metabolic tissues regulate total food consumption
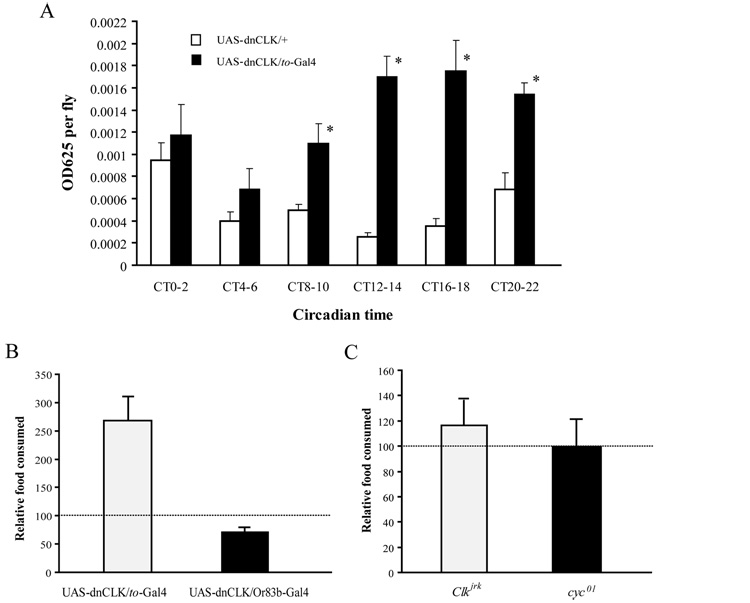
In the feeding rhythm assay, we noticed that food consumption was high in flies with disrupted digestive/metabolic clocks. Thus, we quantified the total amount of food consumed during a 24h period by flies of different genotypes. Food intake measured at different time points was compared across genotypes, either at each time point or pooled over the 24 hour interval (Fig 3A and 3B). We found that when dnCLK was expressed with a to-Gal4 driver, total food consumption was higher (Fig 3B). Similar results were obtained when dominant negative CYC (dnCYC) was expressed under the control of the to-Gal4 driver (data not shown). Furthermore, when we compared the amount of food consumed in 2-hour blocks at different circadian times (CT) between control flies and flies lacking clocks in metabolic tissues, we found that the increase of feeding was more dramatic during the subjective nighttime (Fig 3A). These results suggest that the clock in these tissues regulates food consumption by inhibiting feeding during the nighttime. Interestingly, we found that flies lacking the clock in olfactory receptor neurons showed decreased total food consumption compared to control flies (Fig 3B), suggesting that the clock in olfactory receptor neurons stimulates feeding behavior.
Figure 3. Clocks in metabolic tissues regulate overall food consumption.
(A) Flies in which the fat body clock is disrupted consume more food than control flies. Food consumed during 2 hour intervals at different times of day was determined as described in Figure 1. Asterisks indicate significant differences (p<0.05) in food consumption between flies carrying only UAS-dnCLK and flies carrying both UAS-dnCLK and to-Gal4, at the same time point. The data shown here are the same as in Figure 2B. However, they are plotted to show total rather than relative food consumption at every time point. Note that the increased food consumption of flies carrying both UAS-dnCLK and to-Gal4 is more striking during the nighttime. Statistical significance was determined by two-tailed Student’s t-test with unequal variance. (B) Effect of disrupting the clock in either fat body or in olfactory neurons on total food consumption. The total amount of food consumed in 24h was dramatically increased in flies carrying UAS-dnCLK and to-Gal4 and slightly decreased in flies carrying UAS-dnCLK and Or83b-Gal4, compared to flies carrying only UAS-dnCLK. Total food consumed in 24h was calculated by adding all the normalized OD625values measured during the six time periods. The horizontal line indicates total food consumption in flies carrying only UAS-dnCLK. Data for other lines were normalized to the value for the UAS-dnCLK line. (C) Total amount of food consumed in 24h was not different for Clkjrk or cyc01 flies compared to control ry506 flies. The dashed line indicates total food consumed by ry506 flies. Statistical significance was determined by two-tailed Student’s t-test with unequal variance. Asterisks indicate significant differences (p<0.05).
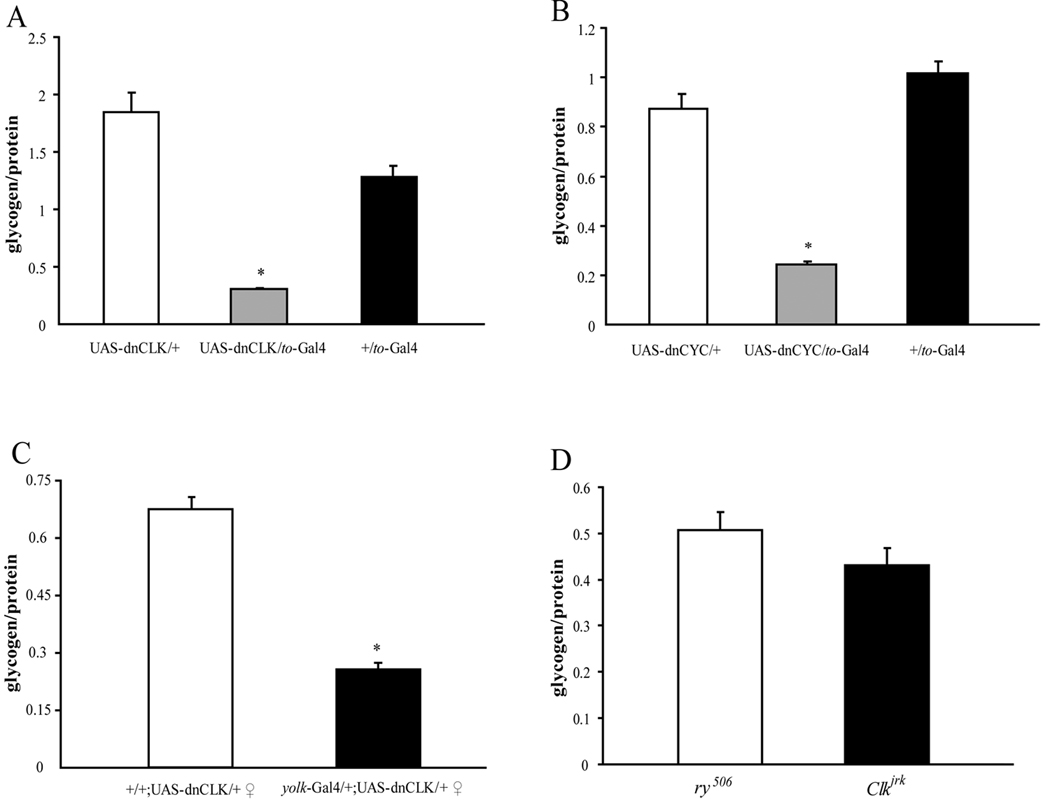
Disruption of the fat body clock increases sensitivity to starvation
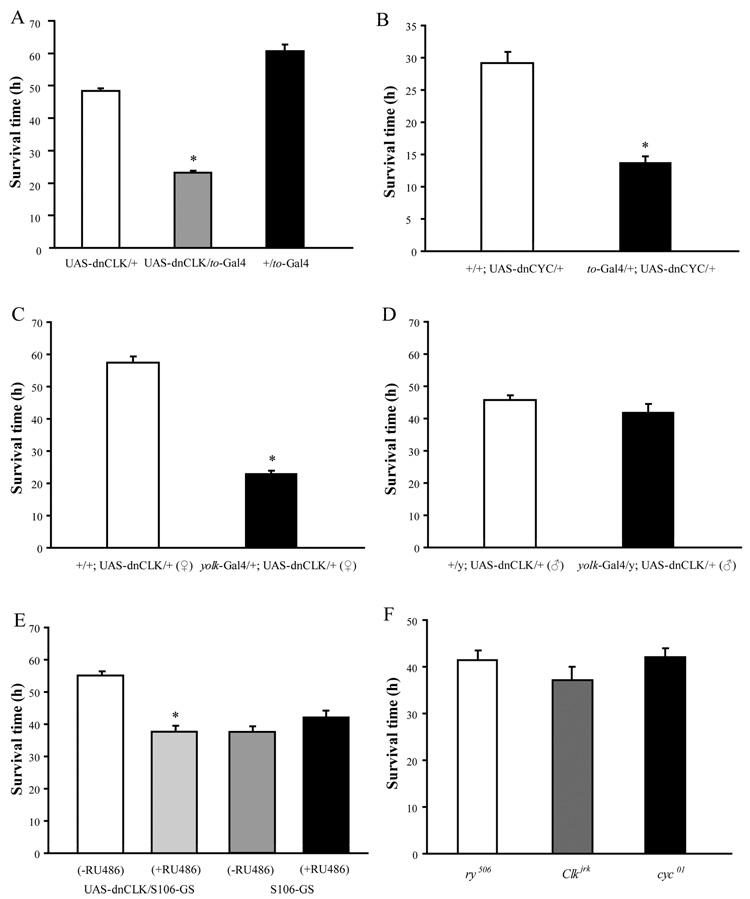
To determine if the increased food consumption in flies with disrupted metabolic clocks made them more resistant to starvation, we recorded the average survival time for food-deprived flies in DD. Contrary to our prediction, we found that flies with disrupted metabolic clocks were more sensitive to starvation than control flies (Fig 4A). Expression of dnCYC with the to-Gal4 driver caused an even more severe phenotype than dnCLK in the response to starvation (Fig 4B). To confirm that the starvation sensitive phenotype was due to the loss of the fat body clock, we used yolk-Gal4, an adult female fat body specific Gal4, to drive the expression of dnCLK (Georgel et al., 2001). Disruption of the fat body clock in this fashion resulted in dramatically reduced survival in response to starvation in virgin female flies (Fig 4C). However, consistent with the female specific expression of yolk-Gal4, male flies carrying UAS-dnCLK and yolk-Gal4 did not display increased sensitivity to starvation (Fig 4D). Finally, to minimize the effect of different genetic backgrounds on the starvation response, we used a drug inducible fat body driver, S106-GS-Gal4 to drive the expression of dnCLK. Flies carrying these constructs showed increased sensitivity to starvation only when they were treated with mifepristone (RU486) from the first instar larval stage onwards (Fig 4E).
Figure 4. Flies with a disrupted fat body clock are more sensitive to starvation.
Starvation resistance was measured by recording the average survival time of 7–14 day old males or females on media containing 0% sucrose and 2% agar. Mortality was defined by the cessation of locomotor activity. (A) – (D) Sensitivity to starvation is increased in adult males carrying both UAS-dnCLK and to-Gal4 (A), adult males carrying UAS-dnCYC and to-Gal4 (B), adult females carrying UAS-dnCLK along with yolk-Gal4 (C), but not in adult males carrying both UAS-dnCLK and yolk-Gal4 (D). (E) Adult males in which the fat body clock is inducibly disrupted with a S106-GS-Gal4 driver are more sensitive to starvation when they are treated with the inducer, RU486. RU486 has no effect on control flies carrying the driver alone. (F) Male Clkjrk or cyc01 flies have similar survival times under starvation conditions compared to control ry506 flies. Statistical significance was determined by two-tailed Student’s t-test with unequal variance. Asterisks indicate significant differences (p<0.05).
Since CLK and CYC are transcriptional activators, and therefore positive regulators of the clock, their effects on clock targets are likely to be different from those of the negative regulators, PER and TIM. One would predict that over-expression of PER or TIM would yield the same effect as knockdown of CLK and CYC. Indeed, we found that over-expression of TIM with the to-Gal4 driver also increased sensitivity to starvation (Supplemental Figure 4). Based on these data, we conclude that the normal function of the fat body clock is important for flies to survive an environmental stress such as starvation.
The fat body clock is important for energy storage
Because the level of energy stores is critical for survival during starvation conditions, we hypothesized that the increased sensitivity to starvation of flies with a disrupted fat body clock was caused by decreased energy storage. To test this hypothesis, we measured levels of one of the major energy stores, glycogen, in control and experimental flies. As predicted, glycogen levels were extremely low in flies in which the fat body clock was disrupted by expression of either dnCLK (Fig 5A) or dnCYC (Fig 5B) with to-Gal4. Disruption of the fat body clock in female flies with a yolk-Gal4 driver also led to a similar low glycogen phenotype (Fig 5C). In contrast, male flies carrying yolk-Gal4 and UAS-dnCLK did not display such a phenotype (data not shown). The other major form of energy storage, lipid, was only slightly decreased in flies with a disrupted fat body clock as compared to control flies (Supplement Fig 5A and 5B). These results suggest that although flies lacking a functional clock in the fat body consume more food than control flies, they have deficits in energy storage, especially in glycogen storage, which is most likely, the basis of their starvation sensitivity.
Figure 5. The clock in the fat body affects glycogen storage.
Glycogen levels were measured in 1-week-old male flies of different genotypes. At least three independent groups of 10 adult male flies each were analyzed for each genotype. Glycogen levels were normalized to the protein concentration. (A) – (C) Glycogen levels are lower in ,males carrying both UAS-dnCLK and to-Gal4 (A), male flies carrying UAS-dnCYC and to-Gal4 (B) and Female flies carrying UAS-dnCLK and yolk-Gal4 (C). (D) Glycogen levels are normal in Clkjrk flies and equivalent to levels in control ry506 flies. Asterisks indicate significant differences (p<0.05). Statistical significance was determined by two-tailed Student’s t-test with unequal variance.
Effects of dnCLK in the fat body require the expression of CYC
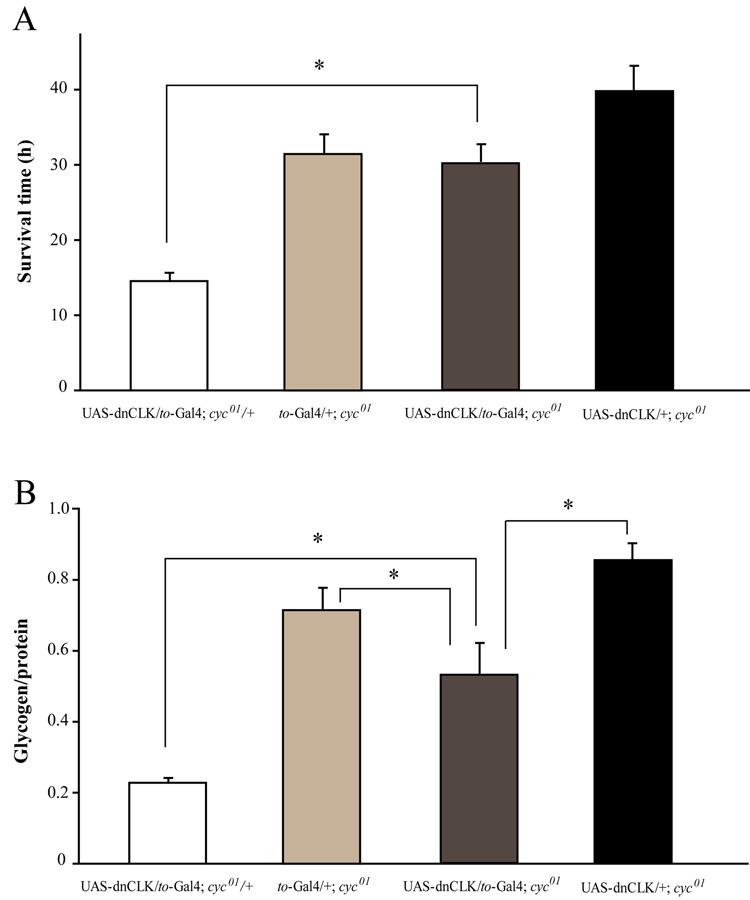
dnCLK most likely acts by interfering with clock function. However, since CLK is a bHLH transcription activator, it is possible that it interacts with, and affects the function of, other bHLH transcriptional factors. To determine if this was the case, we introduced the UAS-dnCLK and to-Gal4 transgenes into a cyc01 background. If the starvation sensitivity and low glycogen level were caused by interactions between dnCLK and non-circadian factors, then these phenotypes would not be affected by the absence of CYC protein. However, in cyc01 flies, overexpression of dnCLK in the fat body failed to affect starvation sensitivity (Fig 6A). Glycogen levels were lower than those of control cyc01 flies carrying only UAS-dnCLK or to-Gal4, but the decrease was less than that seen in heterozygous cyc01 flies with a disrupted fat body clock (Fig 6B). These data suggest that the starvation sensitivity phenotype is CYC dependent most like due to a direct interaction between dnCLK and CYC in the fat body.
Figure 6. Effects of dnCLK in the fat body require expression of CYC.
(A) and (B) Overexpression of dnCLK in the fat body of cyc01 flies does not have any effect on starvation resistance (A), and a less severe effect on glycogen storage (B). Asterisks indicate significant differences (p<0.05). Statistical significance was determined by two-tailed Student’s t-test with unequal variance. There was no significant difference in survival time between UAS-dnCLK/+;cyc01 and UAS-dnCLK/to-Gal4;cyc01 flies.
Neuronal clocks oppose effects of the fat body clock
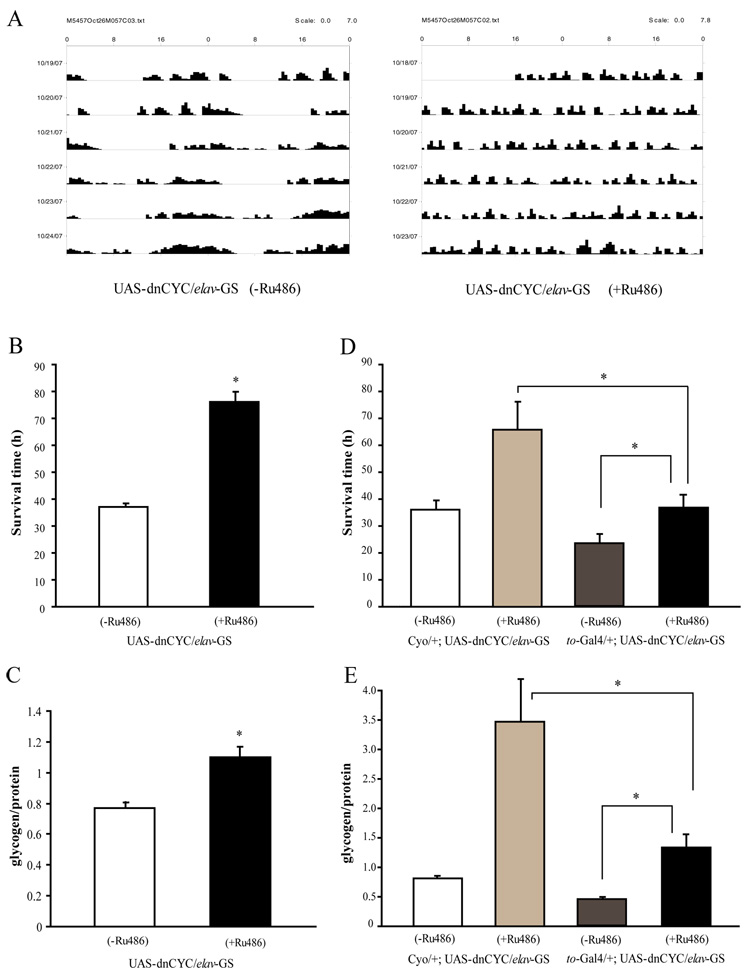
Although flies lacking a functional clock in the fat body display clear phenotypes for both starvation sensitivity and energy storage, we failed to identify any phenotypes in the clock mutants, Clkjrk and cyc01, related to either survival time under starvation conditions (Fig 4F) or glycogen storage (Fig 5D). In addition, total food consumption was unchanged in Clkjrk and cyc01 flies, as compared to control ry506 flies (Fig 3C). Given the complex nature of metabolic regulation, it is possible that clocks in other tissues oppose effects of the fat body clock. As mentioned above, we found that flies lacking a functional clock in olfactory receptor neurons had a small decrease in food consumption i.e. their phenotype was the reverse of that of flies lacking a fat body clock. However, flies expressing UAS-dnCLK under the control of Or83b-Gal4 had similar survival times under starvation as control flies (data not shown), suggesting that the effect of olfactory neurons on metabolic activity is not sufficient to result in a starvation phenotype. Thus we targeted the clock in all neuronal cells using a drug inducible panneuronal driver, elav-GS-Gal4 (Osterwalder et al., 2001), and assayed effects upon starvation and upon energy stores. UAS-dnCYC/elav-GS-Gal4 flies treated with RU486 were arrhythmic for locomotor activity in DD (Fig 7A), indicating a disruption of neuronal clocks. Interestingly, these flies were more resistant to starvation than those that did not receive RU486 treatment (Fig 7B). On the other hand, control flies carrying only UAS-dnCYC were slightly more sensitive to starvation (data not shown). We also measured nutrients in flies carrying UAS-dnCYC and elav-GS-Gal4 and found that glycogen levels were higher in the presence of RU486 (Fig 7C). These results suggest that the effects of neuronal clocks on glucose metabolism and starvation sensitivity oppose those of the clock in the fat body.
Figure 7. Clocks in neuronal cells oppose effects of the fat body clock on starvation and energy stores.
(A) Expression of dnCYC in neuronal cells abolishes the locomotor activity rhythm. Adult male flies expressing dnCYC in neuronal cells display arrhythmic locomotor activity in DD. (B) Flies expressing dnCYC in neuronal cells are less sensitive to starvation than control male flies. Asterisks indicate significant differences (p<0.05). (C) Glycogen storage is significantly higher in flies expressing dnCYC in neuronal cells as compared to control male flies. (D) and (E) Flies with both neuronal and fat body clocks disrupted have a normal response to starvation and normal glycogen stores. Flies carrying UAS-dnCYC and elav-GS-Gal4 with or without to-Gal4 were treated with RU486 to induce expression of elav-GS. In the absence of RU486, to-Gal4/+;UAS-dnCYC/elav-GS flies are more sensitive to starvation (D) and have less glycogen (E) than controls that lack to-Gal4. This is because disruption of the fat body clock increases sensitivity to starvation. However, after treatment with RU486 to induce additional disruption of neuronal clocks, the starvation response and glycogen levels are similar to those in wild type. Note that for the experiments in Figure 7D and 7E flies were treated with RU484 for ~2 weeks as compared to a 5–6 day treatment in Figure 7A –7C; this most likely accounts for the difference in the glycogen increase. Asterisks indicate significant differences (p<0.05). Statistical significance was determined by two-tailed Student’s t-test with unequal variance.
If the neuronal clocks and fat body clocks oppose each other in the control of metabolic activity, then disruption of clocks in both tissues may be expected to produce a phenotype that is close to wild type. Indeed, we found that disrupting neuronal clocks rescued both the starvation sensitivity and low glycogen level phenotypes of flies lacking a functional fat body clock (Fig 7D and 7E). These data further support the idea that clocks in neuronal cells counter metabolic effects of the clock in the fat body.
DISCUSSION
A rhythm of feeding in Drosophila
We have identified metabolic functions of circadian clocks in Drosophila. We first investigated the effect of the circadian clock on feeding behavior because of the close relationship of this behavior with metabolism. In the wild, animals tend to eat at specific times of day that may vary from one species to another. This behavioral pattern is often maintained in animals kept in experimentally controlled laboratory conditions. Using two different methods to quantify the amount of food flies eat at different times of day, we confirmed that flies tend to eat during the daytime in the presence of LD cycles. This feeding rhythm is driven by two mechanisms-one is the circadian clock, because even in constant darkness (DD) flies maintain a rhythmic feeding pattern, and moreover the DD rhythm is eliminated in clock mutant flies; the other process that can drive this rhythm is light, as Clkjrk flies display a rhythmic feeding pattern in LD cycles similar to that of wild type flies.
Feeding is one of the most essential activities of animals and so its optimization is fundamentally important. A number of environmental stimuli such as temperature, light and food availability exhibit rhythmic patterns which vary in a circadian or seasonal fashion. Animals that adapt their behaviors to these stimuli probably have a much better survival rate; this includes adopting feeding patterns that minimize exposure to predators or harsh environments while taking advantage of food availability. Although feeding rhythms have been reported in mammals (Strubbe et al., 1986), and a recent study also established the importance of a clock protein in maintaining feeding rhythms (Turek et al., 2005), they have not been dissociated from activity rhythms. Ours is the first report of a feeding rhythm in Drosophila and while we cannot exclude the possibility that the activity rhythm affects the feeding rhythm, the feeding profile of flies lacking a clock in digestive/metabolic tissues indicates that the two rhythms are dissociable (see also below).
Circadian clocks in peripheral tissues regulate feeding rhythms
Several peripheral clocks in Drosophila may be relevant for feeding behavior. For example, olfactory receptor neurons sense the attractive odor of the food, and metabolic tissues such as the fat body monitor the body energy status (Colombani et al., 2003), which would determine if the fly needs more food. To determine if clocks in these tissues underlie the feeding rhythm, we generated flies lacking clocks in each of these two tissue types and tested their feeding behavior. Flies lacking a clock in digestive/metabolic tissues tend to eat more during the subject nighttime than daytime in DD, resulting in an 8-hour shift in the peak of the feeding rhythm as compared to wild type flies. This phenotype is not caused by a shift in locomotor activity, as the locomotor activity rhythm remains unchanged (Supplemental Fig 3). In addition, the feeding rhythm is not eliminated which could, in part, be due to incomplete disruption of clock function by dominant negative CLK or CYC. However, a more likely possibility is that clocks in other tissues contribute to the regulation of the feeding rhythm, just like clocks in multiple tissues including the LNs and the pupal prothoracic gland control eclosion rhythms (Myers et al., 2003). The feeding rhythm, in turn, may regulate other aspects of energy homeostasis. For instance, we have found that the response to starvation varies in the presence of LD cycles, based upon the time of day food deprivation is initiated (Supplemental Fig 6). Since flies are most resistant if starvation is initiated in the mid-day, shortly after the time of peak feeding activity, it is likely that this rhythmic response derives from the rhythm of feeding.
The fat body clock regulates energy metabolism and total food consumption
Another phenotype of flies lacking clocks in metabolic tissues is dramatically increased consumption of food, especially during the night. This suggests that clocks in these tissues not only regulate the timing of feeding, but also affect overall food intake. Because feeding provides calories necessary for body function and helps maintain a relatively stable fat mass in the animal, we originally hypothesized that flies that eat more, due to the lack of metabolic clocks, would be resistant to starvation. However, our results showed that these flies are extremely sensitive to starvation. Locomotor activity assays did not indicate any hyperactivity phenotype of these flies, making it unlikely that the starvation sensitive phenotype is caused by increased expenditure of energy (data not shown). As the fat body is the major storage site of energy reserves in the fly, and it provides most of the energy during starvation (Scott et al., 2004), the starvation phenotype could be secondary to a defect in energy storage by the fat body after feeding, or a defect in energy release during starvation. To address this question, we measured the levels of two major energy sources, glycogen and lipid, in flies lacking a clock in the fat body, and found a dramatic decrease in glycogen levels and a slight decrease of lipid levels. Because the flies we use for starvation tests are maintained on 2% agar medium containing 5% sucrose for about a week, lipid stores are likely limited in both controls and experimental flies, which was confirmed by our lipid assay (data not shown); thus, we surmise that the hypersensitivity to starvation is caused by the defect in glycogen storage. However, it is possible that lipid stores under starvation conditions are differentially affected in flies lacking a fat body clock.
The deficiency in energy storage may also explain the increased food consumption of flies lacking a clock in the fat body, as the fat body is the tissue responsible for both energy storage and homeostasis control (Colombani et al., 2003). Changes in energy stores may initiate a signaling cascade which involves the release of metabolites like glucose and free fatty acids, or nutrient sensors such as slimfast (Colombani et al., 2003), or even hormones that regulate feeding behavior and nutrient homeostasis, such as AKH and insulin (Lee and Park, 2004; Rulifson et al, 2002). Although the fat body itself does not generate these hormones, it may signal to neurons expressing these peptides.
The clock in the fat body regulates the expression of important metabolic genes
In the mammalian liver, hundreds of genes oscillate diurnally and are likely under clock regulation (Panda et al., 2002, Storch et al., 2002); many of these genes are important for metabolic functions. The Drosophila fat body is a tissue with functions similar to those of the mammalian liver and adipose tissue. To determine whether the clock in the fat body regulates energy metabolism by regulating the expression of metabolic genes, we searched for clock-controlled genes in the fat body using microarray analysis (unpublished data) and identified a number of cyclically expressed metabolic genes. One such gene is zw which encodes glucose-6-phosphate I dehydrogenase, the rate-limiting enzyme in the pentose phosphate pathway, one of the major pathways of carbohydrate metabolism. zw is normally expressed. at high levels at the end of the day, but is high at all times in flies with a disrupted fat body clock (Supplemental Fig 2). Although we do not yet know if zw contributes to the phenotype of flies with a disrupted fat body clock, our findings suggest that the fat body clock regulates metabolic processes by regulating the expression of some key genes.
Neuronal clocks oppose the effects of the fat body clock on metabolism
Our finding that both neuronal and fat body clocks have effects on fly metabolism indicates the intricacy of the circadian network and the complex regulation of energy homeostasis. Flies with disrupted neuronal clocks showed a phenotype opposite that displayed by flies lacking a clock in the fat body- they had high glycogen levels and were resistant to starvation. Blocking clock function in olfactory receptor neurons alone had no effect, indicating that clocks in neurons other than olfactory receptor neurons are responsible for this phenotype. Interestingly, a recent report showed that the circadian LNd neurons in the brain express NPF, a peptide important for the regulation of feeding (Lee et al., 2006), so it is possible that this group of neurons participates in regulating feeding behavior and energy metabolism. However, other clock gene expressing neurons may also have metabolic functions.
Although we did not find energy storage defects in flies lacking olfactory clocks, we found that these flies show slightly decreased food consumption. Olfactory receptor neurons are the major chemosensory cells in Drosophila. They send olfactory signals through the antennal lobe to the calyx of the mushroom body (reviewed by Davis, 2001), which is also the site where a major food-intake regulating peptide, sNPF, is secreted (Lee et al., 2004). Since disruption of the clock in olfactory neurons results in reduced food intake, we speculate that normally olfactory clocks increase the fly’s sensitivity to food and thus counterbalance the regulatory effect of the clock in the fat body. Of course, since feeding behavior appears to be under multiple controls, clocks in other tissues may also be involved in regulating total food consumption.
Interaction between clocks in different tissues
In this study we discovered an interesting phenomenon, namely that some metabolic phenotypes can only be observed in flies that lack clocks in certain tissues. Moreover, in some cases, blocking clocks in different tissues yields totally different phenotypes. As a result, clock mutant flies do not show any difference compared to wild type flies. If this is a common phenomenon then traditional circadian studies, which focus on the phenotype of clock mutant animals, could miss some important clock functions. Also, our findings indicate the importance of connections between different clocks in regulating behavioral rhythms and other physiological functions. Given that circadian clocks are located in many tissues with varying biological functions, it is extremely important for animals to have a well-organized network to coordinate these different clocks and ensure a consistent output. Mammals use a clock in the SCN to synchronize peripheral clocks to light (Liu et al., 2007). In Drosophila, although the central clock is required for the functioning of some peripheral clocks like the prothoracic gland clock (Myers et al., 2003), many others are not dependent on the central clock at all, and little is known about how these different clocks communicate (Tanoue et al., 2004). Our studies show how the feeding rhythm and glycogen storage are regulated by different clock systems; clocks in the fat body and in neurons regulate glycogen storage and likely the feeding rhythm, but neither is necessary. Considering the likely importance of a feeding rhythm to flies in the wild, multiple levels of regulation may increase their chances of survival. Although we do not know the signals used by these two clock systems to communicate with each other, at least at the physiological and behavioral level the two systems are functionally related. This relationship in Drosophila may have evolved to an even more complex circadian system in mammals.
EXPERIMENTAL PROCEDURES
Adult fat body collection
Flies (about 1 week old) were entrained for at least 3 days in 12h light:12 h dark cycles (LD) at 25°C before they were collected on dry ice. To collect abdominal fat bodies, the abdomen of the fly was cut open and internal organs were removed; the remaining epidermis, including the fat body attached to it, was collected for either RNA or protein preparation.
Real-Time Quantitative PCR
For each time point, fat bodies from 20 fly bodies were collected for RNA preparation. Total RNA was extracted using Trizol reagent (Invitrogen). All RNA samples were treated with RNase-free DNase (Promega). cDNA was synthesized using a high capacity cDNA reverse transcription kit (Applied Biosystems). The levels of mRNA for different genes were measured by using SYBR-GREEN PCR Mix (Applied Biosystems) on an ABI 7000 Sequence Detection System (Applied Biosystems). The following primers were designed using ABI PrimerExpress software- for actin mRNA: 5’ primer, gcg cgg tta ctc ttt cac ca; 3’ primer, atg tca cgg acg att tca cg; for tim mRNA: 5’ primer, tgg ctg cac tga tgg act tg; 3’ primer, ccc agc gat tgc att gg. The level of actin mRNA was used as a control for the total RNA content in each sample. The values for other RNAs were normalized to those of actin.
Western blot analysis
1 week old adult female flies were entrained to 12 hr:12hr light:dark cycles for 3 days and abdominal fat bodies along with the attached epidermis were dissected at the time points indicated in Figure 2. Western blot analysis was performed as described previously (Sathyanarayanan et al. 2004). Rat anti-TIM and Rabbit anti-VRI antibodies were used at 1:1000 dilutions. Following ECL, blots were exposed to film and images were scanned into Adobe Photoshop.
Feeding assay
The feeding assay was modified from Robert Edgecomb et al (Edgecomb et al., 1994). Briefly, male flies aged 3–10 days were entrained at 25°C in LD for three days, and then transferred to constant darkness (DD). On the third day in DD, flies were switched from normal food (cornmeal/yeast/molasses/agar) to blue-color food (15% sucrose/1% agar diets containing 1% FD&C Blue No.1 (McCormick)) for 2 hours at different time points. As described by Robert S.E. (Edgecomb et al., 1994), FD&C Blue No.1 remains in the digestive tract and is not affected by gut pH and enzymes. The feeding profile for each line was determined in at least three independent experiments. Each independent experiment consisted of 1–2 groups (of 30 flies each) assayed at each of six time points over a 24 hour cycle. After the feeding, the flies were frozen on dry ice immediately. To prevent eye pigment from interfering with the absorbance spectrum of the dye, fly bodies were separated from heads, homogenized in PBS buffer and centrifuged (13000 rpm) for 25min. The supernatants were transferred to a new tube and, following a 25-minute spin at 13000 rpm, were transferred to cuvettes. Absorbance was measured at 625 nm. The absorbance measured for supernatants from flies fed with normal food was subtracted from the absorbance of the supernatant from blue food-fed flies. The net absorbance reflected the amount of food ingested.
For the CAFÉ assay, we simplified the original version (Ja et al., 2007) by using normal vials with 1% agar as a water source. The assay was performed on the third day in DD after LD entrainment. 12 hours before the assay, about 10 flies were transferred from normal food vials to agar vials and fed 5% sucrose solution maintained in calibrated glass micropipettes (5 µl, catalog no. 53432-706; VWR, West Chester, PA). At the start of the assay, the old micropipettes were replaced by new experimental ones containing the same type of food; after 2 hours, the old micropipettes were placed back in. The amount of liquid consumed from the experimental micropipette was recorded, and the evaporation effect was measured by measuring the change in liquid volume in a micropipette placed in a vial without a fly.
Starvation resistance assay
~1 week old flies raised on normal food were loaded into locomotor activity tubes containing 5% sucrose/2% agar food, and entrained for 3 days in LD conditions. To determine if the resistance to starvation is rhythmic, starvation was initiated at different times for w1118 flies maintained in an LD cycle. Thus, every two hours, a group of 16 male flies was transferred to tubes containing agar without sucrose. For other starvation tests, flies were moved to DD and starvation was initiated at different circadian times on the third day in DD. Since no clear rhythm of starvation resistance was detected in DD, the data from different time points were later pooled. For each independent experiment, 16 control and 16 experimental flies were tested at each time point, and the experiment was repeated at least three times. Prior to starvation, locomotor activity was recorded in LD and DD to determine the behavior of flies of different genotypes. Fly locomotor activity was monitored using the Trikinetics DAM system. Fly mortality was defined by the cessation of locomotor activity.
Glycogen and Triglyceride measurement
For each independent experiment, 20–30 male flies were divided into 2–3 groups, each consisting of 10 flies. For experiments involving drug induction, flies were raised on 5% sucrose agar with or without drug for at least 5 days or as indicated in the figure legends; in other cases, flies were raised on normal food. Flies were collected without entrainment since no clear rhythm of glycogen levels was detected in the presence of LD cycles, After homogenization on ice in 250ul of homogenization buffer [10mM KH2PO4, 1mM EDTA (pH7.4)], the fly homogenates were centrifuged at 2000 rpm for 2 min in a 4°C room. The supernatant was moved into a 1.5-ml tube. Protein measurement was performed using Quick-Start Bradford Reagent 500-0205 and BSA Std. 500-0007 from Bio-Rad. Samples were loaded onto a 96-well plate (Corning 3595, 360ul) and the absorbance at 595nm was recorded using a 1420 Multilabel counter (Perkin-Elmer). Protein concentrations were used to normalize glycogen and triglyceride levels in different samples. Triglyceride levels were measured using Kit TR100-1KT and standard G7793 (Sigma). The absorbance at 540nm was recorded using the 1420 Multilabel counter. To measure glycogen levels in the homogenates, glycogen was first reduced to glucose using A1602-25MG amyloglucosidase (Sigma), then glucose levels were measured by a method similar to that used for determining blood glucose (Raabo and Terkildsen, 1960). Briefly, glucose oxidase and peroxidase (PGO Enzymes) solution was made by adding the contents of one capsule of P7119-10CAP PGO enzyme (Sigma) into 100 ml water, and the o-Dianisidine solution was made by dissolving 50mg of o-Dianisidine hydrochloride in 20 ml of water. To make the PGO enzyme reaction solution, 100 ml PGO Enzymes solution was mixed with 1.6 ml of the o-Dianisidine solution. The final reaction system contained 1.5ul homogenate, 2.5ul 0.1 unit/µl amyloglucosidase and 221µl PGO enzyme reaction solution. After incubating for 30 min at 37°C, absorbance at 450nm was recorded using a 1420 Multilabel counter and the glucose level was determined from the glucose standard curve. A G0885-1G glycogen (Sigma) standard was used to determine the amount of glycogen reduced in the homogenate. Each experiment was repeated at least three times.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Paul Hardin for UAS-dnCLK, UAS-dnCYC and Or83b-Gal4 flies and helpful discussions. We thank Karen Ho and Susan Harbison for invaluable suggestions on our experimental design, and other members of the laboratory for comments on the manuscript. This work was supported in part by National Institutes of Health Grant NS048471.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Ando H, Yanagihara H, Hayashi Y, Obi Y, Tsuruoka S, Takamura T, Kaneko S, Fujimura A. Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology. 2005;146:5631–5636. doi: 10.1210/en.2005-0771. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Herzog ED. Come together right…now synchronization of rhythms in amammalian circadian clock. Neuron. 2005;48:531–534. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A. Clock genes in mammalian peripheral tissues. Cell Tissue Res. 2002;309:193–199. doi: 10.1007/s00441-002-0585-0. [DOI] [PubMed] [Google Scholar]
- Boden G, Chen X, Polansky M. Disruption of circadian insulin secretion is associated with reduced glucose uptake in first-degree relatives of patients with type 2 diabetes. Diabetes. 1999;48:2182–2188. doi: 10.2337/diabetes.48.11.2182. [DOI] [PubMed] [Google Scholar]
- Bray MS, Young ME. Circadian rhythms in the development of obesity: potential role for the circadian clock within the adipocyte. Obes. Rev. 2007;8:169–181. doi: 10.1111/j.1467-789X.2006.00277.x. [DOI] [PubMed] [Google Scholar]
- Colles SL, Dixon JB, O'Brien PE. Night eating syndrome and nocturnal snacking: association with obesity, binge eating and psychological distress. Int. J. Obes. (Lond) 2007;31:1722–1730. doi: 10.1038/sj.ijo.0803664. [DOI] [PubMed] [Google Scholar]
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwalder B, Tsujimoto S, Moss J, Mattox W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002;16:2879–2892. doi: 10.1101/gad.1010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL. Mushroom bodies, Ca(2+) oscillations, and the memory gene amnesiac. Neuron. 2001;30:653–656. doi: 10.1016/s0896-6273(01)00329-4. [DOI] [PubMed] [Google Scholar]
- Edgecomb RS, Harth CE, Schneiderman AM. Regulation of feeding behavior in adult Drosophila melanogaster varies with feeding regime and nutritional state. J. Exp. Biol. 1994;197:215–235. doi: 10.1242/jeb.197.1.215. [DOI] [PubMed] [Google Scholar]
- Fukada Y, Okano T. Circadian clock system in the pineal gland. Mol. Neurobiol. 2002;25:19–30. doi: 10.1385/MN:25:1:019. [DOI] [PubMed] [Google Scholar]
- Fuller PM, Lu J, Saper CB. Differential rescue of light- and food-entrainable circadian rhythms. Science. 2008;320:1074–1077. doi: 10.1126/science.1153277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- Giebultowicz JM. Molecular mechanism and cellular distribution of insect circadian clocks. Annu. Rev. Entomol. 2000;45:769–793. doi: 10.1146/annurev.ento.45.1.769. [DOI] [PubMed] [Google Scholar]
- Giebultowicz JM, Ivanchenko M, Vollintine T. Organization of the insect circadian system: spatial and developmental expression of clock genes in peripheral tissues of Drosophila melanogaster. In: Denlinger DL, Giebultowicz JM, Saunders DS, editors. In Insect Timing: Circadian rhythmicity to seasonality. Amsterdam, Netherlands: Elsevier Science B.V.; 2001. pp. 31–42. [Google Scholar]
- Hara R, Wan K, Wakamatsu H, Aida R, Moriya T, Akiyama M, Shibata S. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr. Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Prandiology of Drosophila and the CAFE assay. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Bass J. A sense of time: how molecular clocks organize metabolism. Trends Endocrinol. Metab. 2007;18:4–11. doi: 10.1016/j.tem.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Lee G, Bahn JH, Park JH. Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12580–12585. doi: 10.1073/pnas.0601171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, You KH, Choo JK, Han YM, Yu K. Drosophila short neuropeptide F regulates food intake and body size. J. Biol. Chem. 2004;279:50781–50789. doi: 10.1074/jbc.M407842200. [DOI] [PubMed] [Google Scholar]
- Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat. Chem. Biol. 2007;3:630–639. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
- Myers EM, Yu J, Sehgal A. Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr. Biol. 2003;13:526–533. doi: 10.1016/s0960-9822(03)00167-2. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Ptitsyn AA, Zvonic S, Conrad SA, Scott LK, Mynatt RL, Gimble JM. Circadian clocks are resounding in peripheral tissues. PLoS Comput. Biol. 2006;2:e16. doi: 10.1371/journal.pcbi.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabo E, Terkildsen TC. On the enzymatic determination of blood glucose. Scand. J. Clin. Lab. Invest. 1960;12:402–407. doi: 10.3109/00365516009065404. [DOI] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Shibata S. Neural regulation of the hepatic circadian rhythm. Anat. Rec. A.Discov. Mol. Cell. Evol. Biol. 2004;280:901–909. doi: 10.1002/ar.a.20095. [DOI] [PubMed] [Google Scholar]
- Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Strubbe JH, Keyser J, Dijkstra T, Prins AJ. Interaction between circadian and caloric control of feeding behavior in the rat. Physiol. Behav. 1986;36:489–493. doi: 10.1016/0031-9384(86)90320-3. [DOI] [PubMed] [Google Scholar]
- Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr. Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, Ivanova E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Sehgal A. Molecular components of the circadian system in Drosophila. Annu. Rev. Physiol. 2001;63:729–755. doi: 10.1146/annurev.physiol.63.1.729. [DOI] [PubMed] [Google Scholar]
- Young ME. The circadian clock within the heart: potential influence on myocardial gene expression, metabolism, and function. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H1–H16. doi: 10.1152/ajpheart.00582.2005. [DOI] [PubMed] [Google Scholar]
- Young ME, Bray MS. Potential role for peripheral circadian clock dyssynchrony in the pathogenesis of cardiovascular dysfunction. Sleep Med. 2007;8:656–667. doi: 10.1016/j.sleep.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonic S, Floyd ZE, Mynatt RL, Gimble JM. Circadian rhythms and the regulation of metabolic tissue function and energy homeostasis. Obesity (Silver Spring) 2007;15:539–543. doi: 10.1038/oby.2007.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.