Abstract
The major histocompatibility complex (MHC) on chromosome 6 is associated with susceptibility to more common diseases than any other region of the human genome, including almost all disorders classified as autoimmune. In type 1 diabetes the major genetic susceptibility determinants have been mapped to the MHC class II genes HLA-DQB1 and HLA-DRB1 (refs 1-3), but these genes cannot completely explain the association between type 1 diabetes and the MHC region4-11. Owing to the region’s extreme gene density, the multiplicity of disease-associated alleles, strong associations between alleles, limited genotyping capability, and inadequate statistical approaches and sample sizes, which, and how many, loci within the MHC determine susceptibility remains unclear. Here, in several large type 1 diabetes data sets, we analyse a combined total of 1,729 polymorphisms, and apply statistical methods—recursive partitioning and regression—to pinpoint disease susceptibility to the MHC class I genes HLA-B and HLA-A (risk ratios>1.5; Pcombined=2.01×10-19 and 2.35×10-13, respectively) in addition to the established associations of the MHC class II genes. Other loci with smaller and/or rarer effects might also be involved, but to find these, future searches must take into account both the HLA class II and class I genes and use even larger samples. Taken together with previous studies4-8,10-16, we conclude that MHC-class-I-mediated events, principally involving HLA-B*39, contribute to the aetiology of type 1 diabetes.
The MHC spans 4 megabases (Mb) and contains 149 genes, of which eight (the class II loci HLA-DRB1, HLA-DQB1, HLA-DQA1, HLA-DPB1, HLA-DPA1; the class I loci HLA-A, HLA-B and HLA-C) are the highly polymorphic immune response genes. There are many other candidate genes with common variants—any one of which or a combination thereof—that might also be involved in disease susceptibility. We studied 850 type-1-diabetes-affected sibling-pair (ASP) families from the United Kingdom and the United States and a first set of 2,049 type 1 diabetes patients and 1,912 controls from across Great Britain, in which we genotyped a combined total of 254 polymorphic MHC loci, including HLA-DQB1, HLA-DRB1, HLA-A and HLA-B (Table 1 and Supplementary Table 1). A second independent set of 1,050 type 1 diabetes cases and 1,125 controls was used for validation. After these analyses were completed, 1,475 additional single nucleotide polymorphisms (SNPs) in 1,964 of our type 1 diabetes cases and 2,923 controls became available as part of our collaboration with the Wellcome Trust Case Control Consortium (WTCCC)17 (Table 1).
Table 1.
Data sets used to test for unconditional single locus and MHC-class-II-independent type 1 diabetes associations in the MHC
| Data set | Size of data set | Classical MHC genes genotyped in data set | SNPs and microsatellite markers genotyped in data set |
|---|---|---|---|
| Family set | 850 affected sibling-pair (ASP) families from the United Kingdom and the United States |
HLA-DQB1, HLA-DRB1, HLA-DQA1, HLA-DPB1, HLA-A, HLA-B and HLA-C* |
27 microsatellites; 46 SNPs in candidate genes† |
| First case–control set | 2,049 type 1 diabetes patients and 1,912 controls from Great Britain |
HLA-DQB1, HLA-DRB1, HLA-A and HLA-B* |
7 SNPs in the candidate genes‡; 169 nsSNPs from the GWA scan§ |
| Second case–control set (for validation of the HLA-A association) |
Additional 1,050 type 1 diabetes patients and 1,125 controls from Great Britain |
HLA-DQB1, HLA-DRB1 and HLA-A | Not applicable |
| WTCCC case–control set | 1,964 type 1 diabetes patients and 2,923 controls—overlaps with the main case– control set in 1,281 type 1 diabetes cases and 860 controls |
HLA-DQB1, HLA-DRB1, HLA-A and HLA-B: available for 1,281 type 1 diabetes patients and 860 controls |
1,475 SNPs from WTCCC GWA scan |
| WTCCC follow-up case–control set |
2,484 type 1 diabetes patients and 2,019 controls |
HLA-DQB1, HLA-DRB1, HLA-A and HLA-B |
The eight most class-II-independently- associated SNPs from the WTCCC scan |
Forty-six newly typed SNPs from fifteen candidate immune genes in or near the MHC (ITPR3, HLA-DPB1, HLA-DMA, HLA-DMB, PPP1R2P1, TAP1, TAP2, HLA-DOB, BTNL2, C6orf25, LY6G6C, NCR3, TNFA, LTA and NFKBIL1).
SNPs rs241447 and rs241448 from TAP2, rs1800750 from TNFA, and rs2296336, rs3131020, rs1233478 and rs389419 in the recently reported type-1-diabetes-associated genes ITPR3 (ref. 23), UBD and MAS1L (ref. 24).
One-hundred-and-sixty-nine nsSNPs were part of a genome-wide association (GWA) scan of over 12,000 nsSNPs30, spanning the entire 10-Mb extended MHC region. These SNPs were analysed in the case–control set and an additional 2,077 type 1 diabetes patients and 2,482 controls from Great Britain that were not typed at the MHC class II loci.
As expected1-3,18, the strongest type 1 diabetes associations mapped to the MHC class II genes HLA-DQB1 and HLA-DRB1 (P=10-117 and P=10-124, respectively, for the genotype model in the families, and similarly, P<10-300 and P=10-300, respectively, in the first case–control set; Fig. 1 and Supplementary Table 2, see also http://dil.t1dbase.org/page/poster/mhc_association). The data did not fit the multiplicative model owing to the known epistatic interactions between alleles and dominance effects of HLA-DRB1 and HLA-DQB1 genotypes2,19.
Figure 1. Association analyses across the MHC.
a, b, -log10(P) versus chromosome position. Unconditional single locus analyses are presented for loci typed in up to 850 families (a) and in up to 2,049 cases and 1,125 controls (b, first case–control set). c, d, Analyses conditional on HLA-DRB1 and HLA-DQB1 in the families (c) and in the first case–control set (d). Results are listed in Supplementary Tables 1 and 2.
There was evidence for a secondary peak of type 1 diabetes association around HLA-B (P=3.44×10-30 and 3.59×10-42, in the families and the case–control set, respectively; Fig. 1 and Supplementary Table 2). To test whether these were MHC-class-II-independent effects, or merely reflected linkage disequilibrium with class II, we had to use a method that takes into account the complex multi-allelic effects of the highly disease-associated HLA-DQB1 and HLA-DRB1 genes. We compared three strategies for grouping class II genotypes in our families (Supplementary Results). The P-value for the test locus, conditional on the class II genotypes, was, at some loci, dependent on the method adopted for grouping the class II loci (Supplementary Results and Supplementary Table 3). Hence, these methods were unsatisfactory and we adopted a classification tree approach, namely, recursive partitioning20-22 (http://cran.r-project.org). This is a risk-categorization method of grouping that differs from other risk-based grouping methods because it does not require the risk to be known a priori. The method classifies individuals as affected or unaffected using their class II genotypes by carrying out a series of binary splits on the basis of those class II genotypes, such that homogeneity with respect to disease status (risk) is maximized for each group while retaining good statistical power (see Methods). In contrast to other grouping methods considered, the recursive partitioning model provided consistent results (Supplementary Table 3).
In the families, using the optimized tree model, we found evidence of an additional effect of HLA-B (P=4.19×10-7), HLA-DPB1 (P=2.21×10-5) and of a SNP in the TAP2 gene (rs241448, P=5.29×10-5; Fig. 1 and Supplementary Table 2). In the case–control set, we again found evidence of an independent effect of HLA-B (P=1.74×10-7) over and above the combined effect of HLA-DQB1 and HLA-DRB1, as well as an independent effect of HLA-A (P=1.67×10-10). No evidence was obtained for independent effects of the 169 non-synonymous (ns)SNPs, or the seven candidate SNPs (P>0.001, Fig. 1 and Supplementary Table 2). Specifically, no evidence was found for association of the TAP2 SNP (rs241448, P=0.074), the recently reported ITPR3 SNP23 (rs2296336), nor for the UBD and MAS1L gene regions24 (Supplementary Results and Supplementary Table 2).
After conditioning on HLA-B, HLA-DRB1 and HLA-DQB1, we had 49% power to find an effect of odds ratio 2.0 in the first case–control set, assuming a minor allele frequency of 0.1 at α=1×10-5 with HLA-B, HLA-DRB1 and HLA-DQB1 in the model (Supplementary Methods). Hence, we conditioned on HLA-B, HLA-DRB1 and HLA-DQB1, obtaining evidence that HLA-A was independently associated with type 1 diabetes (P=2.31×10-7), as was rs4151651 (P=8.13×10-5), a nsSNP in the complement factor B (CFB) gene. However, we only had 13% power to test for additional associations to HLA-B, HLA-DRB1 and HLA-DQB1 in our 850 families, probably accounting for our failure to detect the HLA-A association in these families. So we sought to replicate the HLA-A result in an independent 1,050 cases and 1,125 controls (Table 1, second case–control set), obtaining convincing confirmatory evidence at P=1.77×10-5 after conditioning on both HLA-DRB1 and HLA-DQB1 (Supplementary Table 4).
Having taken into account the combined effect of HLA-DQB1 and HLA-DRB1, as above, we found that the HLA-B*39 allele (where * represents the allele) was consistently associated with type 1 diabetes susceptibility (relative risk=3.55 (95% confidence interval 2.21–5.72) in the families; odds ratio=2.41 (95% confidence interval 1.49–3.89) in the first case–control set; Table 2 and Supplementary Table 5). Moreover, HLA-B*39 was also associated with a lower age-at-diagnosis of type 1 diabetes in the families (P=0.0022) and in the cases from the first case–control set (P=0.0021; Supplementary Table 5). Once the association of HLA-B*39 was taken into account, there was no association of other HLA-B alleles in the families (P=0.047). Nevertheless, in the first case–control set HLA-B*18 conferred susceptibility to and HLA-B*27 protection from type 1 diabetes (Table 2 and Supplementary Table 5). These HLA-B allele associations were still present after conditioning on HLA-A as well as HLA-DRB1 and HLA-DQB1 combined (Table 2).
Table 2.
Type 1 diabetes association of the HLA-B alleles (with frequencies >0.015) conditioned on the MHC class II genes and HLA-A
| HLA-B allele | Allele frequency* |
Families (RR (95% CI)) |
Case–control set (OR (95% CI)) |
||
|---|---|---|---|---|---|
| Number of cases (%) |
Number of controls (%) |
Conditioning on HLA-DQB1 and HLA-DRB1 |
Conditioning on HLA-DQB1 and HLA-DRB1 |
Conditioning on HLA-DQB1, HLA-DRB1 and HLA-A |
|
| HLA-B*39 | 143 (4.7) | 79 (2.4) | 3.55 (2.21–5.72) | 2.41 (1.49–3.89) | 1.92 (1.16–3.19) |
| HLA-B*18 | 202 (6.7) | 128 (3.8) | 1.77 (1.24–2.53) | 1.83 (1.19–2.82) | 1.95 (1.20–3.15) |
| HLA-B*13 | 43 (1.4) | 56 (1.7) | 1.17 (0.65–2.12) | 1.94 (0.98–3.85) | 1.88 (0.90–3.90) |
| HLA-B*08 | 788 (26.1) | 461 (13.8) | 1.26 (0.96–1.65) | 0.95 (0.72–1.27) | 1.24 (0.87–1.76) |
| HLA-B*55 | 37 (1.2) | 71 (2.1) | 0.73 (0.38–1.39) | 1.28 (0.64–2.56) | 1.19 (0.58–2.47) |
| HLA-B*07 | 249 (8.2) | 465 (13.9) | 1.14 (0.84–1.55) | 1.25 (0.88–1.78) | 1.11 (0.76–1.61) |
| HLA-B*44 | 344 (11.4) | 567 (17.0) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| HLA-B*51 | 78 (2.6) | 109 (3.3) | 0.96 (0.61–1.52) | 0.93 (0.54–1.62) | 0.93 (0.52–1.66) |
| HLA-B*15 | 388 (12.8) | 267 (8.0) | 1.24 (0.91–1.69) | 0.98 (0.70–1.36) | 0.87 (0.61–1.23) |
| HLA-B*35 | 130 (4.3) | 197 (5.9) | 1.00 (0.70–1.43) | 0.82 (0.55–1.22) | 0.85 (0.55–1.31) |
| HLA-B*40 | 257 (8.5) | 232 (6.9) | 1.17 (0.86–1.61) | 0.87 (0.61–1.22) | 0.85 (0.59–1.22) |
| HLA-B*37 | 21 (0.69) | 51 (1.5) | 1.03 (0.51–2.08) | 0.67 (0.28–1.58) | 0.71 (0.28–1.79) |
| HLA-B*14 | 54 (1.8) | 159 (4.8) | 1.16 (0.75–1.80) | 0.66 (0.39–1.12) | 0.67 (0.38–1.17) |
| HLA-B*57 | 26 (0.86) | 143 (4.3) | 0.76 (0.37–1.55) | 0.50 (0.25–1.00) | 0.52 (0.26–1.07) |
| HLA-B*27 | 113 (3.7) | 145 (4.3) | 1.02 (0.69–1.52) | 0.52 (0.34–0.80) | 0.51 (0.33–0.80) |
Alleles are ordered by risk in the case–control set (once HLA-DQB1, HLA-DRB1 and HLA-A have been accounted for). The most common allele, HLA-B*44, gives the tightest 95% confidence intervals, so is used as a reference. Results are given for the families (736) and the first case–control set (1,451 type 1 diabetes patients and 1,628 controls) that were successfully typed at all four classical HLA loci. Note that HLA-B*08 is not a primary effect in type 1 diabetes and is only elevated in frequency in type 1 diabetes cases because of its strong linkage disequilibrium with HLA-DRB1*03 (D′ 0.8). CI, confidence interval; OR, odds ratios; RR, relative risks.
Allele frequencies in the families are shown in Supplementary Table 1.
In the first case–control set, having conditioned on HLA-DQB1, HLA-DRB1 and HLA-B using allele HLA-A*02 as a reference, HLA-A*01, HLA-A*11 and HLA-A*31 were protective and HLA-A*24 was predisposing for type 1 diabetes; HLA-A*03 was more predisposing than HLA-A*11 and HLA-A*31 (Supplementary Table 4). Once these alleles were accounted for, there was no further detectable HLA-A effect in the case–control set (P=0.15). In the second case–control set, having conditioned on HLA-DRB1 and HLA-DQB1, both HLA-A*01 and HLA-A*11 were again more protective than HLA-A*02. HLA-A*24 was still the most predisposing for type 1 diabetes and may also be associated with an earlier age-at-diagnosis (P=0.01; Supplementary Tables 4 and 5).
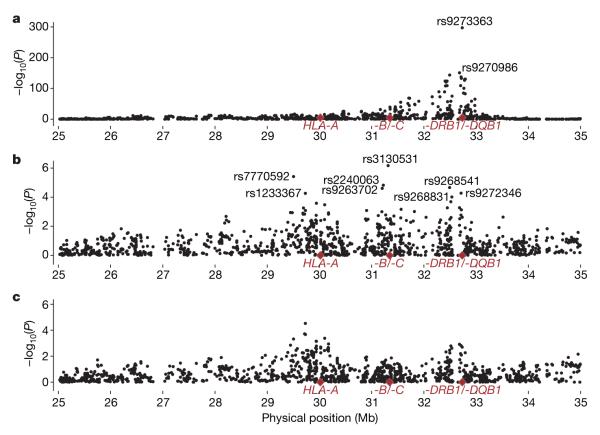
Finally, the SNPs from the WTCCC17 scan were analysed for association with type 1 diabetes. The 20 most associated SNPs all lay within the MHC class II region, with the most associated locus, rs9273363, close to HLA-DQB1 (P=4.29×10-298 in 1,964 cases and 2,923 controls; Fig. 2 and Supplementary Table 6; see also http://dil.t1dbase.org/page/poster/mhc_association). Once the effects of HLA-DRB1 and HLA-DQB1 are accounted for, the polymorphisms in the MHC class I region provide the strongest signals of association in the 1,281 cases and 860 controls genotyped at HLA-DRB1, HLA-DQB1 and the WTCCC SNPs (Fig. 2 and Supplementary Table 6). The most associated locus became rs3130531, located ~40 kilobases telomeric of HLA-C (P=6.74×10-7 compared with P=0.0056 before conditioning). Once HLA-DRB1, HLA-DQB1 and HLA-B were conditioned on, however, none of the WTCCC SNPs were convincingly associated with type 1 diabetes (Fig. 2 and http://dil.t1dbase.org/page/poster/mhc_association).
Figure 2. Association analyses of 1,475 SNPs across the MHC.
a, Unconditional single locus analysis in up to 1,964 cases and 2,923 controls—the limits of the association are at 25.9 Mb (rs1324088 P=4.65×10-6) and 34.0 Mb (rs6941621 P=9.95×10-6). b, c, Results are presented in up to 1,281 cases and 860 controls for analyses conditioned on HLA-DRB1 and HLA-DQB1 combined (annotated SNPs were followed up in a larger case–control set; Supplementary Table 6) (b), and for analyses conditioned on HLA-DRB1 and HLA-DQB1 combined and the alleles of HLA-B with frequency >0.01 (c).
Nevertheless, to increase our statistical power, we genotyped the eight WTCCC SNPs most associated with type 1 diabetes, after HLA-DRB1 and HLA-DQB1 conditioning, in a larger set of 2,484 cases and 2,019 controls with complete HLA-DRB1 and HLA-DQB1 genotyping. The most associated locus was rs9268831 (P=6.95×10-8; Supplementary Table 6); note that this was less significant than either HLA-B or HLA-A, which remained the most associated loci in the same data set after class II genes were accounted for (P=3.80×10-17 and 4.59×10-15, respectively; Supplementary Table 6). This SNP, located ~15 kb centromeric of HLA-DRA, was still associated once HLA-B was included in the model (P=5.44×10-6). In contrast, the SNP rs3130531 at 31.3 Mb was not associated in this data set after conditioning on HLA-DRB1, HLA-DQB1 and HLA-B (P=0.16; Supplementary Table 6).
Our results indicate that, once the effect of the MHC class II genes has been accounted for, most of the detectable residual association is attributable to HLA-B and HLA-A (combining all data sets, Pcombined = 2.01×10-19 and 2.35×10-13, respectively). We conclude that the existence of other major type 1 diabetes genes in the extended MHC is unlikely. Smaller independent effects, however, might still exist, necessitating future studies including analysis of rs9268831 (HLA-DRA), rs4151651 (CFB), HLA-C, HLA-DQA1 and the HLA-DP loci. The HLA-B and HLA-A alleles have previously been associated with type 1 diabetes4,5,8,10,11, but unlike these previous studies, our results localize the effects to these specific loci and alleles, thereby implicating them directly in disease aetiology.
In the nonobese diabetic mouse model of type 1 diabetes, MHC class I molecules and class-I-restricted CD8+ T cells are central to the development of autoimmune diabetes12-14. This correlates with the observations that in type 1 diabetes patients, cells infiltrating pancreatic islets are predominantly CD8+ and islet cells hyperexpress MHC class I molecules15,16. Taken together with our results, we conclude that class-I-mediated anti-islet β-cell responses are critical in type 1 diabetes and may accelerate disease onset. This might involve both the innate and adaptive immune system25,26. The HLA-A*02 allotype has been functionally and directly linked to T-cell autoreactivity to insulin27, and our results now justify investigation of the naturally processed peptides that bind the HLA-B*39 allotype as a first step towards future evaluation of inducing tolerance to such peptides in attempts to prevent type 1 diabetes.
METHODS SUMMARY
A detailed description of the methods is given in Methods and Supplementary Information. All subjects were of white ethnicity: 850 families came from established collections; 4,126 cases were Juvenile Diabetes Research Foundation/Wellcome Trust type 1 diabetes cases and 4,394 were British 1958 Birth Cohort controls. The classical loci were typed using Dynal RELI SSO assays. All loci conformed to Hardy–Weinberg equilibrium in unaffected subjects. Family data were analysed using cases with matched pseudo-controls in regression models. Stepwise logistic regression28 was used to test for effects independent of HLA-DRB1 and HLA-DQB1. The class II genotypes (rather than the alleles of HLA-DRB1 and HLA-DQB1 which do not behave multiplicatively in conferring type 1 diabetes risk) were modelled using a recursive partitioning approach20-22 (http://cran.r-project.org). These groups created by recursive partitioning, define strata within which additional loci can be tested. Pruning of the tree, that is, assessing how much of the tree to retain, was done by cross-validation29. Although when to stop pruning the tree was unclear, the number of leaves (HLA-DQB1/HLA-DRB1 groups) must be sufficient to prevent residual confounding, but not so great as to become inestimable. The recursive partitioning model was assessed using a third locus. The non-class-II loci were modelled as multiplicative effects of alleles. All analyses of the case–control set were stratified by broad geographical region17,30. Power to detect effects independent of HLA-DRB1 and HLA-DQB1 was assessed using data sets simulated from the case–pseudo-control data (and separately from the case–control set). Crucially the groupings from the recursive partitioning model were retained. A total of 100,000 replicates were performed. The Wald test was used to assess significance and subsequently the power of the study. We had 49% power to detect odds ratio=2.0 for an allele frequency of 0.1 at α=1×10-5 in the case–control set with HLA-DRB1, HLA-DQB1 and HLA-B in the model. We set a threshold of P<0.0001 for rejection of H0.
Supplementary Material
Acknowledgements
This work was funded by the Wellcome Trust and the Juvenile Diabetes Research Foundation International. We thank all of the patients, control subjects and family members for their participation. The Human Biological Data Interchange and Diabetes UK Warren repositories and UK GRID project are acknowledged for the collection of the type 1 diabetes patients and families. We acknowledge use of DNA from the British 1958 Birth Cohort collection (D. Strachan, S. Ring, W. McArdle, P. Burton, R. Jones and M. Pembrey), funded by the Medical Research Council and Wellcome Trust. S.N. is a Diabetes Research and Wellness Foundation Non-Clinical Fellow. R.D.C. and G.R. were funded by the Medical Research Council.
APPENDIX
METHODS
Subjects
The family set comprised 850 type 1 diabetes families of white ethnicity with both parents and at least two affected children in each family, including 472 Warren type 1 diabetes families from the United Kingdom (11 with only one affected offspring) and 378 Human Biological Data Interchange (HBDI) type 1 diabetes families from the United States (five with only one affected offspring). The case–control sample comprised 4,126 type 1 diabetes patients collected as part of the JDRF/WT DIL British type 1 diabetes case collection (http://www-gene.cimr.cam.ac.uk/ucdr/grid.shtml) and 4,394 controls selected from the British 1958 Birth Cohort of people born in England, Scotland and Wales during 1 week in 1958 (http://www.b58cgene.sgul.ac.uk). Of these individuals, 2,049 cases and 1,912 controls were typed at the classical MHC loci. A further 1,445 controls were from the WTCCC’s UK Blood Service samples17. The relevant research ethics committees approved the study, and written informed consent was obtained from the participants, or their parents/guardian for those too young to consent.
Grouping of HLA-DRB1 and HLA-DQB1 alleles and genotypes: recursive partitioning
In the families, cases and matched pseudo-controls were generated28 and the matching discarded so as to be able to run recursive partitioning in the recursive partitioning library in R (http://cran.r-project.org; refs 20-22). Cases and pseudo-controls were matched when doing later analyses. The alleles of HLA-DRB1 and HLA-DQB1 do not behave multiplicatively in conferring type 1 diabetes risk, so, to allow for dominance effects, the genotypes of the individual loci were modelled, as this does not assume a specific mode of inheritance. All possible binary splits of the data corresponding to presence or absence of various different genotypes at HLA-DRB1 and HLA-DQB1 were considered. The split that best categorized the data as cases and pseudo-controls, which corresponds to the split that maximizes the reduction in impurity (or maximizes the homogeneity of the cases or pseudo-controls within groups), was chosen. We chose the information index, which has the form f(p) = -2plog(p) (where p = the proportion of observations in a node that for future samples belong to a different class) as the impurity measure because it is likelihood based. This process was repeated until no further improvement could be made or the minimum group size was met. The terminal ‘leaves’ of the tree represent optimized groups of the HLA-DQB1/HLA-DRB1 genotypes and so define strata within which additional MHC loci can be tested. However, the trees generated are often complex and need to be pruned. Pruning of the tree, that is, assessing how much of the tree to retain, was done by cross-validation29. Nevertheless, when to stop pruning the tree was unclear, the number of leaves (HLA-DQB1/-DRB1 groups) must be sufficient to prevent residual confounding, but not so great as to become inestimable.
Pruning and evaluation of the HLA-DQB1/HLA-DRB1 trees
Initially, we used the TAP2 SNP rs241448 to assess the appropriateness of the HLA-DRB1 and HLA-DQB1 tree models and subsequently used an additional 14 loci (Supplementary Table 3). As the number of terminal leaves (HLA-DRB1/-DQB1 groups) in the model increases, we expect the effect size of the additional locus to decrease as confounding is reduced, while the 95% confidence interval will become larger (Supplementary Fig. 1). This information can be used to optimize the choice of tree by considering the best compromise between number of terminal leaves (groups of HLA-DRB1 and HLA-DQB1 genotypes), effect size and 95% confidence interval. By adding the TAP2 SNP rs241448, we tested each of the possible six pruned HLA-DRB1/HLA-DQB1 trees (that corresponded to different complexity parameters) in the family data set. The greatest disparity in effect size was seen between the two models with the minimum number of terminal leaves (four and seven groups) with relative risk = 0.7 (95% confidence interval 0.5–0.9) and the remainder with between 12 and 22 groups, relative risk = 0.6 (95% confidence interval 0.5–0.8). The effect size and 95% confidence interval at this locus was stable with respect to the number of terminal leaves (that is, HLA-DRB1/-DQB1 groups). Therefore, using the TAP2 SNP only, the 12 group model appeared to be the best compromise between effect size, 95% confidence interval and number of groups (that is, complexity of the model). Nevertheless, to verify that the number of groups used for the MHC class II model would not affect the interpretation of results, we tested the remaining 14 loci used to evaluate other grouping methods (Supplementary Results) for association, conditioning on class II effects using each of the four tree models with 12 or greater terminal leaves (Supplementary Table 3). The 12 group model exhibited one inconsistent result at MICA compared to all the other recursive partitioning models (P = 0.0005 versus P > 0.05), otherwise all loci tested were stable with respect to the number of groups in the model (Supplementary Table 3). Hence, the model with 16 terminal leaves was chosen to model the confounding effects of HLA-DRB1 and HLA-DQB1: this model had the minimum number of groups that gave results consistent with both the 18 group model and the 22 group model at all loci.
We did not use this 16 group model for the case–control collection because the way in which the two sample sets were ascertained could affect their MHC associations. The ASP families are likely to be enriched for HLA susceptibility haplotypes compared to isolated cases. They were also collected over 10 years earlier, during which time the incidence of type 1 diabetes has increased, and have a higher average age-at-diagnosis (12 years) compared to the British cases (7 years). We did, however, use the same approach to choosing the optimal tree model for the cases and controls as for the families. The MHC class II genotypes were put into the recursive partitioning library. The maximum number of groups obtained was 14 (which corresponded to a complexity parameter of 0). Of the five pruned HLA-DRB1/HLA-DQB1 trees possible (corresponding to different complexity parameters) the model with 12 terminal leaves was the best compromise between effect size and 95% confidence interval, gave consistent results across loci and was very similar to the tree with the maximum 14 terminal leaves.
The effect sizes with corresponding 95% confidence intervals are given in Supplementary Table 7 for the case–control model and the model used for the families, using an approximately neutral group as reference. Note that although the tree used for the families has 16 groups, one of these groups only contains pseudo-controls and so is not used for the analysis. Similarly one of the 12 groups used for the case–control set consisted of just 13 cases, which are dropped from the association analysis.
We then assessed the effectiveness of the HLA-DRB1/HLA-DQB1 tree model. Hence, we generated 1,000 bootstrap sample data sets, with replacement, within geographical and case–control strata. Four loci were used as the non-class-II test locus and analysed in each data set: the TAP2 SNP rs241448; the UBD SNP rs389419; the HLA-DRA SNP rs9268831; and the HLA-B Bw4/Bw6 epitope polymorphism. Supplementary Fig. 1 shows a plot of the regression coefficient for the test locus rs9268831 against number of groups for each bootstrap data set. Notably, although the effect size decreases (that is, the regression coefficient increases) with number of groups, this decrease was very modest, indicating that our effect size estimates are good. We then used the bootstrap samples to calculate 95% confidence intervals for each test locus. Reassuringly, all bootstrap 95% confidence intervals were consistent with the original 95% confidence intervals (rs241448, 95% CIorig = 0.66–1.02 and 95% CIboot = 0.71–1.17; rs389419, 95% CIorig = 1.13–1.59 and 95% CIboot = 1.17–1.84; rs9268831, 95% CIorig = 0.62–0.83 and 95% CIboot = 0.57–0.80; HLA-B Bw4/Bw6, 95% CIorig = 1.02–1.45 and 95% CIboot = 1.003–1.45) and do not lead to a different interpretation of results. Hence, we believe that the trees are effective models for the HLA-DRB1/HLA-DQB1 effects.
Testing for associations at non-class-II loci in families
We specifically wished to test the hypothesis that loci within the MHC were associated with type 1 diabetes independently of the highly associated class II genes HLA-DRB1 and HLA-DQB1. Owing to the complex relationship between these two genes, extensive linkage disequilibrium and epistatic interaction effects2, we believed that a joint model was required to explain the observed association. This approach was justified because both loci were necessary to partition the data within recursive partitioning.
Forward stepwise conditional logistic regression was used to test whether any of the 83 loci typed in the MHC had an effect in addition to the HLA class II DRB1/DQB1 effect28. Only individuals typed at both the class II loci and the test locus were used for the stepwise analysis. The HLA-DRB1/HLA-DQB1 loci (modelled using the recursive partitioning method described above) were placed in the regression model as confounders and other loci added; whether or not a non-HLA-DRB1/HLA-DQB1 locus improved on the model was tested by a Wald test where robust variance estimates could be applied, or else by a likelihood ratio test. The non-HLA-DRB1/HLA-DQB1 loci were modelled as alleles when the multiplicative model was appropriate, and genotypes otherwise.
Testing for non-HLA-DRB1/HLA-DQB1 loci in the case–control collection
As with the family data set, HLA-DRB1 and HLA-DQB1 were grouped by recursive partitioning (detailed above) and placed in the logistic regression model as confounders. A likelihood ratio test was used to test whether other loci added to the regression model. P < 0.0001 was considered significant. The analysis was stratified both by broad geographical region30 and by the HLA-DRB1/HLA-DQB1 groups. The most significant locus was added as alleles or genotypes to the grouped class II loci and other loci added to them to test for additional effects.
The Wellcome Trust Case Control Consortium
Management committee Paul R. Burton1, David G. Clayton2, Lon R. Cardon3, Nick Craddock4, Panos Deloukas5, Audrey Duncanson6, Dominic P. Kwiatkowski3,5, Mark I. McCarthy3,7, Willem H. Ouwehand8,9, Nilesh J. Samani10, John A. Todd2 & Peter Donnelly (Chair)11
Analysis committee Jeffrey C. Barrett3, Paul R. Burton1, Dan Davison11, Peter Donnelly11, Doug Easton12, David Evans3, Hin-Tak Leung2, Jonathan L. Marchini11, Andrew P. Morris3, Chris C. A. Spencer11, Martin D. Tobin1, Lon R. Cardon (Co-chair)3 & David G. Clayton (Co-chair)2
UK blood services and University of Cambridge controls Antony P. Attwood5,8, James P. Boorman8,9, Barbara Cant8, Ursula Everson13, Judith M. Hussey14, Jennifer D. Jolley8, Alexandra S. Knight8, Kerstin Koch8, Elizabeth Meech15, Sarah Nutland2, Christopher V. Prowse16, Helen E. Stevens2, Niall C. Taylor8, Graham R. Walters17, Neil M. Walker2, Nicholas A. Watkins8,9, Thilo Winzer8, John A. Todd2 & Willem H. Ouwehand8,9
1958 birth cohort controls Richard W. Jones18, Wendy L. McArdle18, Susan M. Ring18, David P. Strachan19 & Marcus Pembrey18,20
Bipolar disorder Gerome Breen21, David St Clair21 (Aberdeen); Sian Caesar22, Katherine Gordon-Smith22,23, Lisa Jones22 (Birmingham); Christine Fraser23, Elaine K. Green23, Detelina Grozeva23, Marian L. Hamshere23, Peter A. Holmans23, Ian R. Jones23, George Kirov23, Valentina Moskvina23, Ivan Nikolov23, Michael C. O’Donovan23, Michael J. Owen23, Nick Craddock23 (Cardiff); David A. Collier24, Amanda Elkin24, Anne Farmer24, Richard Williamson24, Peter McGuffin24 (London); Allan H. Young25 & I. Nicol Ferrier25 (Newcastle)
Coronary artery disease Stephen G. Ball26, Anthony J. Balmforth26, Jennifer H. Barrett26, D. Timothy Bishop26, Mark M. Iles26, Azhar Maqbool26, Nadira Yuldasheva26, Alistair S. Hall26 (Leeds); Peter S. Braund10, Paul R. Burton1, Richard J. Dixon10, Massimo Mangino10, Suzanne Stevens10, Martin D. Tobin1, John R. Thompson1 & Nilesh J. Samani10 (Leicester)
Crohn’s disease Francesca Bredin27, Mark Tremelling27, Miles Parkes27 (Cambridge); Hazel Drummond28, Charles W. Lees28, Elaine R. Nimmo28, Jack Satsangi28 (Edinburgh); Sheila A. Fisher29, Alastair Forbes30, Cathryn M. Lewis29, Clive M. Onnie29, Natalie J. Prescott29, Jeremy Sanderson31, Christopher G. Mathew29 (London); Jamie Barbour32, M. Khalid Mohiuddin32, Catherine E. Todhunter32, John C. Mansfield32 (Newcastle); Tariq Ahmad33, Fraser R. Cummings33 & Derek P. Jewell33 (Oxford)
Hypertension John Webster34 (Aberdeen); Morris J. Brown35, David G. Clayton2 (Cambridge); G. Mark Lathrop36 (Evry, France); John Connell37, Anna Dominiczak37 (Glasgow); Nilesh J. Samani10 (Leicester); Carolina A. Braga Marcano38, Beverley Burke38, Richard Dobson38, Johannie Gungadoo38, Kate L. Lee38, Patricia B. Munroe38, Stephen J. Newhouse38, Abiodun Onipinla38, Chris Wallace38, Mingzhan Xue38, Mark Caulfield38 (London); Martin Farrall39 (Oxford)
Rheumatoid arthritis Anne Barton40, The Biologics in RA Genetics and Genomics Study Syndicate (BRAGGS) Steering Committee*, Ian N. Bruce40, Hannah Donovan40, Steve Eyre40, Paul D. Gilbert40, Samantha L. Hider40, Anne M. Hinks40, Sally L. John40, Catherine Potter40, Alan J. Silman40, Deborah P. M. Symmons40, Wendy Thomson40 & Jane Worthington40
Type 1 diabetes David G. Clayton2, David B. Dunger2,41, Sarah Nutland2, Helen E. Stevens2, Neil M. Walker2, Barry Widmer2,41 & John A. Todd2
Type 2 diabetes Timothy M. Frayling42,43, Rachel M. Freathy42,43, Hana Lango42,43, John R. B. Perry42,43, Beverley M. Shields43, Michael N. Weedon42,43, Andrew T. Hattersley42,43 (Exeter); Graham A. Hitman44 (London); Mark Walker45 (Newcastle); Kate S. Elliott3,7, Christopher J. Groves7, Cecilia M. Lindgren3,7, Nigel W. Rayner3,7, Nicholas J. Timpson3,46, Eleftheria Zeggini3,7 & Mark I. McCarthy3,7 (Oxford)
Tuberculosis Melanie Newport47, Giorgio Sirugo47 (Gambia); Emily Lyons3, Fredrik Vannberg3 & Adrian V. S. Hill3 (Oxford)
Ankylosing spondylitis Linda A. Bradbury48, Claire Farrar49, Jennifer J. Pointon48, Paul Wordsworth49 & Matthew A. Brown48,49
Autoimmune thyroid disease Jayne A. Franklyn50, Joanne M. Heward50, Matthew J. Simmonds50 & Stephen C. L. Gough50
Breast cancer Sheila Seal51, Breast Cancer Susceptibility Collaboration (UK)*, Michael R. Stratton51,52 & Nazneen Rahman51
Multiple sclerosis Maria Ban53, An Goris53, Stephen J. Sawcer53 & Alastair Compston53
Gambian controls David Conway47, Muminatou Jallow47, Melanie Newport47, Giorgio Sirugo47 (Gambia); Kirk A. Rockett3 & Dominic P. Kwiatkowski3,5 (Oxford)
DNA, genotyping, data QC and informatics Claire Bryan5, Suzannah J. Bumpstead5, Amy Chaney5, Kate Downes2,5, Jilur Ghori5, Rhian Gwilliam5, Sarah E. Hunt5, Michael Inouye5, Andrew Keniry5, Emma King5, Ralph McGinnis5, Simon Potter5, Rathi Ravindrarajah5, Pamela Whittaker5, David Withers5, Panos Deloukas5 (Wellcome Trust Sanger Institute, Hinxton); Hin-Tak Leung2, Sarah Nutland2, Helen E. Stevens2, Neil M. Walker2 & John A. Todd2 (Cambridge)
Statistics Doug Easton12, David G. Clayton2 (Cambridge); Paul R. Burton1, Martin D. Tobin1 (Leicester); Jeffrey C. Barrett3, David Evans3, Andrew P. Morris3, Lon R. Cardon3, Niall J. Cardin11, Dan Davison11, Teresa Ferreira11, Joanne Pereira-Gale11, Ingeleif B. Hallgrimsdóttir11, Bryan N. Howie11, Jonathan L. Marchini11, Chris C. A. Spencer11, Zhan Su11, Yik Ying Teo3,11, Damjan Vukcevic11 & Peter Donnelly11 (Oxford)
Primary investigators David Bentley5†, Matthew A. Brown48,49, Lon R. Cardon3, Mark Caulfield38, David G. Clayton2, Alistair Compston53, Nick Craddock23, Panos Deloukas5, Peter Donnelly11, Martin Farrall39, Stephen C. L. Gough50, Alistair S. Hall26, Andrew T. Hattersley42,43, Adrian V. S. Hill3, Dominic P. Kwiatkowski3,5, Christopher G. Mathew29, Mark I. McCarthy3,7, Willem H. Ouwehand8,9, Miles Parkes27, Marcus Pembrey18,20, Nazneen Rahman51, Nilesh J. Samani10, Michael R. Stratton51,52, John A. Todd2 & Jane Worthington40
*See Supplementary Information for details.
Footnotes
Genetic Epidemiology Group, Department of Health Sciences, University of Leicester, Adrian Building, University Road, Leicester LE1 7RH, UK.
Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory, Department of Medical Genetics, Cambridge Institute for Medical Research, University of Cambridge, Wellcome Trust/MRC Building, Cambridge CB2 0XY, UK.
Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK.
Department of Psychological Medicine, Henry Wellcome Building, School of Medicine, Cardiff University, Heath Park, Cardiff CF14 4XN, UK.
The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK.
The Wellcome Trust, Gibbs Building, 215 Euston Road, London NW1 2BE, UK.
Oxford Centre for Diabetes, Endocrinology and Medicine, University of Oxford, Churchill Hospital, Oxford OX3 7LJ, UK.
Department of Haematology, University of Cambridge, Long Road, Cambridge CB2 2PT, UK.
National Health Service Blood and Transplant, Cambridge Centre, Long Road, Cambridge CB2 2PT, UK.
Department of Cardiovascular Sciences, University of Leicester, Glenfield Hospital, Groby Road, Leicester LE3 9QP, UK.
Department of Statistics, University of Oxford, 1 South Parks Road, Oxford OX1 3TG, UK.
Cancer Research UK Genetic Epidemiology Unit, Strangeways Research Laboratory, Worts Causeway, Cambridge CB1 8RN, UK.
National Health Service Blood and Transplant, Sheffield Centre, Longley Lane, Sheffield S5 7JN, UK.
National Health Service Blood and Transplant, Brentwood Centre, Crescent Drive, Brentwood CM15 8DP, UK.
The Welsh Blood Service, Ely Valley Road, Talbot Green, Pontyclun CF72 9WB, UK.
The Scottish National Blood Transfusion Service, Ellen’s Glen Road, Edinburgh EH17 7QT, UK.
National Health Service Blood and Transplant, Southampton Centre, Coxford Road, Southampton SO16 5AF, UK.
Avon Longitudinal Study of Parents and Children, University of Bristol, 24 Tyndall Avenue, Bristol BS8 1TQ, UK.
Division of Community Health Services, St George’s University of London, Cranmer Terrace, London SW17 0RE, UK.
Institute of Child Health, University College London, 30 Guilford Street, London WC1N 1EH, UK.
University of Aberdeen, Institute of Medical Sciences, Foresterhill, Aberdeen AB25 2ZD, UK.
Department of Psychiatry, Division of Neuroscience, Birmingham University, Birmingham B15 2QZ, UK.
Department of Psychological Medicine, Henry Wellcome Building, School of Medicine, Cardiff University, Heath Park, Cardiff CF14 4XN, UK.
SGDP, The Institute of Psychiatry, King’s College London, De Crespigny Park Denmark Hill, London SE5 8AF, UK.
School of Neurology, Neurobiology and Psychiatry, Royal Victoria Infirmary, Queen Victoria Road, Newcastle upon Tyne NE1 4LP, UK.
LIGHT and LIMM Research Institutes, Faculty of Medicine and Health, University of Leeds, Leeds LS1 3EX, UK.
IBD Research Group, Addenbrooke’s Hospital, University of Cambridge, Cambridge CB2 2QQ, UK.
Gastrointestinal Unit, School of Molecular and Clinical Medicine, University of Edinburgh, Western General Hospital, Edinburgh EH4 2XU, UK.
Department of Medical & Molecular Genetics, King’s College London School of Medicine, 8th Floor Guy’s Tower, Guy’s Hospital, London SE1 9RT, UK.
Institute for Digestive Diseases, University College London Hospitals Trust, London NW1 2BU, UK.
Department of Gastroenterology, Guy’s and St Thomas’ NHS Foundation Trust, London SE1 7EH, UK.
Department of Gastroenterology & Hepatology, University of Newcastle upon Tyne, Royal Victoria Infirmary, Newcastle upon Tyne NE1 4LP, UK.
Gastroenterology Unit, Radcliffe Infirmary, University of Oxford, Oxford OX2 6HE, UK.
Medicine and Therapeutics, Aberdeen Royal Infirmary, Foresterhill, Aberdeen, Grampian AB9 2ZB, UK.
Clinical Pharmacology Unit and the Diabetes and Inflammation Laboratory, University of Cambridge, Addenbrooke’s Hospital, Hills Road, Cambridge CB2 2QQ, UK.
Centre National de Genotypage, 2, Rue Gaston Cremieux, Evry, Paris 91057, France.
BHF Glasgow Cardiovascular Research Centre, University of Glasgow, 126 University Place, Glasgow G12 8TA, UK.
Clinical Pharmacology and Barts and The London Genome Centre, William Harvey Research Institute, Barts and The London, Queen Mary’s School of Medicine, Charterhouse Square, London EC1M 6BQ, UK.
Cardiovascular Medicine, University of Oxford, Wellcome Trust Centre for Human Genetics, Roosevelt Drive, Oxford OX3 7BN, UK.
arc Epidemiology Research Unit, University of Manchester, Stopford Building, Oxford Road, Manchester M13 9PT, UK.
Department of Paediatrics, University of Cambridge, Addenbrooke’s Hospital, Cambridge CB2 2QQ, UK.
Genetics of Complex Traits, Institute of Biomedical and Clinical Science, Peninsula Medical School, Magdalen Road, Exeter EX1 2LU, UK.
Diabetes Genetics, Institute of Biomedical and Clinical Science, Peninsula Medical School, Barrack Road, Exeter EX2 5DU, UK.
Centre for Diabetes and Metabolic Medicine, Barts and The London, Royal London Hospital, Whitechapel, London E1 1BB, UK.
Diabetes Research Group, School of Clinical Medical Sciences, Newcastle University, Framlington Place, Newcastle upon Tyne NE2 4HH, UK.
The MRC Centre for Causal Analyses in Translational Epidemiology, Bristol University, Canynge Hall, Whiteladies Road, Bristol BS2 8PR, UK.
MRC Laboratories, Fajara, The Gambia.
Diamantina Institute for Cancer, Immunology and Metabolic Medicine, Princess Alexandra Hospital, University of Queensland, Woolloongabba, Queensland 4102, Australia.
Botnar Research Centre, University of Oxford, Headington, Oxford OX3 7BN, UK.
Department of Medicine, Division of Medical Sciences, Institute of Biomedical Research, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK.
Section of Cancer Genetics, Institute of Cancer Research, 15 Cotswold Road, Sutton SM2 5NG, UK.
Cancer Genome Project, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK.
Department of Clinical Neurosciences, University of Cambridge, Addenbrooke’s Hospital, Hills Road, Cambridge CB2 2QQ, UK. †Present address: Illumina Cambridge, Chesterford Research Park, Little Chesterford, Nr Saffron Walden, Essex CB10 1XL, UK.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Todd JA, Bell JI, McDevitt HO. HLA-DQβ gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 2.Cucca F, et al. A correlation between the relative predisposition of MHC class II alleles to type 1 diabetes and the structure of their proteins. Hum. Mol. Genet. 2001;10:2025–2037. doi: 10.1093/hmg/10.19.2025. [DOI] [PubMed] [Google Scholar]
- 3.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nature Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fennessy M, et al. A gene in the HLA class I region contributes to susceptibility to IDDM in the Finnish population. Childhood Diabetes in Finland (DiMe) Study Group. Diabetologia. 1994;37:937–944. doi: 10.1007/BF00400951. [DOI] [PubMed] [Google Scholar]
- 5.Nejentsev S, et al. Non-class II HLA gene associated with type 1 diabetes maps to the 240-kb region near HLA-B. Diabetes. 2000;49:2217–2221. doi: 10.2337/diabetes.49.12.2217. [DOI] [PubMed] [Google Scholar]
- 6.Lie BA, et al. The predisposition to type 1 diabetes linked to the human leukocyte antigen complex includes at least one non-class II gene. Am. J. Hum. Genet. 1999;64:793–800. doi: 10.1086/302283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valdes AM, et al. Extended DR3–D6S273-HLA-B haplotypes are associated with increased susceptibility to type 1 diabetes in US Caucasians. Tissue Antigens. 2005;65:115–119. doi: 10.1111/j.1399-0039.2005.00348.x. [DOI] [PubMed] [Google Scholar]
- 8.Valdes AM, Erlich HA, Noble JA. Human leukocyte antigen class I B and C loci contribute to Type 1 Diabetes (T1D) susceptibility and age at T1D onset. Hum. Immunol. 2005;66:301–313. doi: 10.1016/j.humimm.2004.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aly TA, et al. Extreme genetic risk for type 1A diabetes. Proc. Natl Acad. Sci. USA. 2006;103:14074–14079. doi: 10.1073/pnas.0606349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noble JA, et al. The HLA class I A locus affects susceptibility to type 1 diabetes. Hum. Immunol. 2002;63:657–664. doi: 10.1016/s0198-8859(02)00421-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honeyman MC, Harrison LC, Drummond B, Colman PG, Tait BD. Analysis of families at risk for insulin-dependent diabetes mellitus reveals that HLA antigens influence progression to clinical disease. Mol. Med. 1995;1:576–582. [PMC free article] [PubMed] [Google Scholar]
- 12.Wicker LS, et al. β2-microglobulin-deficient NOD mice do not develop insulitis or diabetes. Diabetes. 1994;43:500–504. doi: 10.2337/diab.43.3.500. [DOI] [PubMed] [Google Scholar]
- 13.Utsugi T, et al. Major histocompatibility complex class I-restricted infiltration and destruction of pancreatic islets by NOD mouse-derived beta-cell cytotoxic CD8+ T-cell clones in vivo. Diabetes. 1996;45:1121–1131. doi: 10.2337/diab.45.8.1121. [DOI] [PubMed] [Google Scholar]
- 14.Marron MP, Graser RT, Chapman HD, Serreze DV. Functional evidence for the mediation of diabetogenic T cell responses by HLA-A2.1 MHC class I molecules through transgenic expression in NOD mice. Proc. Natl Acad. Sci. USA. 2002;99:13753–13758. doi: 10.1073/pnas.212221199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh N, et al. Mononuclear cell infiltration and its relation to the expression of major histocompatibility complex antigens and adhesion molecules in pancreas biopsy specimens from newly diagnosed insulin-dependent diabetes mellitus patients. J. Clin. Invest. 1993;92:2313–2322. doi: 10.1172/JCI116835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foulis AK, Liddle CN, Farquharson MA, Richmond JA, Weir RS. The histopathology of the pancreas in type 1 (insulin-dependent) diabetes mellitus: a 25-year review of deaths in patients under 20 years of age in the United Kingdom. Diabetologia. 1986;29:267–274. doi: 10.1007/BF00452061. [DOI] [PubMed] [Google Scholar]
- 17.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herr M, et al. Evaluation of fine mapping strategies for a multifactorial disease locus: systematic linkage and association analysis of IDDM1 in the HLA region on chromosome 6p21. Hum. Mol. Genet. 2000;9:1291–1301. doi: 10.1093/hmg/9.9.1291. [DOI] [PubMed] [Google Scholar]
- 19.Noble JA, et al. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am. J. Hum. Genet. 1996;59:1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 20.Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression Trees. Wadsworth; Belmont: 1984. [Google Scholar]
- 21.R Development Core Team . A Language and Environment for Statistical Computing. R Foundation for statistical computing; Vienna: 2006. [Google Scholar]
- 22.Therneau TM, Atkinson EJ. An Introduction to Recursive Partitioning Using the rpart Routine. Mayo Clinic, section of statistics; Minnesota: 1997. Technical report no. 61. [Google Scholar]
- 23.Roach JC, et al. Genetic mapping at 3-kilobase resolution reveals inositol 1,4,5-triphosphate receptor 3 as a risk factor for type 1 diabetes in Sweden. Am. J. Hum. Genet. 2006;79:614–627. doi: 10.1086/507876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aly TA, et al. High density SNP analysis of the MHC region reveals multiple loci for type 1A diabetes. Clin. Immunol. 2007;123:S133. [Google Scholar]
- 25.de Jersey J, et al. Beta cells cannot directly prime diabetogenic CD8 T cells in nonobese diabetic mice. Proc. Natl Acad. Sci. USA. 2007;104:1295–1300. doi: 10.1073/pnas.0610057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirot L, Benoist C, Mathis D. Natural killer cells distinguish innocuous and destructive forms of pancreatic islet autoimmunity. Proc. Natl Acad. Sci. USA. 2004;101:8102–8107. doi: 10.1073/pnas.0402065101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinkse GG, et al. Autoreactive CD8 T cells associated with beta cell destruction in type 1 diabetes. Proc. Natl Acad. Sci. USA. 2005;102:18425–18430. doi: 10.1073/pnas.0508621102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am. J. Hum. Genet. 2002;70:124–141. doi: 10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stone M. Cross-validation choice and assessment of statistical predictions. J. R. Stat. Soc. B. 1974;36:111–147. [Google Scholar]
- 30.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nature Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.