Abstract
Insulin negatively regulates expression of the insulin-like growth factor binding protein 1 (IGFBP-1) gene by means of an insulin-responsive element (IRE) that also contributes to glucocorticoid stimulation of this gene. We find that the Caenorhabditis elegans protein DAF-16 binds the IGFBP-1⋅IRE with specificity similar to that of the forkhead (FKH) factor(s) that act both to enhance glucocorticoid responsiveness and to mediate the negative effect of insulin at this site. In HepG2 cells, DAF-16 and its mammalian homologs, FKHR, FKHRL1, and AFX, activate transcription through the IGFBP-1⋅IRE; this effect is inhibited by the viral oncoprotein E1A, but not by mutants of E1A that fail to interact with the coactivator p300/CREB-binding protein (CBP). We show that DAF-16 and FKHR can interact with both the KIX and E1A/SRC interaction domains of p300/CBP, as well as the steroid receptor coactivator (SRC). A C-terminal deletion mutant of DAF-16 that is nonfunctional in C. elegans fails to bind the KIX domain of CBP, fails to activate transcription through the IGFBP-1⋅IRE, and inhibits activation of the IGFBP-1 promoter by glucocorticoids. Thus, the interaction of DAF-16 homologs with the KIX domain of CBP is essential to basal and glucocorticoid-stimulated transactivation. Although AFX interacts with the KIX domain of CBP, it does not interact with SRC and does not respond to glucocorticoids or insulin. Thus, we conclude that DAF-16 and FKHR act as accessory factors to the glucocorticoid response, by recruiting the p300/CBP/SRC coactivator complex to an FKH factor site in the IGFBP-1 promoter, which allows the cell to integrate the effects of glucocorticoids and insulin on genes that carry this site.
Insulin signaling via the phosphatidylinositol 3-kinase (PI 3-kinase)/protein kinase B (PKB) pathway has diverse effects on cellular metabolism and apoptosis (1, 2). A major role of insulin is to act in opposition to the catabolic effects of cAMP and glucocorticoids, agents that stimulate liver gluconeogenesis. The rate-limiting step in gluconeogenesis is catalyzed by the phosphoenolpyruvate carboxykinase (GTP) (PEPCK; EC 4.1.1.32) gene. The insulin-like growth factor binding protein 1 (IGFBP-1) gene indirectly promotes gluconeogenesis by binding insulin-like growth factor (IGF)-I and -II and inhibiting their insulin-like effects. Expression of the PEPCK and IGFBP-1 genes is controlled at the transcriptional level by a complex regulatory mechanism in which glucocorticoids activate and insulin inhibits gene expression (3–6).
In the case of the PEPCK gene, the response to both glucocorticoids and insulin is mediated by the accessory factor II (AFII) site, located upstream of the glucocorticoid-response element (GRE); this site is also referred to as the PEPCK insulin-response sequence, IRS-1 (7). Similarly the response of the IGFBP-1 promoter to glucocorticoids and insulin is mediated by one site, the insulin-response element (IRE) site located upstream of its GRE (5, 8). Biochemical evidence first showed that the forkhead (FKH) hepatic nuclear factor (HNF)3β binds the IRE in the PEPCK and IGFBP-1 genes and enhances the effect of glucocorticoids on gene transcription (9, 10). Thus, early efforts to identify the mediator of glucocorticoid and insulin action at this site focused on HNF3β. The effect of HNF3β to enhance the action of glucocorticoids can be mimicked by GAL4-HNF3β, if a GAL4 DNA-binding site is substituted for the AFII site in the PEPCK gene. This observation provides strong support for a role of HNF3β as an accessory factor for the glucocorticoid response (11). However, if a single protein mediates the response to both glucocorticoids and insulin at this site, it is unlikely to be HNF3β, inasmuch as certain HNF3 sites fail to mediate the negative effect of insulin and other HNF3 sites actually confer insulin inducibility to a reporter gene (12, 13).
Genetic evidence identifying the transcriptional outputs of insulin-like factors in Caenorhabditis elegans indicates that the FKH transcription factor DAF-16 is the major target downstream of the C. elegans daf-2 (insulin receptor), age-1 (PI 3-kinase), PKB/Akt (AKT1/AKT2)-dependent pathway (14, 15). The effect of mutations in daf-2 and age-1 is reversed by loss of function mutations in C. elegans daf-16. Thus, insulin signaling via this pathway negatively regulates the activity of DAF-16 (14–17). Several groups simultaneously showed that close relatives of DAF-16, including FKHRL1 (18), AFX (19), and FKHR (13, 20–23) are direct targets of insulin/insulin-like growth factor signaling to PI 3-kinase and PKB. These factors can activate transcription via the IGFBP-1⋅IRE. Phosphorylation of FKHRL1 (18), FKHR/FKHR1 (23), and AFX (24) by insulin/insulin-like growth factor signaling or overexpression of PKB promotes export of these proteins from the nucleus, thereby preventing their transcriptional effect. Therefore DAF-16-like factors are likely candidates for the protein that integrates the response to glucocorticoids and insulin at the IGFBP-1⋅IRE.
While the three mammalian homologs of DAF-16 show up to 60% homology to DAF-16 within the FKH DNA-binding domain and marked conservation of their AKT/PKB phosphorylation sites, these homologs are not conserved compared with DAF-16 outside of these regions (14, 15). Thus there is uncertainty as to which of the mammalian FKH proteins is functionally most similar to DAF-16. Therefore, we compared the hormone response of DAF-16 and its mammalian homologs in HepG2 cells, hoping that DAF-16 would behave as an ortholog of the mammalian DAF-16-like factors and that we could proceed to examine the effect of nonfunctional DAF-16 homologs on hormone-regulated IGFBP-1 gene expression.
We find that DAF-16 and FKHR are most similar in their ability to activate gene transcription and modulate the response of the IGFBP-1 promoter to glucocorticoids and insulin. Both DAF-16 and FKHR bind to the KIX domain of CREB-binding protein (CBP) and to steroid receptor coactivator (SRC)-1. A C-terminal deletion mutant of DAF-16 (15), which fails to interact with the KIX domain of CBP, is transcriptionally inactive in mammalian cells and prevents the effect of glucocorticoids to stimulate IGFBP-1 gene expression. Furthermore, we find that AFX, a homolog of DAF-16 that fails to bind SRC-1, also fails to respond to glucocorticoids and insulin. Thus we have uncovered a link between DAF-16 homologs and their ability to recruit the p300/CBP/SRC coactivator complex that could explain their ability to integrate complex hormonal signals.
Materials and Methods
Constructs.
The DAF-16a1 HindIII/NheI insert from pGEM-FLAG-DAF-16a1 was ligated into the HindIII/XbaI site of pcDNA3.1(+) (Invitrogen) to generate pcDNA3.1+FLAG-DAF-16a1. The DAF-16a1 BstYI insert from pGEM-FLAG-DAF-16a1 was ligated into the BamHI site of pGEX-4T-1 (Pharmacia) to generate pGEX4T-DAF-16a1. Phosphorylation site mutants were prepared by using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). Primers were T54A (so164/165): GAT CGG TGC AAT GCT TGG CCA ATG CGT/ACG CAT TGG CCA AGC ATT GCA CCG ATC; S240A/S242A (so200/201): CGT ACA CGT GAA CGA GCC AAT GCT ATT GAG ACG ACT AC/GTA GTC GTC TCA ATA GCA TTG GCT CGT TCA CGT GTA CG; and S314A (so168/169): CCC CGA ACT CAA GCT AAC CTC TCG ATT/AAT CGA GAG GTT AGC TTG AGT TCG GGG.
Sources of plasmids: The rat IGFBP-1 promoter (nucleotides −925 to +79) cloned in PGL3-LUC was a gift from Matthew Rechler, National Institutes of Health, Bethesda, MD). FKHR, FKHRL1, and AFX were obtained from K. Arden (Univ. of California, San Diego). pCMV⋅HNF3α was a gift from J. Darnell (Rockefeller University, New York). Constitutively active pCMV6⋅gag-PKB was a gift from J. R. Woodgett (British Columbia Cancer Agency, Jack Bell Research Centre, Vancouver, Canada). CMV⋅HA-tagged p300 was a gift from Marc Montminy (Joslin Diabetes Center, Boston). Plasmids encoding glutathione S-transferase (GST)-SRC (25) and GST-CBP constructs containing the C/H1 domain (amino acids 312–450), the KIX domain (amino acids 450–684), and the C/H3 domain (amino acids 1890–2441) were gifts from Tony Hollenberg and Fred Wondisford (Beth Israel Hospital, Boston) (26). The pcDNA⋅SRC-1 (27) plasmid was a gift from William Chin (Brigham and Women's Hospital, Boston). CMV⋅E1A and CMV⋅E1A Δ2–36 (28), pCMX·VP-16 and pCMX·VP-16 p300, which are driven by a cytomegalovirus (CMV) promoter, were gifts from D. Livingston (Dana–Farber Cancer Institute, Boston).
Tissue Culture, Transfection, and Reporter Assays.
Hepatoma (HepG2) cells were purchased from the American Type Culture Collection. HepG2 cells were cultured in minimal essential medium (MEM) supplemented with nonessential amino acids, glutamine, sodium pyruvate, penicillin/streptomycin, and 10% fetal bovine serum (FBS).
HepG2 cells (passages 2–6) were seeded on 30-mm six-well plates at 50% confluence. Twenty-four hours later, the cells were incubated with Dulbecco's modified Eagle's medium (DMEM) supplemented with charcoal-treated 10% FBS. Two hours later, the cells were exposed to a DNA/calcium phosphate precipitate for 4 h and then shocked with 20% (vol/vol) dimethyl sulfoxide (DMSO) in PBS for 1 min. HepG2 cells were cotransfected with 10 μg/ml IGFBP-1⋅IRE or the IGFBP-1⋅luciferase plasmid, and 1 μg/ml pcDNA expression vector alone, or vectors encoding DAF-16, FKHR, mFKHRL1, AFX, or pCMV⋅HNF3α per ml of precipitate. RSV⋅GH or RSV-β-galactosidase (RSV, Rous sarcoma virus; GH, growth hormone) was included as a cotransfected control. In some experiments a CMV expression vector alone (0.2 μg/ml) or expression vector encoding E1A wild type, E1A Δ2–36, or active pcDNA-PKB was included. The cells were washed twice with PBS before the addition of serum-free DMEM supplemented with 0.1% crystalline BSA. Luciferase gene activity was measured 24 h after the transfection by using a luciferase assay kit (Promega). Each transfection was performed in triplicate and repeated at least three times. Cells were harvested 22 h after the addition of insulin (1 milliunit/ml) or dexamethasone (0.5 μM). The GH RIA and β-galactosidase assays (29) were performed as previously described. HEK293 cells were transfected by using the modification of the calcium phosphate precipitation protocol described above except that the cells were not shocked with 20% DMSO.
Protein Interaction Assay.
The pcDNA⋅DAF-16, pCMV⋅p300, and pcDNA⋅SRC1 plasmids were used for in vitro synthesis of proteins in rabbit reticulocyte lysate by using protocols described by the supplier (Promega). GST-fusion proteins were prepared as described previously (29). The quality of each preparation was examined by SDS/PAGE, and the GST proteins were matched for molar content in the crude preparation. GST pull-down assays were performed by using a method described by Lai and Herr (30) with modifications described previously (29). The amount of GST-fusion protein absorbed to the beads was quantitated by subjecting a fraction of the proteins released to SDS/PAGE followed by staining with Coomassie blue.
Yeast Two-Hybrid Screen.
In the yeast two-hybrid screen, a fusion between GAL4 DNA-binding domain and amino acid 397–683 of C. elegans CBP-1 was constructed by using the PAS2–1 vector (CLONTECH) and used as bait to screen a mixed-stage C. elegans library (kindly provided by Robert Barstead, Oklahoma Medical Research Foundation, Oklahoma City, OK). The library was screened by using the reagents and protocols provided in the Matchmaker Two-Hybrid System 2 kit (CLONTECH).
Results
DAF-16 Homologs Modulate the Effect of Glucocorticoids and Insulin on the IGFBP-1 Gene.
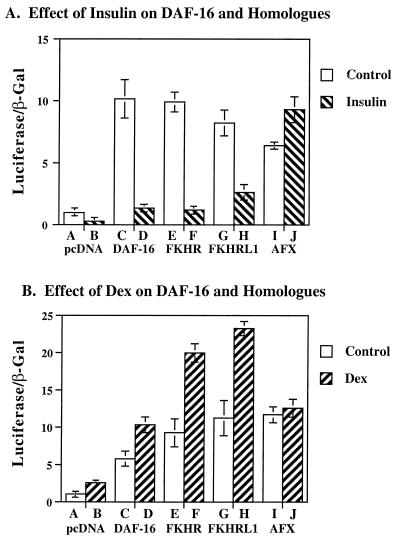
As previously shown for its mammalian homolog FKHR (31), DAF-16 binds the wild-type IGFBP-1⋅IRE and not a mutant that eliminates the effect of glucocorticoids and insulin on this gene (data not shown). To determine which of three mammalian homologs of DAF-16—FKHR, FKHRL1, or AFX—was most similar in function to DAF-16, we compared their effects on glucocorticoid- and insulin-responsive gene transcription. In three independent experiments, DAF-16 stimulated IGFBP-1 promoter activity by 8- to 10-fold, and insulin inhibited this effect by 90% (Fig. 1A, compare bar A to bars C and D). The abilities of DAF-16 and FKHR to activate IGFBP-1 gene expression were identical in magnitude (Fig. 1A, bars C and E), as were the effects of insulin to inhibit DAF-16 and FKHR by 90% (Fig. 1A, bars D and F). The effect of insulin on FKHRL1 was consistently less pronounced than its effect on FKHR (Fig. 1A, bars G, H and E, F). In contrast, insulin did not inhibit activation of the IGFBP-1 promoter by AFX at all (bars I and J). Thus, in HepG2 cells, FKHR is the mammalian homolog that functions most like its C. elegans homolog, DAF-16.
Figure 1.
Effect of DAF-16 homologs on insulin- and glucocorticoid-responsive gene transcription. HepG2 were cotransfected with a construct encoding the native IGFBP-1 promoter (10 μg/ml) driving luciferase gene expression and the pcDNA expression vector alone (1 μg/ml), or the expression vectors pcDNA⋅DAF-16, pcDNA⋅FKHR, pcDNA⋅FKHRL1, and pcDNA⋅AFX (1 μg/ml). In A, insulin (1 milliunit/ml) was added to serum-starved cells during the last 18 h of incubation. In B, cells were exposed to dexamethasone (0.5 μM) for 18 h. The effect of these agents on endogenous proteins, pcDNA (bars A and B), or the exogenous proteins encoded by pcDNA⋅DAF-16 (bars C and D) and its three mammalian homologs, pcDNA⋅FKHRL1 (bars E and F), pcDNA⋅FKHR (bars G and H), and pcDNA⋅AFX (bars I and J) is shown. Luciferase activity was corrected for β-galactosidase gene expression. The data shown are representative of three experiments.
The effect of DAF-16 and its homologs on glucocorticoid-responsive gene transcription was assessed by using a concentration of each designed to achieve less than maximal stimulation of basal activity. Again DAF-16 homologs stimulated basal gene expression by 6- to 10-fold. Under these conditions, dexamethasone enhanced the effect of DAF-16 by 70%, and the effect of FKHR and FKHRL1 by 100% (Fig. 1B, bars D, F, and H, respectively). Thus we conclude that in the absence of insulin, FKHR and FKHRL1 facilitate glucocorticoid activation of the IGFBP-1 promoter. In contrast, although AFX stimulated basal transcription of the IGFBP-1 promoter by 10-fold, this homolog failed to show glucocorticoid-stimulated gene expression under any conditions. Thus we conclude that individual DAF-16 homologs mediate distinct regulatory functions.
E1A Interacts with p300/CBP and Blocks Activation of IGFBP-1 Gene Transcription by DAF-16.
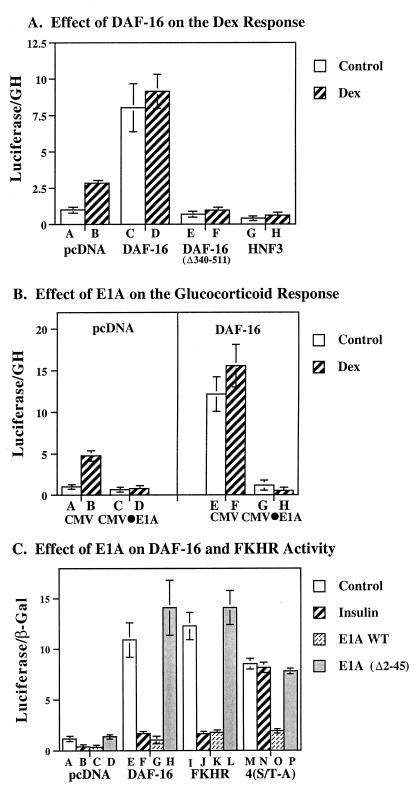
Mutations of the IGFBP-1⋅IRE impair glucocorticoid-stimulated gene expression, pointing to an enhancing effect of the protein complex bound at the IRE site on glucocorticoid-induced transcription. To determine whether binding of DAF-16-like proteins to the IRE modulates the effect of glucocorticoids, we overexpressed wild-type and mutant DAF-16 in HepG2 cells. In the absence of exogenous factors, IGFBP-1 promoter activity was stimulated 3-fold by glucocorticoids (Fig. 2A, bars A and B). Wild-type DAF-16 increased basal promoter activity by 8-fold, and there was no additional effect of glucocorticoids (bars C and D). Mutant DAF-16 (Δ340–511) had no effect on basal activity, and it prevented the effect of glucocorticoids to stimulate IGFBP-1 gene expression by means of endogenous factors (compare bars E, F to A, B). Thus the C-terminal domain of DAF-16 appears to be required for basal- and glucocorticoid-responsive activation of the IGFBP-1 promoter. The effect of HNF3α, a FKH family DNA-binding protein (bars G and H) was similar to that of DAF-16 (Δ340–511) in that occupation of the site by HNF3α also prevented the effect of endogenous factors.
Figure 2.
(A) Effect of DAF-16 and HNF3α on insulin-responsive gene transcription. HepG2 cells were incubated in the presence or absence of insulin (1 milliunit/ml) for 16 h before harvesting. Luciferase activity recovered in the presence of pcDNA (bars A and B), pcDNA⋅DAF-16 (bars C and D), pcDNA⋅DAF-16 (Δ340–511) (bars E and F), and HNF3α (bars G and H) is shown. (B) Effect of E1A on dexamethasone-responsive gene transcription. Cells were transfected with the native IGFBP-1 promoter driving luciferase gene expression (15 μg/ml), and expression vectors including pcDNA alone (bars A–D) or pcDNA⋅DAF-16 (bars E–H); and CMV alone (1 μg/ml) (bars A, B, E, and F); or CMV⋅E1A (bars C, D, G, and H). Cells were inoculated with (bars B, D, F, and H) and without (bars A, C, E, and G) dexamethasone (0.5 μM) for 18 h. Luciferase activity is shown corrected for growth hormone (GH) and normalized to the control value for pcDNA alone. (C) DAF-16 gene expression is inhibited by insulin and by wild-type E1A but not by E1A Δ2–36. HepG2 cells were transiently cotransfected with the native IGFBP-1 promoter-luciferase gene (10 μg/ml), and the pcDNA expression vector alone (1 μg/ml) (bars A–D), or wild-type pcDNA⋅DAF-16 (bars E–H), or pcDNA⋅FKHR (bars I–L) or pcDNA⋅DAF-16 4(S/T-A) (bars M–P). Control and insulin-stimulated activity was assessed in the presence of the expression vector CMV alone (0.2 μg/ml; bars A, B, E, F, I, J, M, and N). The effect of wild-type CMV⋅E1A (bars C, G, K, and O) or a derivative of E1A that fails to interact with CBP, CMV⋅E1A Δ2–36 (bars D, H, L, and P) is shown.
We speculated that the proposed ability of FKH proteins to act as accessory factors for the stimulatory effect of glucocorticoids by means of the IGFBP-1⋅IRE might result from recruitment of the p300/CBP coactivator complex to this site, as is seen with the glucocorticoid receptor (32, 33). If so, one interpretation of the observation that glucocorticoids do not further stimulate the IGFBP-1 gene in the presence of DAF-16 (compare bars A, B and C, D in Fig. 2A) would be that both DAF-16 and glucocorticoids act by the same mechanism. Therefore, we examined whether E1A, a viral oncoprotein that interacts with and sequesters p300/CBP, could inhibit the stimulatory effect of DAF-16 or glucocorticoids on IGFBP-1 gene expression. In the absence of DAF-16, basal IGFBP-1 promoter activity was stimulated 4-fold by glucocorticoids (Fig. 2B, bars A and B), and this effect was inhibited by wild-type E1A (Fig. 2B, compare bars A and B to C and D). In the presence of DAF-16, IGFBP-1 gene expression was stimulated 8-fold, and no further activation was observed in the presence of glucocorticoids (compare bars A and B to E and F in Fig. 2B). E1A inhibited the effect of DAF-16, independent of the addition of glucocorticoids (Fig. 2B, compare bars E and F to bars G and H).
Next we compared the effect of wild-type E1A and a derivative of E1A missing the N-terminal p300/CBP-interaction domain, E1A Δ2–36 (28, 34), on IGFBP-1 promoter activity. In the absence of DAF-16, wild-type E1A inhibited the effect of endogenous factors on IGFBP-1 promoter activity by 80% (Fig. 2C, bars A and C), whereas no inhibition was seen by E1A Δ2–36 (Fig. 2C, bars A and D). Thus, in HepG2 cells, basal activation of the IGFBP-1 promoter by endogenous factors is likely to depend on p300/CBP. DAF-16 activated expression of the IGFBP-1 promoter 8-fold (Fig. 2C, compare A to E), and its homolog FKHR activated expression 10-fold (Fig. 2C, compare A to I). The effect of DAF-16 and FKHR on IGFBP-1 promoter activity was inhibited 90% by wild-type E1A (Fig. 2C, compare bars E to G and I to K). Again, E1A Δ2–36 had no effect on DAF-16- or FKHR-stimulated IGFBP-1 promoter activity (Fig. 2C, compare bars E to H and I to L). A similar pattern of inhibition by wild-type E1A, but not mutant E1A Δ2–36 was observed with all of the mammalian homologs of DAF-16 (data not shown). This observation suggested that, in HepG2 cells, binding of the p300/CBP coactivator complex to DAF-16 and FKHR is essential for the ability of DAF-16 to activate IGFBP-1 gene transcription.
We compared the effect of insulin and E1A on the activity of wild-type DAF-16 and an insulin-insensitive mutant DAF-16 4(S/T-A), which carries an alanine substitution within its four consensus AKT/PKB phosphorylation sites (Thr-54, Ser-240, Thr-242, and Ser-314). While wild-type DAF-16 was inhibited 85% by insulin (Fig. 2C, bars E and F), the activity of the DAF-16 4(S/T-A) site mutant was not affected by insulin (bars M and N). This observation confirms previous reports that the inhibitory effect of insulin on the wild-type DAF-16 protein is dependent on phosphorylation of DAF-16 at one or more of its putative AKT/PKB sites in HepG2 cells (13). Although insulin had no effect on the activity of DAF-16 4(S/T-A), wild-type E1A, but not mutant E1A Δ2–36, inhibited the activity of this mutant by 70% (Fig. 2C, compare bar M to bar O and P). Thus, in HepG2 cells, phosphorylation of the AKT sites on DAF-16 is not required for the interaction of p300/CBP with DAF-16 and activation of the IGFBP-1 promoter or for E1A inhibition of this effect. In contrast, insulin inhibits DAF-16 activity by a mechanism that requires the AKT sites in DAF-16.
p300/CBP Interacts with DAF-16 in Vitro and in Vivo.
In an independent series of experiments aimed at identifying important CBP interactors in C. elegans we obtained further evidence that CBP and DAF-16 interact in cellular systems. Using the N-terminal region of C. elegans CBP-1 (amino acids 397–683), which contains the C/H1 and KIX domains as bait, we recovered 22 C. elegans transcription factors that interact with CBP-1, 4 of which were related to mammalian factors (Table 1). In addition to DAF-16, C. elegans clones F38A6.3, TO1B10.4, and 2K1290.4 were recovered from the screen and found to bear striking homology to hepatocyte nuclear factor 4 (HNF4), hypoxia-inducible factor-1α (HIF-1α), and nuclear factor 1 (NF-1), respectively. The KIX domain of mammalian CBP has previously been shown to interact with HNF4 and HIF-1α in mammalian systems (35, 36). Thus, we conclude that the interaction of DAF-16 with CBP-1 is comparable to that for other known CBP-interacting proteins (Table 1).
Table 1.
CBP interacts with DAF-16 in yeast and mammalian two-hybrid system
| Yeast two-hybrid system
|
Mammalian two-hybrid system
|
||||||
|---|---|---|---|---|---|---|---|
| Clone | Amino acids | No. of clones | lacZ+/HIS | GAL4⋅DAF-16 derivative | Activity (Luc/β-gal)
|
-Fold effect of p300 | |
| VP16 | VP16·p300 | ||||||
| DAF-16 | 176–508 | 1 | ++/+ | GAL4⋅BD | 0.9 ± 0.09 | 0.91 ± 0.03 | 1 |
| F38A6.3 (HIF-1α) | 104–343 | 5 | ++/+ | GAL4⋅DAF-16 wild type | 31 ± 1.89 | 284 ± 5.03 | 9 |
| T01B10.4 (HNF4) | 51–450 | 2 | +/+ | GAL4⋅DAF-16 4(S/T-A) | 57 ± 2.23 | 328 ± 22.4 | 6 |
| ZK1290.4 (NF-1) | 525–1026 | 1 | ++/+ | GAL4⋅DAF-16 (Δ340–511) | 4.5 ± 0.14 | 12.5 ± 0.93 | 3 |
Yeast two-hybrid system: Two million independent colonies were screened and 92 positive clones were isolated by using the reporter genes lacZ and HIS3. Yeast plasmids encoding the 92 “preys” were rescued into Escherichia coli HB101 and used in new yeast transformation experiments to confirm the two-hybrid interaction. On the second round of screening 46 clones were positive with GAL4⋅CBP-1, but not with the GAL4 DNA-binding domain alone or GAL4LAM5′-1 (CLONTECH). Twenty-two of the 46 clones encoded putative C. elegans transcription factors, some of which have known mammalian homologs. A partial list is shown in this table. The predicted open reading frames (ORFs) of clones F38A6.3, T01B10.4, and ZK1290.4 encode proteins related to mammalian HIF-1α, HNF4, and NF-1, respectively. The length of the ORFs of the isolated clones, the number of independent clones identified for each interacting molecule, and the relative strength of the interactions is shown in columns 2, 3, and 4, respectively. Mammalian two-hybrid system: GAL4⋅DAF-16 derivatives (2 μg) were cotransfected into HEK 293 cells with VP-16 alone or VP-16⋅p300 (2.5 μg) and the luciferase (15 μg) or β-galactosidase (2.5 μg) reporter genes as described in the text. Cells were assayed for luciferase (Luc) and β-galactosidase (β-gal) activity. Luciferase activity was corrected for coexpression of β-galactosidase activity (±SEM). The fold effect indicates the specific interaction of the GAL4⋅DAF-16 derivative with VP-16⋅p300 compared to VP-16 alone.
We used the mammalian two-hybrid system to confirm that CBP interacts with DAF-16 in mammalian cells. DAF-16 was expressed as a fusion protein with the GAL4 DNA-binding domain and full-length CBP as a fusion protein with the VP-16 activation domain. GAL4⋅DAF-16 derivatives and the VP-16 derivatives were cotransfected into HEK293 cells in the presence of a luciferase reporter gene driven by a GAL4 DNA-binding site. There was no effect of the VP-16⋅CBP fusion gene on the GAL4 DNA-binding domain construct alone. In the presence of GAL4⋅DAF-16 the CBP⋅VP16 fusion gene activated gene transcription by 9-fold compared with VP-16 alone. Mutation of the AKT sites in DAF-16 had little effect on the ability of CBP⋅VP16 to activate gene transcription; activation by CBP⋅VP16 was 6-fold over that of VP-16 alone. In contrast, the ability of the C-terminal DAF-16 mutant to interact with CBP⋅VP16 was markedly decreased compared with wild-type DAF-16. The fact that the interaction of CBP with mutant DAF-16 was not abolished can be explained by the fact that both the KIX domain and the E1A/SRC domain of p300 interact with DAF-16; thus, the C-terminal mutant can still interact with the E1A/SRC domain of p300 (see Fig. 3).
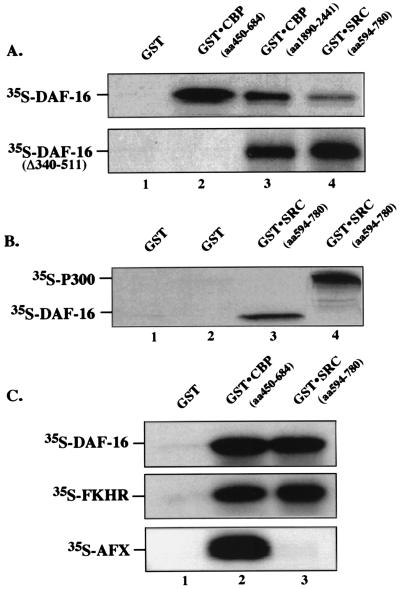
Figure 3.
DAF-16 interacts with p300 and SRC. (A) Interaction of [35S]methionine-labeled DAF-16 with the KIX and C/H3 interaction domains of GST⋅CBP. In vitro translated [35S]methionine-labeled DAF-16 or DAF-16 (Δ340–511) was incubated with GST (lane 1); or GST⋅CBP (KIX), which encodes amino acids 450–684 of CBP (lane 2); or GST⋅CBP (C/H3), which encodes amino acids 1890–2441 of CBP (lane 3); or GST⋅SRC, which encodes amino acids 594–780 (lane 4). The bound proteins were washed, eluted, and subjected to SDS/PAGE as described in the text. The autoradiograph of a dried gel is shown. (B) Interaction of GST⋅SRC with [35S]methionine-labeled p300 and DAF-16. In vitro translated [35S]methionine-labeled p300 (lanes 2 and 4) or DAF-16 (lanes 1 and 3) was incubated with bacterially produced GST (lanes 1 and 2) or GST⋅SRC (amino acids 594–780) (lanes 3 and 4) bound to glutathione-Sepharose beads. Eluted proteins are shown. (C) Interaction of [35S]methionine-labeled DAF-16, FKHR, and AFX with GST⋅CBP (KIX) and GST⋅SRC. [35S]Methionine-labeled DAF-16, FKHR, and AFX were incubated with GST (lane 1), GST⋅KIX (lane 2), or GST⋅SRC (lane 3) bound to glutathione-Sepharose beads. Eluted proteins are shown.
To determine whether CBP could interact directly with DAF-16 and to map the domain involved in vitro, the interaction of CBP with DAF-16 was confirmed by using the GST pull-down assay. Crude GST fusion proteins that include three major interaction domains of CBP⋅C/H1 (amino acids 312–450), GST-CBP⋅KIX (amino acids 450–684), and GST-CBP⋅C/H3 (amino acids 1890–2441) (25), respectively, were bound to glutathione-Sepharose columns and incubated with [35S]methionine-labeled DAF-16. DAF-16 did not bind GST alone (Fig. 3A, lane 1), nor did it bind the C/H1 domain of CBP (data not shown). As expected, however, DAF-16 interacts with the KIX domain of CBP (Fig. 3A, lane 2); approximately 10% of the applied proteins were recovered (data not shown). DAF-16 also interacts with the C/H3 domain of GST⋅CBP (Fig. 3A, lane 3), the domain that interacts with E1A and SRC. Thus, there are two domains within CBP that interact with DAF-16.
In mammalian cells p300/CBP is known to interact with SRC and certain acetyltransferases to form a coactivator complex that is essential for activation of gene transcription by members of the nuclear receptor superfamily (37, 38). To determine whether recruitment of p300/CBP by DAF-16 might be reinforced by an indirect interaction with SRC, we incubated [35S]methionine-labeled DAF-16 with GST⋅SRC and found that DAF-16 could indeed interact with SRC (Fig. 3A, lane 4). When we compared the ability of p300/CBP and DAF-16 to bind GST⋅SRC, we found that binding of in vitro translated DAF-16 to GST⋅SRC (Fig. 3B, lane 3) was comparable to binding of p300 with GST⋅SRC (Fig. 3B, lane 4). Thus, we conclude that DAF-16 interacts with p300/CBP and SRC.
In contrast to wild-type DAF-16, the transcriptionally inactive DAF-16 (Δ340–511) failed to bind the KIX domain of CBP (amino acids 450–684) (Fig. 3A, lane 2), but it did bind the E1A/SRC interaction domain of GST⋅CBP (amino acids 1890–2441) (lane 3) and GST⋅SRC (lanes 4). Thus wild-type DAF-16 interacts with two distinct sites on CBP, whereas the inactive C-terminal truncation mutant of DAF-16 interacts with the E1A/SRC domain of CBP, but not the KIX domain. Therefore, we conclude that basal and glucocorticoid-mediated activation of the IGFBP-1 promoter requires an interaction of DAF-16 with the KIX domain of DAF-16.
Binding of in vitro translated DAF-16, FKHR, and AFX to the KIX domain of CBP and the interaction domain of SRC is compared in Fig. 3C. DAF-16 and FKHR bind to both the KIX domain of CBP and the interaction domain of SRC (Fig. 3C, lanes 2 and 3), whereas AFX binds only CBP⋅KIX, and not SRC (lanes 2 and 3). We conclude that the interaction of AFX with the KIX domain of CBP is sufficient for basal transcriptional activity but not for hormone-regulated activity.
Discussion
We show that in HepG2 cells, DAF-16, a member of the FKH family of transcriptional regulators, can recruit the p300/CBP coactivator complex to the IGFBP-1⋅IRE, a site that both enhances the positive effect of glucocorticoids and mediates the negative effect of insulin on the IGFBP-1 gene. Consistent with previously published findings that insulin inhibits the activity of FKHR, a mammalian homolog of DAF-16 (13), we show that the inhibitory effects of insulin on DAF-16 and FKHR were identical, ranging from 70% to 90%. However, in contrast to previously published findings where insulin inhibited the activity of AFX by 40% in NIH 3T3 cells (19), insulin did not inhibit activation of the IGFBP-1 gene by AFX in HepG2 cells. Thus we conclude that the mammalian homologs of DAF-16 are not functionally equivalent.
In the absence of insulin, we find that DAF-16-like proteins recruit the p300/CBP coactivator complex to the IGFBP-1 gene. Recruitment of the p300/CBP coactivator complex is essential for cAMP- and glucocorticoid-responsive gene transcription (32, 39). In the presence of dexamethasone, the glucocorticoid receptor can interact with the coactivators SRC/GRIP/CBP and their associated histone acetyltransferases (40, 41) to its target genes. Thus our observation that DAF-16 can recruit the coactivator complex to the site that enhances glucocorticoid responsiveness implies that one function of DAF-16-like proteins is to provide additional binding sites for the coactivator complex on the IGFBP-1 promoter. Two findings support this view. First, the nonfunctional mutant of DAF-16 lacking the CBP–KIX interaction domain acts as a dominant inhibitor of the glucocorticoid response. Second, in the presence of saturating amounts of wild-type DAF-16, glucocorticoids have no further stimulatory effect on IGFBP-1 gene expression, suggesting that the glucocorticoid receptor and DAF-16 act by a common mechanism. Accordingly, at subsaturating concentrations of DAF-16 and FKHR, we were able to demonstrate an enhancing effect of these factors on glucocorticoid-responsive gene transcription.
Although AFX interacts with the KIX domain of CBP, it does not respond to glucocorticoids or insulin. Thus the interaction with the KIX domain of CBP appears to be necessary, but not sufficient, for glucocorticoid regulation of IGFBP-1 gene transcription. We conclude that the interaction of DAF-16-like proteins with the KIX domain of CBP is essential for basal activation of IGFBP-1 gene transcription in HepG2 cells, but it is not sufficient for hormone-regulated gene expression. Examination of the DAF-16 amino acid sequence shows several regions that carry variations on the interaction motif found in the accessory factor II domain (42) of nuclear receptors that interact with SRC (33, 43). SRC can, in turn, interact with the C/H3 domain of p300/CBP (37); thus CBP is indirectly recruited to nuclear receptors by SRC. We find that DAF-16 and FKHR can bind directly to SRC as well as CBP. The ability of DAF-16-like factors to mediate the effect of both glucocorticoids and insulin on the IGFBP-1 gene correlates with this interaction as opposed to the interaction of DAF-16-like factors with the KIX domain of CBP. Specifically, the DAF-16 homolog AFX fails to bind SRC in vitro and fails to mediate the effect of glucocorticoids and insulin on IGFBP-1 gene transcription. Thus, distinct DAF-16 family members appear to play distinct roles on their target genes.
The physiologic relevance of the interaction of DAF-16 with SRC and CBP as it pertains to hormone-regulated gene expression is not yet known. However, a precedent for the suggestion that recruitment of CBP is essential for hormone-regulated stimulation of PIT-1 gene transcription by agents such as cAMP and insulin, which usually show opposing effects on gene transcription, has been established in GH4 cells (44). For example, the recruitment of CBP is essential for positive regulation of PIT-1 by cAMP, whereas recruitment of CBP by SRC is critical for positive regulation of PIT-1 by insulin. Thus, it seems reasonable to suggest that recruitment of distinct coactivator or corepressor complexes may play a role in mediating multihormonal regulation of IGFBP-1 gene transcription.
The PEPCK gene carries two weak GREs and three well-described accessory factor binding sites that are essential for the effect of glucocorticoids to activate gene expression (7, 11). The accessory factor I (AFI) site binds HNF4 and the accessory factor II (AFII) site binds FKH domain factors such as HNF3 (11) and FKHR (31). In mammalian cells, HNF4 interacts with the CH/1 domain of CBP and with SRC/GRIP (45). Using the yeast two-hybrid system to find targets of CBP in C. elegans, we found that the N terminus of CBP selected a clone that encodes a protein related to HNF4 as well as DAF-16. Our observation that DAF-16 and FKHR can interact with CBP and SRC just as the nuclear receptor HNF4 can suggests a common mechanism whereby either family of factors could enhance glucocorticoid-responsive gene expression by recruiting the CBP coactivator complex to promoters with relatively weak GREs.
We propose then that in HepG2 cells, DAF-16 and its mammalian homologs activate basal transcription of the IGFBP-1 gene by recruiting p300/CBP to the promoter. Neither a nonfunctional mutant of DAF-16 that fails to bind the KIX domain of CBP nor a nonresponsive homolog of DAF-16 that fails to bind SRC can inhibit glucocorticoid-responsive gene transcription. From this we conclude that, in the absence of insulin, a major role of DAF-16-like factors may be to enhance glucocorticoid stimulation of its target genes. We show that the DAF-16-like protein FKHR is most similar to DAF-16 in its ability to mediate the negative effect of insulin on transcription of the IGFBP-1 gene. Our findings suggest that insulin may alter the activity of certain DAF-16-like proteins by preventing their association with coactivator proteins in addition to promoting association with 14-3-3 and retention in the cytoplasm as previously proposed (18, 23, 24). Thus, dependence of certain glucocorticoid-responsive genes on an FKH accessory factor site would allow DAF-16-like proteins to integrate the opposing effects of glucocorticoids and insulin on specific target genes.
Acknowledgments
We thank Hank Kronenberg, Joseph Avruch, and Joseph Bonventre for critical reading of the manuscript. This work was supported by Grants CA73818-1 from the National Cancer Institute to M.C.A.-B. and Joseph Avruch, as well as by institutional support from the Massachusetts General Hospital. N.N., S.O., G.R., and Y.S. were supported by funds from National Institutes of Health Grants T32 DK07028-24, AG05790, AG14161, and GM58012, respectively. D.C. is a recipient of a Taplin Research Fellowship.
Abbreviations
- CBP
CREB-binding protein
- SRC
steroid receptor coactivator
- GRE
glucocorticoid-response element
- IRE
insulin-response element
- IGFBP-1
insulin-like growth factor binding protein 1
- PEPCK
phosphoenolpyruvate carboxykinase
- PI 3-kinase
phosphatidylinositol 3-kinase
- PKB
protein kinase B
- GST
glutathione S-transferase
- FKH
forkhead
- HNF
hepatic nuclear factor
- CMV
cytomegalovirus
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190326997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190326997
References
- 1.Alessi D R, Cohen P. Curr Opin Genet Dev. 1998;8:55–62. doi: 10.1016/s0959-437x(98)80062-2. [DOI] [PubMed] [Google Scholar]
- 2.Avruch J. Mol Cell Biochem. 1998;183:31–48. [PubMed] [Google Scholar]
- 3.Hanson R W, Reshef L. Annu Rev Biochem. 1997;66:581–611. doi: 10.1146/annurev.biochem.66.1.581. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki K, Cripe T P, Koch S R, Andreone T L, Petersen D D, Beale E G, Granner D K. J Biol Chem. 1984;259:15242–15251. [PubMed] [Google Scholar]
- 5.Suwanickul A, Allander S V, Morris S L, Powell D R. J Biol Chem. 1994;269:30835–30841. [PubMed] [Google Scholar]
- 6.Suh D S, Ooi G T, Rechler M M. Mol Endocrinol. 1994;8:794–805. doi: 10.1210/mend.8.6.7523864. [DOI] [PubMed] [Google Scholar]
- 7.O'Brien R M, Granner D K. Physiol Rev. 1996;76:1109–1161. doi: 10.1152/physrev.1996.76.4.1109. [DOI] [PubMed] [Google Scholar]
- 8.Unterman T G, Fareedudin A, Harris M A, Goswami R G, Porcella A, Costa R H, Lacson R G. Biochem Biophys Res Commun. 1994;203:1835–1841. doi: 10.1006/bbrc.1994.2401. [DOI] [PubMed] [Google Scholar]
- 9.O'Brien R M, Noisin E L, Suwanichkul A, Yamasaki T, Lucas P C, Wang J, Powell D R, Granner D K. Mol Cell Biol. 1995;15:1747–1758. doi: 10.1128/mcb.15.3.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J-C, Strömstedt P-E, O'Brien R M, Granner D K. Mol Endocrinol. 1996;10:794–800. doi: 10.1210/mend.10.7.8813720. [DOI] [PubMed] [Google Scholar]
- 11.Wang J-C, Strömstedt P-E, Sugiyama T, Granner D K. Mol Endocrinol. 1999;13:604–618. doi: 10.1210/mend.13.4.0269. [DOI] [PubMed] [Google Scholar]
- 12.Allander S V, Durham S K, Scheimann A O, Wasserman R M, Suwanichkul A, Powell D R. Endocrinology. 1997;138:4291–4300. doi: 10.1210/endo.138.10.5268. [DOI] [PubMed] [Google Scholar]
- 13.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 14.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. Nature (London) 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 15.Lin K, Dorman J B, Rodan A, Kenyon C. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 16.Paradis S, Ruvkun G. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottlieb S, Ruvkun G. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunet A, Bonni A, Zigmond M, Lin M, Juo P, Hu L, Anderson M, Arden K, Blenis J, Greenberg M. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 19.Kops G, de Ruiter N, De Vries-Smits A, Powell D, Bos J, Burgering B. Nature (London) 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 20.Rena G, Guo S, Cichy S, Unterman T, Cohen P. J Biol Chem. 1999;274:17179–17183. doi: 10.1074/jbc.274.24.17179. [DOI] [PubMed] [Google Scholar]
- 21.Nakae J, Park B-C, Accili D. J Biol Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 22.Tang E, Nuñez G, Barr F, Guan K-L. J Biol Chem. 1999;274:16741–16746. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- 23.Biggs W H, Meisenhelder J, Hunter T, Cavenee W K, Arden K C. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takaishi H, Konishi H, Matsuzaki H, Ono Y, Shirai Y, Saito N, Kitamura T, Ogawa W, Kasuga M, Kikkawa U, Nishizuka Y. Proc Natl Acad Sci USA. 1999;96:11836–11841. doi: 10.1073/pnas.96.21.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safer J D, Cohen R N, Hollenberg A N, Wondisford F E. J Biol Chem. 1998;273:30175–30182. doi: 10.1074/jbc.273.46.30175. [DOI] [PubMed] [Google Scholar]
- 26.Cohen R N, Wondisford F E, Hollenberg A N. Mol Endocrinol. 1998;12:1567–1581. doi: 10.1210/mend.12.10.0188. [DOI] [PubMed] [Google Scholar]
- 27.Takeshita A, Yen P M, Misiti S, Cardona G R, Liu Y, Chin W W. Endocrinology. 1996;137:3594–3597. doi: 10.1210/endo.137.8.8754792. [DOI] [PubMed] [Google Scholar]
- 28.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Nature (London) 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 29.Buggs C, Nasrin N, Mode A, Tollet P, Zhao H-F, Gustafsson J-A, Alexander-Bridges M. Mol Endocrinol. 1998;12:1294–1309. doi: 10.1210/mend.12.9.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai J, Herr B. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durham S, Suwanichkul A, Scheimann A, Yee D, Jackson J, Barr F, Powell D. Endocrinology. 1999;140:3140–3146. doi: 10.1210/endo.140.7.6856. [DOI] [PubMed] [Google Scholar]
- 32.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani H Y, Evans R M. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 33.Dairmont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng X, Reginato M J, Andrews N C, Lazar M A. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida E, Nakajima T, Murakami K, Fukamizu A. Gene. 1998;208:307–314. doi: 10.1016/s0378-1119(98)00008-0. [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharya S, Michels C, Leung M, Arany Z, Kung A, Livingston D. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao T P, Ku G, Zhou N. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freedman L P. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 39.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 40.Wallberg A, Neely K, Gustafsson J, Workman J, Wright A, Grant P. Mol Cell Biol. 1999;19:5952–5959. doi: 10.1128/mcb.19.9.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almlof T, Wallberg A E, Gustafsson J A, Wright A P H. Biochemistry. 1998;37:9586–9594. doi: 10.1021/bi973029x. [DOI] [PubMed] [Google Scholar]
- 42.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Wilson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 43.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 44.Xu L, Lavinsky R, Dasen J, Flynn S, McInerney E, Mullen T-M, Heinzel T, Szeto D, Korzus E, Kurokawa R, Aggarwal A, Rose D, Glass C, Rosenfeld M. Nature (London) 1998;395:310–305. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Stafford J, Granner D. J Biol Chem. 1998;273:30847–30850. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]