Abstract
Purpose:
The CpG island methylator phenotype (CIMP) may be associated with development of malignancy through coordinated inactivation of tumor suppressor and tumor-related genes (TRG) and methylation of multiple noncoding, methylated-in-tumor (MINT) loci. These epigenetic changes create a distinct CIMP pattern that has been linked to recurrence and survival in gastrointestinal cancers. Because epigenetic inactivation of TRGs also has been shown in malignant melanoma, we hypothesized the existence of a clinically significant CIMP in cutaneous melanoma progression.
Experimental Design:
The methylation status of the CpG island promoter region of TRGs related to melanoma pathophysiology (WIF1, TFPI2, RASSF1A, RARβ2, SOCS1, and GATA4) and a panel of MINT loci (MINT1, MINT2, MINT3, MINT12, MINT17, MINT25, and MINT31) in primary and metastatic tumors of different clinical stages (n = 122) was assessed.
Results:
Here, we show an increase in hypermethylation of theTRGsWIF1,TFPI2, RASSF1A, and SOCS1with advancing clinical tumor stage. Furthermore, we find a significant positive association between the methylation status of MINT17, MINT31, and TRGs. The methylation status of MINT31is associated with disease outcome in stage III melanoma.
Conclusions:
These findings show the significance of a CIMP pattern that is associated with advancing clinical stage of malignant melanoma. Future prospective large-scale studies may determine if CIMP-positive primary melanomas are at high risk of metastasis or recurrence.
Cutaneous malignant melanoma is the sixth most common cancer in the United States and a major public health problem worldwide for which survival depends on both early detection and eradication of disease (1). To date, there have been limited studies addressing the role of epigenetic changes during early melanoma progression, or evaluating differences in the epigenetic patterns of primary versus metastatic tumors. However, epigenetic inactivation of tumor suppressor genes has been implicated in tumorigenesis and progression of a variety of different malignancies (2-4), and recent studies are beginning to show the role of epigenetic events in cutaneous melanoma (5, 6). Existing prognostic factors for primary melanoma include Breslow thickness and ulceration, but the clinical utility of these pathologic characteristics is limited. Delineation of factors involved in the progression of primary tumors may aid in the identification of individuals at high risk for recurrence and may guide the development of future targeted treatment strategies for patients with high-risk resected or metastatic disease.
Although the observation of methylation changes in CpG island promoter regions in a few tumor-related genes and tumor suppressor genes has been reported in the case of malignant cutaneous melanoma (7, 8), the clinical significance of these molecular aberrations is still being defined. For example, it has been well shown in other tumor systems that TFPI2 inhibits tumor growth, invasion, metastasis, and angiogenesis and induces apoptosis (9). Nobeyama et al. (10) noted that TFPI2 was methylated in 5 of 17 (29%) metastatic melanoma lesions but none of the primary tumors examined, suggesting that methylation-induced inactivation of this gene is involved in melanoma metastasis. Silencing of WIF1, a Wnt pathway antagonist, has been implicated in cellular proliferation of a variety of tumor types including non-small cell lung cancer (11), bladder and renal cell cancers (12, 13), and gastrointestinal cancers (14), and restoration of WIF1 expression has been shown to inhibit growth of melanoma in vitro and in vivo (15). SOCS1 is a known tumor suppressor gene that has been found circulating in the methylated form in melanoma patients (7). Expression of GATA4, a gene encoding a transcription factor thought to act like a tumor suppressor gene through its activation of several other genes with antitumor effects, has been found to be epigenetically silenced in gastrointestinal cancers (16) and lung cancer (17), although there are no reports to date of its role in melanoma development. RARβ2 methylation has been shown previously by our laboratory to be present in a high percentage of clinical melanoma specimens and to be associated with increased Breslow depth of primary melanomas (5), implicating its role in tumori-genesis. Our laboratory and others have shown the significance of RASSF1A and RARβ2 hypermethylation in predicting nonresponsiveness to biochemotherapy in American Joint Committee on Cancer (AJCC) stage IV melanoma patients (6).
Translational Relevance.
This study was designed to profile multiple tumor-related genes (TRG) and members of the methylated-in-tumor loci family to identify a CpG island methylator phenotype (CIMP) pattern in malignant cutaneous melanoma. We now describe, for the first time in cutaneous melanoma, that the CIMP is associated with tumor progression. In addition, we describe several key TRGs that become progressively hypermethylated with progression of primary melanoma. By knowing the epigenetic biomarkers associated with advancing tumor stage, it is conceivable that their identification in primary tumors may help to identify those tumors at high risk of metastasis or recurrence. Thus, the epigenetic biomarker phenotype of a primary melanoma could be used, in addition to currently used clinical and histopathologic features, to determine which patients may derive the most benefit from adjuvant therapy. Furthermore, the identification of epigenetic biomarkers may also be used to design future targeted therapeutics that act to reverse hypermethylation of selected TRGs.
In gastric and colorectal cancer, the existence of a CpG island methylator phenotype (CIMP) has been described and found to be associated with tumor development through coordinated inactivation of multiple tumor suppressor and mismatch repair genes (4, 18). The CIMP is marked by methylation of multiple noncoding, methylated-in-tumor (MINT) loci, which have been shown to underlie epigenetic changes in gastrointestinal tumors. Methylation of MINT loci is thought to be associated with a high degree of hypermethylation of tumor-related genes (TRG) as observed, for example, with the high prevalence of p16 and THBS1 hypermethylation in CIMP-positive colorectal tumors (18). The CIMP has also been shown to be a predictive marker of survival benefit from adjuvant 5-fluorouracil-based chemotherapy in patients with colorectal carcinoma metastatic to regional lymph nodes (19).
In this study, we examined the methylation status of CpG islands in the promoter region of the above-described six TRGs known to exhibit epigenetic aberrations associated with malignancy, and seven MINT loci to determine whether there exists a clinically significant CIMP related to melanoma progression.
Materials and Methods
Cell lines
HeMnMP, a moderately pigmented human melanocyte strain, was obtained from Cascade Biologics and maintained in medium 254 with human melanocyte growth supplement. A dermal fibroblast cell line originating from a healthy donor was established and kindly donated by the Osaka University Department of Dermatology and maintained in DMEM supplemented with 10% heat-inactivated FCS. Twelve melanoma cell lines were established from metastatic tumors at the John Wayne Cancer Institute and maintained in RPMI 1640 supplemented with penicillin, streptomycin, and 10% heat-inactivated FCS. All cultures were maintained at 37°C, 5% CO2 in a humidified incubator.
Clinical specimens
Approval for the use of human tissues was obtained from the John Wayne Cancer Institute/Saint John's Health Center Institutional Review Board before study initiation. Analysis was undertaken of 122 paraffin-embedded archival tissue specimens from 107 patients diagnosed with malignant melanoma by the Division of Surgical Pathology at Saint John's Health Center. Specimens were classified using the 2002 AJCC staging criteria for cutaneous melanoma (20). Of the 122 paraffin-embedded archival tissue melanoma specimens, 35 were from primary tumors associated with AJCC stage I (n = 18) and stage II (n = 17) disease. A total of 25 stage III patients were included. Of these, 7 had only primary tumor specimens available for analysis and 8 had only specimens from nodal metastases. For 10 patients, both primary and nodal specimens were available.
Normal skin control samples were obtained from tumor-free areas of primary melanoma tissue blocks. Clinical characteristics of the enrolled patients are summarized in Table 1. The patients consisted of 39 females and 68 males between ages 12 and 88 years. Breslow thickness data were available for 48 of 52 patients with primary tumor specimens and 41 of 55 patients with regional lymph node or distant organ metastases. Mean clinical follow-up was 38.9 months (range, 0-328).
Table 1.
Clinical characteristics of melanoma patients and tissue samples
| Characteristics | n (%) |
|---|---|
| Total patients | 107 |
| Age | |
| Mean ± SD | 59.4 ± 16.64 |
| Median, min-max | 60, 12-88 |
| <50 | 27 (25.2) |
| ≥50 | 80 (74.8) |
| Gender | |
| Female | 39 (36.4) |
| Male | 68 (63.6) |
| Breslow thickness | |
| ≤1.0 | 19 (17.8) |
| 1.01-2.0 | 22 (20.6) |
| 2.01-4.0 | 32 (29.9) |
| >4.0 | 16 (15) |
| Unknown | 18 (16.8) |
| Total tissue samples | 122 |
| AJCC stage | |
| I | 18 (14.8) |
| II | 17 (13.9) |
| III (primary tumor only) | 7 (5.7) |
| III (lymph node metastasis only) | 8 (6.6) |
| III (primary tumor and lymph node metastasis) | 10 (8.2) |
| IV (metastasis) | 52 (42.6) |
| Skin/soft tissue | 12 (23.1) |
| Lung | 11 (21.2) |
| Adrenal gland | 10 (19.2) |
| Lymph node | 8 (15.4) |
| Small bowel | 6 (11.5) |
| Other | 5 (9.6) |
Paired early- and advanced-stage specimens were available for 16 patients. Thirteen patients had primary tumor specimens with subsequent nodal (n = 10) or distant (n = 3) metastatic tumor specimens, and 3 patients had nodal metastases followed by distant metastases. Of the 18 stage III patients with specimens from lymph node metastases, 10 had primary tumor specimens available and 3 subsequently developed distant metastatic lesions. These paired tumor specimens were used to examine differences in methylation with stage progression on a per-patient basis.
Sites of distant metastasis for the 52 stage IV patients studied included skin or subcutaneous tissue (n = 12), lung (n = 11), adrenal gland (n = 10), nonregional lymph nodes (n = 8), small bowel (n = 6), and others (n = 5).
DNA isolation
Sections (8 μm) were cut from formalin-fixed, paraffin-embedded archival tissue blocks. A H&E slide was prepared for each sample to confirm tumor location and to assess tissue homogeneity by light microscopy. Tumor tissues were isolated using manual microdissection. To extract DNA, dissected tissues were digested with 100 μL lysis buffer containing 2.4 mAU proteinase K (Qiagen) at 50°C overnight followed by heat inactivation of proteinase K at 95°C for 15 min. DNA was purified with phenol/chloroform/isoamyl alcohol (Fisher Scientific), precipitated by ethanol, and quantified using the PicoGreen Assay (Molecular Probes/Invitrogen). DNA from cell lines was isolated using DNAzol Genomic DNA Isolation Reagent (Molecular Research Center) according to the manufacturer's recommendations and then quantified and assessed for purity by UV spectrophotometry. Extracted DNA was subjected to sodium bisulfite modification (as described previously; refs. 5, 6).
Epigenetic changes detected by methylation-specific PCR
Methylalation status was assessed for each gene using two sets of fluorescent-labeled primers designed to amplify methylated or unmethylated DNA sequences. Methylated and unmethylated primer sequences are summarized in Supplementary Table S1A. Primers were designed using MethPrimer (21). Bisulfite-modified DNA was subjected to PCR amplification in a final reaction volume of 10 μL containing PCR buffer, 2.5 to 4.5 mmol/L MgCl2, 0.8 mmol/L deoxynucleotide triphosphates, 0.3 μmol/L primers, and 0.5 units AmpliTaq Gold DNA polymerase (Applied Biosystems). PCR amplification was done with an initial 10 min incubation at 95°C followed by 36 to 40 cycles of denaturation at 95°C for 30 s, annealing for 30 s, extension at 72°C for 45 s, and a final 7-min hold at 72°C. Lymphocyte DNA obtained from healthy donors and amplified by phi-29 DNA polymerase served as a positive unmethylated control after sodium bisulfite modification (22). SssI methylase (New England Bio Labs)-treated lymphocyte DNA served as a positive methylated control. Unmodified lymphocyte DNA was used as a negative control for methylated and unmethylated reactions.
Capillary array electrophoresis
PCR products were assessed using capillary array electrophoresis (CEQ 8000XL; Beckman Coulter) as described previously (6) using Beckman Coulter WellRED dye-labeled phosphoramidites (Genset Oligos). Forward methylated sequence-specific primers were labeled with D4 dye, and forward unmethylated sequence-specific primers were labeled with D3 dye. Methylated PCR product (1 μL) and unmethylated PCR product (1 μL) were mixed with loading buffer and a dye-labeled size standard (Beckman Coulter) and loaded in a 96-well plate for CEQ peak ratio analysis. Samples showing only a peak for D3 dye (representing unmethylated DNA) were marked as unmethylated. Samples showing a peak for D4 dye (representing methylated DNA) or peaks for both methylated and unmethylated DNA were marked as methylated.
Absolute quantitative analysis of methylated alleles
To quantify the methylation status of seven MINT loci (MINT1, MINT2, MINT3, MINT12, MINT17, MINT25, and MINT31), we employed the absolute quantitative analysis of methylated allele assay as described previously (3). A single set of PCR primers was designed to amplify bisulfite-modified DNA for both methylated and unmethylated sequences using Primer 3 software.4 The methylation status of CpGs was distinguished by two different minor groove binder molecule-containing probes (Applied Biosystems), specific for either methylated or unmethylated sequences, designed with Primer Express software (version 2.0; Applied Biosystems). Methylated and unmethylated probes were labeled with 6-carboxyfluorescein and VIC, respectively. Black hole quenchers were used to silence the probes' fluorescent signals when not hybridized. The sequences of primer sets and minor groove binder probes are listed in Supplementary Table S1B.
Real-time PCR for the absolute quantitative analysis of methylated allele assay was done as described previously (3). The reaction mixture totaling 10 μL for each absolute quantitative analysis of methylated allele PCR consisted of 1 μL modified template DNA, PCR buffer, 0.4 μmol/L of each forward and reverse primer, 1.4 units iTaq DNA polymerase (Bio-Rad Laboratories), 0.6 mmol/L deoxynucleotide triphosphates, 0.025 pmol/L of each minor groove binder probe, and 4.5 mmol/L MgCl2. The mixture was processed by a two-step PCR method using ABI Prism 7900HT Sequence Detection System (Applied Biosystems) with an initial heating at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C (58°C for MINT3 and MINT25) for 60 s. The obtained PCR amplification curves from methylated and unmethylated alleles were analyzed with SDS software version 2.3 (Applied Biosystems). The final data output was reported as “methylation index” (methylation index = methylated copy number / [methylated copy number + unmethylated copy number]). All experiments were done in duplicate; mean values from duplicate measurements were used for calculation of the methylation index. Control DNA from methylated lymphoblastoid cell lines (AGS and Raji) or unmethylated gastric cancer cell lines (RL-0380 and FN-0028 from John Wayne Cancer Institute) was used to verify the reproducibility and accuracy of this assay. To quantify methylated and unmethylated copy numbers, a standard curve was created using high-fidelity and quality-constructed plasmids for methylated and unmethylated sequences as described previously (3). The mean methylation index plus 1 SD obtained from 12 nontumor skin specimens was used as the cutoff point to separate the methylated and unmethylated samples.
Statistical analysis
Categorical data were analyzed using the χ2 test; Fisher's exact test was used in the case of small sample sizes. Two-tailed P values < 0.05 were considered statistically significant. Bonferroni correction was applied to multiple comparisons. Trend analysis of methylation across AJCC stages was done using the Cochran-Armitage test. McNemar's test was used to compare the methylation frequency of different stage samples obtained from the same patient. Cox proportional hazards regression models were created for overall survival and disease-free survival calculations incorporating multiple variables. The factors for multivariate analysis included presence or absence of methylation for each marker, gender, age, Breslow thickness, and presence or absence of ulceration, each as independent variables. Survival curves were constructed using the log-rank method. All statistical calculations were done using SAS software version 8.02 (SAS Institute).
Results
Methylation profiles of cell lines
Methylation-specific PCR and absolute quantitative analysis of methylated allele primers and probes were initially screened using eight melanoma tumor specimens to detect promoter methylation of six TRGs and methylation of seven MINT loci, respectively. Two of the seven MINT loci from these initial screening analyses showed a significant difference in methylation frequency in the tumor specimens compared with tumor-free skin portions of the same patient samples (data not shown). Other MINT loci in the initial screening analysis showed similar high frequencies of methylation (MINT12) or low to absent methylation (MINT1, MINT2, MINT3, and MINT25) in both tumors and normal skin. Therefore, further analyses by absolute quantitative analysis of methylated allele focused on MINT17 and MINT31. Initial screening analysis of the six TRGs similarly showed high frequencies of promoter methylation in the eight tumor tissues tested compared with uniform absence of promoter methylation in the tumor-free skin portions of the same patient samples.
WIF1, TFPI2, RASSF1A, RARβ2, SOCS1, GATA4, MINT17, and MINT31 were each methylated in at least 50% of the 12 melanoma cell lines tested (Table 2). All biomarkers were methylated in cell lines M1 to M3, whereas none were methylated in M11 and M12. All biomarkers were unmethylated in melanocyte and dermal fibroblast cell lines.
Table 2.
Promoter hypermethylation of TRGs and MINT loci in melanoma cell lines
| Cell lines | WIF1 | TFPI2 | RASSF1A | RARβ2 | SOCS1 | GATA4 | MINT17 | MINT31 | Markers methylated |
|---|---|---|---|---|---|---|---|---|---|
| M1 | M | M | M | M | M | M | M | M | 8 |
| M2 | M | M | M | M | M | M | M | M | 8 |
| M3 | M | M | M | M | M | M | M | M | 8 |
| M4 | M | M | M | M | M | U | M | M | 7 |
| M5 | M | M | M | M | M | M | U | M | 7 |
| M6 | U | M | M | M | M | U | M | M | 6 |
| M7 | M | M | U | M | U | M | M | M | 6 |
| M8 | U | U | M | M | M | U | M | M | 5 |
| M9 | M | U | U | M | M | M | U | M | 5 |
| M10 | U | U | U | U | U | U | U | M | 1 |
| M11 | U | U | U | U | U | U | U | U | 0 |
| M12 | U | U | U | U | U | U | U | U | 0 |
| Melanocyte | U | U | U | U | U | U | U | U | 0 |
| Dermal fibroblast | U | U | U | U | U | U | U | U | 0 |
| Methylated lines (%) | 58.3 | 58.3 | 58.3 | 75 | 66.7 | 50 | 58.3 | 83.3 |
Abbreviations: M, methylated; U, unmethylated.
Methylation profiles in melanomas
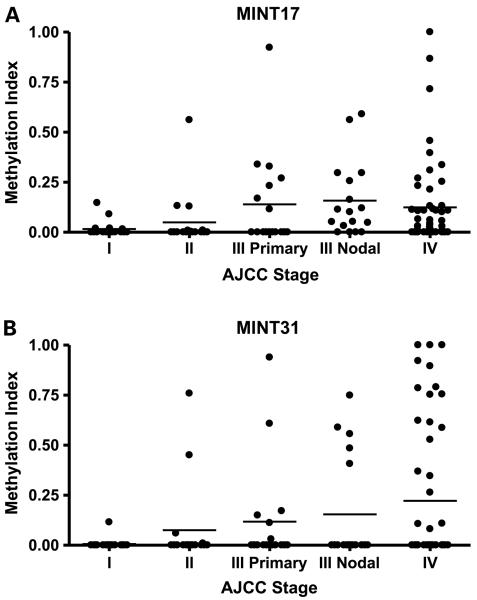
The methylation index obtained for MINT17 and MINT31 loci, stratified by AJCC stage, are depicted in Fig. 1A and B, respectively. Overall methylation percentages stratified by AJCC stage for each of the six TRGs and the two MINT loci are reported in Table 3A. Univariate analysis revealed no significant difference in methylation status by age or gender (data not shown).
Fig. 1.
Methylation of MINT loci increases with advancing AJCC stage. MINT17 (A) and MINT31 (B) methylation indices for each tumor specimen stratified by AJCC stage. Columns, mean for grouping of each stage.
Table 3.
| (A) Percent of melanoma tissues exhibiting methylation of TRGs and MINT loci | ||||||||
|---|---|---|---|---|---|---|---|---|
| AJCC stage (n) | MINT17 | MINT31 | WIF1 | TFPI2 | RASSF1A | RARβ2 | SOCS1 | GATA4 |
| I, P (18) | 11.1 | 5.6 | 5.6 | 0 | 0 | 58.3 | 7.1 | 16.7 |
| II, P (17) | 17.6 | 23.5 | 6.3 | 6.3 | 0 | 66.7 | 23.1 | 8.3 |
| III, P (17) | 41.2 | 35.3 | 31.3 | 17.6 | 26.7 | 64.3 | 25 | 42.9 |
| III, M (18) | 52.9 | 27.8 | 50 | 17.6 | 27.8 | 47.1 | 23.5 | 22.2 |
| IV, M (52) | 38.5 | 36.5 | 44 | 44.9 | 48.9 | 56.3 | 44.9 | 34 |
| Overall (122) | 34.2 | 28.1 | 33.3 | 21.7 | 28.6 | 57.6 | 31.5 | 27.5 |
| (B) Difference in methylation status between AJCC stages | ||||||||
| Stage comparison | MINT17 | MINT31 | WIF1 | TFPI2 | RASSF1A | RARβ2 | SOCS1 | GATA4 |
| Stage I vs II | NS | NS | NS | NS | NS | NS | NS | NS |
| Stage I vs III | 0.049 | 0.036 | NS | NS | NS | NS | NS | NS |
| Stage I vs IV* | 0.021 | 0.005 | 0.003 | 0.006 | <0.0001 | NS | 0.005 | NS |
| Stage II vs III | NS | NS | NS | NS | 0.015 | NS | NS | 0.039 |
| Stage II vs IV* | NS | NS | 0.015 | 0.013 | <0.0001 | NS | NS | 0.055 |
| Stage III vs IV* | NS | NS | NS | NS | NS | NS | NS | NS |
NOTE: Bold entries refers to ≥50%. P value analyzed by Fisher's exact test. NS, not significant. P values < 0.0083 are significant after Bonferroni correction (in bold).
Abbreviations: P, primary; M, metastatic. For additional information on specimen, see Table 1.
For comparisons with stage IV, χ2 test was used.
Advancing AJCC stage was associated with increased methylation of MINT17 (P = 0.0004), MINT31 (P = 0.026), TFPI2 (P = 0.001), WIF1 (P = 0.002), SOCS1 (P = 0.009), and RASSF1A (P < 0.0001), but not GATA4 and RARβ2, as determined by the Cochran-Armitage test. This finding was most pronounced for TFPI2 and RASSF1A, which were uniformly unmethylated (0%) in stage I primary tumor specimens, whereas the methylation frequency of these genes was 45% and 49% in stage IV metastatic specimens, respectively. Conversely, RARβ2 was found to be highly methylated in early-stage primary tumors (58% and 67% for stage I and II specimens, respectively). Similarly, 17% of stage I primary tumors showed GATA4 methylation, which did not reliably or significantly increase with advancing stage. Significant increases in the methylation frequencies of MINT17, MINT31, and the TRGs WIF1, TFPI2, RASSF1A, and SOCS1 were found when comparing stage I primary tumors versus stage IV metastatic tumors, but not stage I versus stage II, or stage III versus stage IV (Table 3B). For MINT17 and WIF1, a decrease in the percentage of hypermethylated specimens was noted from stage III (nodal) to stage IV. There were no significant differences observed in methylation frequency or methylation index between different anatomic sites of distant metastasis (data not shown). Both early- and advanced-stage paired tumors were available for 16 patients in our study group. Examination of these paired tumors showed a significant increase of WIF1 methylation with AJCC stage progression (P = 0.01; data not shown).
Relationship between methylation status of MINT loci and TRGs.
Positive relationships were found for methylation of MINT17 with MINT31, TFPI2, WIF1, and SOCS1 (Table 4). MINT31 methylation was positively associated with methylation of all six TRGs. Methylation of TFPI2 and WIF1 was also associated with methylation of the other TRGs. There was no statistically significant relationship between methylation of MINT17 compared with GATA4, RASSF1A, or RARβ2; methylation of GATA4 was associated with RASSF1A and RARβ2 methylation, however. The absence of a methylation relationship was also noted for SOCS1 compared with GATA4 and RARβ2 as well as RASSF1A with RARβ2.
Table 4.
Relationship between methylation status of MINT loci and TRGs
| Biomarker | Comparison | P* |
|---|---|---|
| MINT17 | MINT31 | 0.033 |
| TFPI2 | 0.014 | |
| WIF1 | 0.002 | |
| SOCS1 | 0.013 | |
| GATA4 | NS | |
| RASSF1A | NS | |
| RARβ2 | NS | |
| MINT31 | TFPI2 | 0.002 |
| WIF1 | 0.009 | |
| SOCS1 | 0.002 | |
| GATA 4 | <0.0001 | |
| RASSF1A | 0.002 | |
| RARβ2 | 0.042 | |
| TFPI2 | WIF1 | <0.0001 |
| SOCS1 | 0.0003 | |
| GATA4 | 0.0001 | |
| RASSF1A | <0.0001 | |
| RARβ2 | 0.008 | |
| WIF1 | SOCS1 | 0.001 |
| GATA4 | 0.002 | |
| RASSF1A | 0.0001 | |
| RARβ2 | 0.005 | |
| SOCS1 | GATA4 | NS |
| RASSF1A | 0.023 | |
| RARβ2 | NS | |
| GATA4 | RASSF1A | <0.0001 |
| RARβ2 | 0.005 | |
| RASSF1A | RARβ2 | NS |
Two-tailed χ2 test. All significant relationships are positive. NS, not significant.
MINT31 hypermethylation predicts improved disease-free and overall survival.
Disease-free and overall survival rates in stage I and II malignant melanoma are very high. Conversely, stage IV disease is marked by a much shorter median survival that is further affected by site of metastasis. Therefore, the assessment of disease outcome in relation to methylation of specific genes and loci was limited to AJCC stage III patients only. Survival analysis was conducted for all stage III patients stratified by biomarker methylation status (n = 25). Stage III patients with primary versus nodal metastatic specimens were compared in a univariate analysis for differences in biomarker methylation, Breslow depth, Clark level, gender, histologic type, and tumor ulceration; no statistically significant differences were found (data not shown). Of the 25 AJCC stage III patients analyzed, clinical treatment consisted of multimodal therapy including surgery, vaccine therapy, chemotherapy, nonspecific or intratumoral Bacillus Calmette-Guerin, cytokine therapy (interleukin-2 and/or IFN), and radiation. After confirming the absence of statistically significant differences in clinicopathologic features and biomarker methylation within the AJCC stage III patient population, a multivariate survival analysis was done.
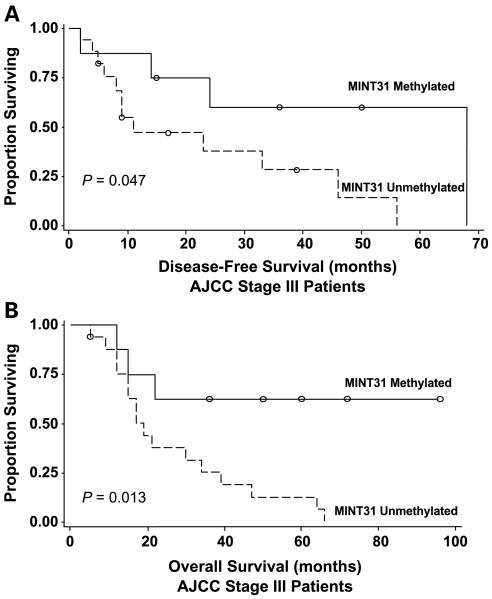
MINT31 methylation was found to be a significant predictor of improved overall survival (Cox proportional hazards regression model; hazard ratio = 0.237; P = 0.024) for all 25 patients in our study with AJCC stage III disease. The log-rank test confirmed both disease-free and overall survival benefits with MINT31 methylation (Fig. 2A and B; P = 0.047 and 0.013, respectively). No adverse or beneficial effects on clinical outcome were noted with methylation of any of the other biomarkers tested.
Fig. 2.
Improved disease-free and overall survival for AJCC stage III patients with MINT31 methylation. Kaplan-Meier curves for disease-free survival (A) and overall survival (B) in stage III patients. Log-rank test confirmed better disease-free survival (P = 0.047) and overall survival (P = 0.013) for patients with tumor samples with MINT31 methylation.
Discussion
Our study investigated the clinical significance of CpG island methylation status in the evolution and progression of malignant melanoma. Analysis of primary and metastatic tumors across different clinical stage groupings provided a unique opportunity to determine whether these epigenetic changes are related to tumor progression. Aberrant hyper-methylation of the genomic markers was not present in normal melanocytes or fibroblasts but was identified to varying degrees in primary and metastatic tumor tissues. Methylation of MINT17, MINT31, TFPI2, WIF1, RASSF1A, and SOCS1 increased significantly with advancing clinical stage, strongly suggesting that inactivation of these genes and loci is associated with tumor progression. These findings in melanoma are consistent with previous reports of MINT methylation as a determinant of a cancer-specific CIMP in gastric and colorectal cancers (4, 18) and of the association of TRG hypermethylation with melanoma and other cancers (6, 9, 10, 15).
For MINT17 and WIF1 in particular, it was interesting to note that lower methylation percentages were found for stage IV (distant) metastatic specimens in contrast with stage III (nodal) metastases. One plausible explanation could be that hyper-methylation of MINT17 and WIF1 is involved with the initiation of the metastatic process, such that tumor clones with a higher degree of hypermethylation are more likely to migrate to and establish metastases in regional lymph nodes, whereas those tumor cells with a lower degree of hyper-methylation are more suited to formation of distant metastases. Alternatively, the tumor microenvironment may select for the establishment of specific tumor cell clones expressing particular methylation patterns.
Paired analyses of patients with tumor specimens from both early- and advanced-stage disease showed significant increases in WIF1 methylation with melanoma stage progression. To our knowledge, this is the first clinical evidence of the role of WIF1 methylation in melanoma progression. These data strongly support the results of earlier in vitro and animal studies of the involvement of Wnt signaling in melanoma tumor growth, the ability to inhibit tumor growth with the restoration of WIF1 expression, and the potential use of Wnt pathway inhibition as a targeted therapy for high-risk or metastatic melanoma (15).
In contrast, RARβ2 methylation was seen in 58% of all tumor specimens tested without a detectable association with AJCC stage, implying that epigenetic inactivation of this particular gene may be a very early event in tumorigenesis. Although there was considerable variability in GATA4 methylation status across tumor stage groupings, GATA4 methylation was identified in a significant percentage of stage I tumors (Table 3A) but not in the melanocyte or dermal fibroblast cell lines (Table 2) or normal skin specimens (data not shown). This implies that GATA4 activation may play a role in tumor suppression, which is consistent with previous reports of its function in other cancers (16).
Of particular interest in this study were the positive relationships between the methylation status of MINT loci and TRGs because a methylator phenotype based on multiple TRGs may have more prognostic clinical value than the methylation status of any one particular TRG. Methylation of MINT31 was positively associated with methylation of all six TRGs, as was methylation of TFPI2 and WIF1. Although methylation of MINT17 was associated with methylation of MINT31, TFPI2, WIF1, and SOCS1, no relation was found between MINT17 methylation and GATA4, RASSF1A, or RARβ2. A pattern emerging from these data suggests that MINT17 methylation is a particularly sensitive marker for disease progression because it is present in conjunction with methylation of the TRGs that are strongly associated with advancing clinical stage. Because MINT31 methylation is associated with methylation of all of the TRGs, it is perhaps more suitable as a biomarker of disease presence or absence. MINT17 and MINT31 methylation may therefore be representative of a CIMP for malignant melanoma. Potential clinical applications of this knowledge include the testing of primary melanomas for MINT17 hypermethylation and, used in conjunction with clinicopathologic factors such as Breslow depth, Clark level, ulceration, and mitotic rate to, offer further treatment such as lymph node biopsy based on the result.
This study included a preliminary analysis of survival in a subgroup of patients with stage III melanoma. Survival plots stratified by methylation status were notable for improved disease-free and overall survival associated with methylation of MINT31 but not of any other biomarkers. It is conceivable that alterations in the activation status of additional genes or gene products other than those examined here may result in phenotypic changes leading to slower disease progression and/or tumor cell doubling times, or perhaps improved recognition of tumor cells by the immune system. Larger validation studies of the role of MINT and TRG methylation in melanoma will be required to answer this question.
Supplementary Material
Acknowledgments
Grant support: NIH, National Cancer Institute project II P0 CA029605 and CA012582 and R33 CA100314 grants (D.S.B. Hoon) and Roy E. Coats Research Laboratories, John Wayne Cancer Institute, Saint John's Health Center; Harold J. McAlister Charitable Foundation, Family of Robert Novick, Weil Family Fund, Wrather Family Foundation, and Samueli Foundation (A.M. Terando).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 3.de Maat MF, Umetani N, Sunami E, Turner RR, Hoon DS. Assessment of methylation events during colorectal tumor progression by absolute quantitative analysis of methylated alleles. Mol Cancer Res. 2007;5:461–71. doi: 10.1158/1541-7786.MCR-06-0358. [DOI] [PubMed] [Google Scholar]
- 4.Kusano M, Toyota M, Suzuki H, et al. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer. 2006;106:1467–79. doi: 10.1002/cncr.21789. [DOI] [PubMed] [Google Scholar]
- 5.Hoon DS, Spugnardi M, Kuo C, Huang SK, Morton DL, Taback B. Profiling epigenetic inactivation of tumorsuppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–22. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mori T, O'Day SJ, Umetani N, et al. Predictive utility of circulating methylated DNA in serum of melanoma patients receiving biochemotherapy. J Clin Oncol. 2005;23:9351–8. doi: 10.1200/JCO.2005.02.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marini A, Mirmohammadsadegh A, Nambiar S, Gustrau A, Ruzicka T, Hengge UR. Epigenetic inactivation of tumor suppressor genes in serum of patients with cutaneous melanoma. J Invest Dermatol. 2006;126:422–31. doi: 10.1038/sj.jid.5700073. [DOI] [PubMed] [Google Scholar]
- 8.Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res. 2003;63:1639–43. [PubMed] [Google Scholar]
- 9.Sierko E, Wojtukiewicz MZ, Kisiel W. The role of tissue factor pathway inhibitor-2 in cancer biology. Semin Thromb Hemost. 2007;33:653–9. doi: 10.1055/s-2007-991532. [DOI] [PubMed] [Google Scholar]
- 10.Nobeyama Y, Okochi-Takada E, Furuta J, et al. Silencing of tissue factor pathway inhibitor-2 gene in malignant melanomas. Int J Cancer. 2007;121:301–7. doi: 10.1002/ijc.22637. [DOI] [PubMed] [Google Scholar]
- 11.Mazieres J, He B, You L, et al. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717–20. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- 12.Urakami S, Shiina H, Enokida H, et al. Epigenetic inactivation of Wnt inhibitory factor-1plays an important role in bladder cancer through aberrant canonical Wnt/h-catenin signaling pathway. Clin Cancer Res. 2006;12:383–91. doi: 10.1158/1078-0432.CCR-05-1344. [DOI] [PubMed] [Google Scholar]
- 13.Urakami S, Shiina H, Enokida H, et al. Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res. 2006;12:6989–97. doi: 10.1158/1078-0432.CCR-06-1194. [DOI] [PubMed] [Google Scholar]
- 14.Taniguchi H, Yamamoto H, Hirata T, et al. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene. 2005;24:7946–52. doi: 10.1038/sj.onc.1208910. [DOI] [PubMed] [Google Scholar]
- 15.Lin YC, You L, Xu Z, et al. Wnt inhibitory factor-1gene transfer inhibits melanoma cell growth. Hum Gene Ther. 2007;18:379–86. doi: 10.1089/hum.2006.005. [DOI] [PubMed] [Google Scholar]
- 16.Akiyama Y, Watkins N, Suzuki H, et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol. 2003;23:8429–39. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo M, Akiyama Y, House MG, et al. Hypermethylation of the GATA genes in lung cancer. Clin Cancer Res. 2004;10:7917–24. doi: 10.1158/1078-0432.CCR-04-1140. [DOI] [PubMed] [Google Scholar]
- 18.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–903. [PubMed] [Google Scholar]
- 20.Balch CM, Buzaid AC, Soong SJ, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19:3635–48. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 21.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 22.Umetani N, de Maat MF, Mori T, Takeuchi H, Hoon DS. Synthesis of universal unmethylated control DNA by nested whole genome amplification with phi29 DNA polymerase. Biochem Biophys Res Commun. 2005;329:219–23. doi: 10.1016/j.bbrc.2005.01.088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.