Abstract
The Meningococcus Genome Informatics Platform (MGIP) is a suite of computational tools for the analysis of multilocus sequence typing (MLST) data, at http://mgip.biology.gatech.edu. MLST is used to generate allelic profiles to characterize strains of Neisseria meningitidis, a major cause of bacterial meningitis worldwide. Neisseria meningitidis strains are characterized with MLST as specific sequence types (ST) and clonal complexes (CC) based on the DNA sequences at defined loci. These data are vital to molecular epidemiology studies of N. meningitidis, including outbreak investigations and population biology. MGIP analyzes DNA sequence trace files, returns individual allele calls and characterizes the STs and CCs. MGIP represents a substantial advance over existing software in several respects: (i) ease of use—MGIP is user friendly, intuitive and thoroughly documented; (ii) flexibility—because MGIP is a website, it is compatible with any computer with an internet connection, can be used from any geographic location, and there is no installation; (iii) speed—MGIP takes just over one minute to process a set of 96 trace files; and (iv) expandability—MGIP has the potential to expand to more loci than those used in MLST and even to other bacterial species.
INTRODUCTION
Epidemiological surveillance of Neisseria meningitidis necessitates molecular typing. Standard methods for molecular typing include, but are not limited to, restriction fragment length polymorphism (1), pulsed field gel electrophoresis (2), multilocus sequence typing (MLST) (3,4), and porA, porB and fetA typing (5–7). MLST is the most modern and widely used of these approaches, and it provides an unambiguous method for typing bacterial strains (8). In MLST, specific regions of seven housekeeping genes are sequenced, their alleles are determined, and then the allele calls are concatenated to produce a profile called the sequence type (ST), which may then be grouped into a larger population called a clonal complex (CC). MLST analysis used in conjunction with the molecular typing methods listed above can provide evidence of possible genetic and epidemiological relatedness of strains identified during outbreak investigations and routine surveillance (9).
Epidemiological surveillance laboratories world-wide perform MLST using PCR and Sanger sequencing. Standard primers are used to amplify each of the seven loci, and the PCR fragments are then characterized using dye-terminator sequencing. The resulting trace files are interpreted by a computer or a human and are converted into unambiguous sequences (base calling). Some computer programs that will make base calls are Phred (10,11) and SeqMan (DNASTAR SeqMan Pro, Madison, WI). If there is more than one sequence read per locus, then those sequences must be assembled to generate a single consensus sequence. Computer programs that can perform assembly include Phrap (12) and SeqMan. The last step in MLST analysis is to determine the allele of the gene by comparing the consensus sequence of the trace files to a database of known allele sequences. In MLST, even a single nucleotide difference is sufficient to define a new allele and thus the comparison between the consensus sequence and the allelic database must be unambiguous.
The current standard software for MLST analysis, STARS (http://sara.molbiol.ox.ac.uk/userweb/mchan/stars/), is no longer supported by the original programmers, only runs on UNIX/Linux systems, and has performance and usability issues. STARS is included in distributions of Bio-Linux, but comprehensive and detailed instructions for its set up are necessary (http://pubmlst.org/software/bio-linux/stars/config/). A commonly used alternative to STARS is to make base calls, assemble sequences manually and then use the BLAST (13) interface at pubmlst.org (14) to ascertain the identity of the MLST alleles. MLST users and laboratories may use a variety of packages to analyze trace files and make base calls, including SeqMan, MEGA (http://www.megasoftware.net), BioNumerics (15) (http://www.applied-maths.com/bionumerics/bionumerics.htm) and the CLC MLST module (http://www.clcbio.com/index.php?id=1018). These alternatives, while viable, either require programming expertise to provide expanded capabilities, accessibility to a Linux system, or are prohibitively expensive. In addition, most of these alternatives represent piecemeal and tedious approaches that require a substantial dedication of time and resources. For all of these reasons, MLST analysis can be burdensome for laboratories in developing countries and/or in laboratories with fewer resources for personnel and computational support. To address these problems, we have developed an integrated suite of MLST analysis tools available at the Meningococcus Genome Informatics Platform (MGIP).
MGIP presents several key advantages over currently existing analytical methods: (i) Ease of use—MGIP runs as a web server, is designed to be user friendly, intuitive and is thoroughly documented. The documentation is on the website itself and covers any topic the user might need to see. If in any case the documentation is not sufficient, there is a conspicuous link to contact the lab for help; (ii) Flexibility—MGIP is compatible with any client computer or operating system and has been tested using Microsoft Internet Explorer, Mozilla Firefox, Safari and Google Chrome. Much of this flexibility is given by the cross-browser compatible JavaScript frameworks Prototype and Scriptaculous; (iii) Speed—MGIP has been shown to take about 1 min per set of 96 sequence trace files, more than five times faster than STARS. The speed of MGIP can be attributed to the fast constituent programs Phred, Phrap and BLAST as well as the server which is an eight-core machine. In addition, MGIP's ability to process multiple loci concurrently is a considerable advantage in comparison to other MLST analysis tools, which can only process one locus at a time; (iv) Expandability—Currently MGIP can analyze sequences from over 15 loci, which substantially increases the resolution of sequence typing. MGIP has the potential to include unlimited loci and has the capacity to include other organisms. We refer to MLST analysis with additional alleles as MLST+. Additional loci can be added in one of two ways: either by the administrator of MGIP or by an individual user. If a locus is added by the administrator, then all users can use the new locus database as both a BLAST database and for analyzing new trace files. If a user adds a database, it will be only visible to that user.
MLST+ ANALYSIS WORKFLOW
The workflow and programs underlying MLST+ analysis with MGIP are shown in Figure 1. Users of MGIP first sequence a set of loci to be used in MLST+. The loci that MGIP can process by default are shown in the Supplementary Table 1. Current protocols for N. meningitidis surveillance laboratories usually yield sequence data from 96-well plates, but the number of wells or traces does not affect the MGIP workflow.
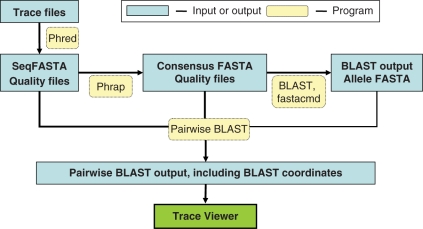
Figure 1.
MLST+ workflow on MGIP. Users upload trace files to the MGIP server for analysis. First, Phred makes base calls on each trace to produce a sequence FASTA file and a quality file. Next, Phrap aligns and produces a consensus sequence FASTA file and other associated files. BLAST is then used to match the consensus sequence against a database of known MLST+ alleles. Allelic FASTA files are extracted from the database using fastacmd and individually aligned to the consensus sequences to determine coordinates, mismatches and indels using pairwise BLAST. Alignments between consensus sequences, called allelic sequences and underlying trace files are displayed using the trace file viewer. The trace file viewer can be used to manually edit consensus sequences based on the aligned trace files (see Figure 3).
MGIP takes two files as input. The first file is a zip file of every trace received from the sequencing machine in one session. The second file is a mapping spreadsheet that assigns the following properties to each trace file: strain name, sequence typing method, locus and primer.
After these two files are submitted to MGIP, the trace files undergo processing: (i) Phred makes base calls on each trace file; (ii) Phrap assembles groups of trace files to make a consensus sequence for each strain/locus; and (iii) MGIP uses BLAST against a database of known MLST+ alleles to determine the allelic identity of the consensus sequence. BLAST results that do not have perfect matches in the database are flagged (perfect matches have 100% identity, 100% subject coverage and no indels). These flags are shown when viewing results and call attention to possibly novel or inaccurate results. MGIP also includes a trace viewer that shows alignments of consensus sequences, allele calls and underlying trace files. The trace viewer can be used to manually edit consensus sequences based on the aligned traces. The results of MGIP analyses are public unless users are registered and logged in at the time analysis is performed.
MGIP EASE-OF-USE AND UNIVERSAL COMPATIBILITY
One of the goals for the development of MGIP was to make a system that is simple and convenient to use. This goal is achieved via (i) an intuitive user interface, (ii) operating system and browser cross-compatibility and (iii) thorough documentation. These features are particularly relevant in the developing world where technical help and systems support may be scarce, but they are also applicable to scientists everywhere who do not wish to devote substantial resources, in both time and hardware, to computation.
As opposed to STARS, which requires machines running Linux, MGIP can be used with any operating system since it is run on a server with a web browser based client interface. MGIP is compatible with most standards-compliant web browsers because it largely conforms to the worldwide standards given by the World Wide Web Consortium (W3C). The Prototype and Scriptaculous frameworks, which are thoroughly tested on many browsers for compatibility, were used in the development of MGIP to ensure JavaScript compatibility. MGIP has been tested on Microsoft Internet Explorer, Mozilla Firefox, Safari and Google Chrome, which together account for almost 99% of all web browsers in use (http://marketshare.hitslink.com/, last accessed November 1, 2008).
USING MGIP
User accounts
MGIP was developed to allow users to upload private or sensitive data. Therefore, MGIP has a user management system, with few administrators and many users. Each user inherently has all of his or her data and user information privatized so that no other user can access them. To this end, MGIP employs standard web server security measures including MD5 password encryption and the use of a firewall (http://www.faqs.org/rfcs/rfc1321). For data that is not sensitive, or for scientists who do not wish to use individual accounts, there is a default public user setting with full functionality except data privatization.
Uploading traces and running analyses
Users upload trace files to MGIP for typing analysis. The user must create a zip file from all sequence trace files and a mapping spreadsheet file which identifies each trace's strain, sequence typing method, locus and primer. This spreadsheet is crucial for assembling the correct sequences together and provides names to each of the final results. An automatic spreadsheet generator is available which will generate the spreadsheet.
Viewing results by set
Fully automated analysis by MLST+ produces results for sets of traces which are viewed on the main page (Figure 2). For each set, the allele calls for each locus are displayed. For each locus, there is a submenu with options to (i) view BLAST results; (ii) view the consensus sequence and quality scores as given by Phrap; (iii) download all files involved in the MLST+ analysis workflow; and (iv) view trace files and edit the consensus sequence, thus allowing the user to manually adjust the results. BLAST results are reported in default format and for each hit show the allele names, bit scores and e-values along with pairwise query-hit sequence alignments. Consensus sequences are shown in FASTA format, with each nucleotide shown in a color corresponding to a range of quality scores. Loci that yield ambiguous results from the MLST+ analysis workflow have flags on the results screen that show the user where to intervene.
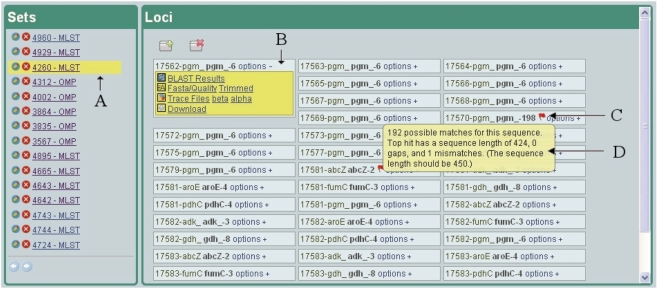
Figure 2.
Viewing MGIP+ results. (A) The user can select a set of results to view. For each set, all strain/loci and their allele calls are shown. (B) Options are shown for each strain/locus that allow the user to view more details. (C) When an allele call is not a perfect match, a flag appears. (D) On mouseover (when the mouse pointer hovers over the flag), a message giving information as to why it is not a perfect match appears.
To view trace files, we have developed a trace viewer Java applet that displays the alignment of all traces involved in the assembly process, the consensus sequence, and the allelic sequence (Figure 3). The applet automatically marks all discrepancies between the trace file base calls, the consensus sequence and the most similar allele, to facilitate scanning for inconsistencies. Trace amplitudes can be adjusted individually, and although the traces are trimmed to show only the aligned regions, there is an option to view trimmed edges. The consensus sequence can be edited by changing, adding, or deleting bases using the trace viewer. Once manual editing is completed, the workflow analysis starting with BLAST can be iterated so that the main page results are updated.
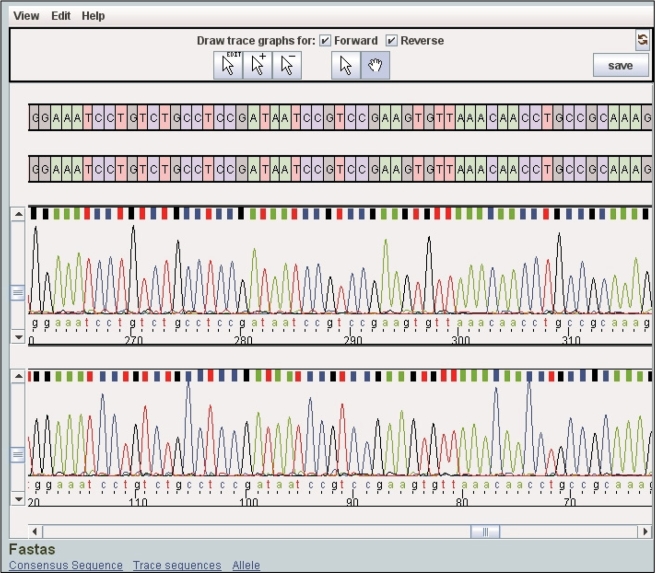
Figure 3.
The trace viewer and editor applet. The consensus sequence acts as a backbone when aligning the allelic sequence and the traces. The applet tools allow users to (1) alter the amplitude of the traces, (2) edit the consensus sequence, (3) insert/delete consensus sequence nucleotides, (4) undo/redo any action and (5) save a modified consensus sequence. Sequences of interest are embedded below the applet so that they can be copied and pasted.
Viewing results in the strain table
The results of MLST analyses are also displayed in the strain table, which shows the allelic profile, ST and CC for each strain that has been analyzed (Supplementary Figure 1). For each individual user account, the strain table is automatically and continually populated with the combined results of all sets that have been analyzed. If fewer than seven known alleles have been unambiguously characterized for any given strain, a list of all possible STs and CCs is shown.
Reference pages
To aid in data analysis, all reference data in the MGIP database has been made transparent and available on the reference pages. The ST reference page allows the user to type in an ST number and to retrieve the alleles associated with that ST. Alternatively, a CC number can be input to retrieve all STs associated with it. The locus reference page shows every locus that can be analyzed using MGIP. For each locus, the sequence typing method and the length of the allele are shown; all sequences in the locus databases can be downloaded. The locus reference page also has a BLAST interface, which accepts one or multiple FASTA query entries for comparison against the locus database.
PERFORMANCE VALIDATION
MGIP was compared against two other methodologies to validate performance in terms of both sensitivity and speed (Table 1). Sensitivity is defined as:
| 1 |
where Sn is sensitivity, TP is the number of true positives and FN is the number false negatives. In this study, a true positive is defined as an unambiguously identical match from the trace file(s) to the allelic database, and a false negative is defined as no match when there should be one. Specificity can not be measured because all methodologies in this study filter out false positives before they are reported. Speed is calculated simply as the time elapsed from upload to the end of sample processing.
Table 1.
MGIP is more sensitive and faster than other commonly used methods
| TP | FN | Sn (%) | Speed (s)a | |
|---|---|---|---|---|
| MGIP versus STARSb | ||||
| MGIP | 660 | 18 | 97.4 | 63 ± 0.58 |
| STARS | 653 | 35 | 94.9 | 323 ± 30 |
| MGIP versus SeqMan methodc | ||||
| MGIP | 323 | 6 | 98.2 | 75 ± 2 |
| SeqMan | 319 | 8 | 97.6 | 1520 ± 173 |
TP: true positives; FN: false negatives; Sn: sensitivity.
aSpeed is shown as an average per trace file set (84 traces in the STARS comparison, 96 traces in the SeqMan comparison), plus or minus standard deviation. The speed tests were performed over a 1 Gigabyte per second network connection and therefore the upload time was negligible. However, the upload time from a slower connection will understandably increase the time to process a set of trace files. Approximate times for uploading a set of traces is given in Supplementary Table 2.
bFor the MGIP versus STARS comparison, 17 sets of MLST data were tested which were composed of trace files over 691 strain/loci. The speed test was performed on three randomly selected sets, composed of 126 strain/loci.
cFor the MGIP versus SeqMan method comparison, 10 sets of fetA were tested in the SeqMan comparison, totaling 331 loci. The speed test was performed on three randomly selected sets, composed of 103 strain/loci.
The first methodology compared to MGIP was STARS, which is the current standard for MLST analysis. For comparison to STARS, 17 MLST sets were analyzed totaling 691 strain/locus combinations. MGIP showed 97.4% sensitivity compared to 94.9% sensitivity for STARS (Table 1). In addition to being more sensitive, MGIP is also substantially faster than STARS. On average, MGIP finished analyzing a set of 84 trace files in 63 s compared to 323 s for STARS (Table 1).
The second methodology compared to MGIP was the ‘SeqMan method’, where a consensus sequence is created from trace files using SeqMan and used as a query in the Pubmlst BLAST interface. SeqMan is used when non-standard MLST alleles are being analyzed, and requires substantial manual intervention by the user. Ten sets totaling 331 fetA traces were analyzed for the comparison of MGIP to the SeqMan method. MGIP showed 98.2% sensitivity compared to 97.6% for the SeqMan method (Table 1). MGIP showed an even greater relative increase in speed over SeqMan. MGIP completed the ten fetA trace sets in 75 s compared to 1520 s for the SeqMan method (Table 1).
FURTHER DEVELOPMENT OF MGIP
There are several lines of further development of MGIP planned. MGIP allows for the discovery of novel alleles and/or STs, which can not be named or curated until they are sent to one of the central repositories of MLST data such as Pubmlst. We have been collaborating with the developers of Pubmlst to design an application programming interface that will allow MGIP users to directly submit new alleles and new STs. In addition to N. meningitidis, there are many more bacterial pathogens that are analyzed using MLST and MGIP will add the capacity to analyze additional organisms in the near future. The MGIP website is being translated to other languages, starting with French, to facilitate collaboration with non-English speakers.
CONCLUSION
The web-based design and implementation of MGIP helps to ensure that it stands alone among MLST analysis methods in terms of cost, speed of processing, ease-of-use, cross-compatibility and expandability. These features are particularly relevant to laboratories in the developing world, many of which may lack access to the level of computational infrastructure and support currently needed for MLST analysis. The use of simple web-based analytical platform should allow any investigator with Internet access to rapidly analyze his or her MLST data. Furthermore, MGIP is designed to be scalable to accommodate MLST+ analysis of multiple non-standard alleles. This feature should enable the expansion of current MLST based surveillance approaches.
The development of MGIP has been done in close contact with typing centers around the world to ensure that it will emerge as the global standard for MLST+ analysis. Labs that have been testing MGIP include the Meningitis Laboratory of CDC in the USA, the Health Protection Agency in England, Martin Maiden's research group at the University of Oxford and the National Institute for Communicable Diseases in South Africa. MGIP has been tested on over 1000 different strain/locus combinations, and the results show that it is 1–3% more sensitive and an order of magnitude faster than existing methods for MLST+ analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centers for Disease Control and Prevention (1 R36 GD000075-1 to L.S.K.); Alfred P. Sloan Research Fellowship in Computational and Evolutionary Molecular Biology (BR-4839 to I.K.J.); Georgia Research Alliance (GRA.VAC09.O to I.K.J. and L.W.M.). Funding for open access charge: Centers for Disease Control and Prevention.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Ahsan Huda and Maryanne Ku for their help in making the site more intuitive and easier to use. Also, many thanks go to Keith Jolley for his technical expertise and encouragement. Lastly, we would like to thank Troy Hilley for maintaining the MGIP server.
REFERENCES
- 1.Arreaza L, Vázquez JA. In: Meningococcal Disease. Methods and Protocols. Pollard AJ, Maiden MCJ, editors. Totowa, NJ: Humana Press; 2001. pp. 107–119. [Google Scholar]
- 2.Achtman M, Morelli G. In: Meningococcal Disease. Methods and Protocols. Pollard AJ, Maiden MCJ, editors. Totowa, NJ: Humana Press; 2001. pp. 147–155. [Google Scholar]
- 3.Jolley KA, Gray SJ, Suker J, Urwin R. In: Handbook of Meningococcal Disease. Infection Biology, Vaccination, Clinical Management. Frosch M, Maiden MCJ, editors. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co.; 2006. pp. 37–51. [Google Scholar]
- 4.Jolley KA. In: Meningococcal Disease. Methods and Protocols. Pollard AJ, Maiden MCJ, editors. Totowa, NJ: Humana Press; 2001. pp. 173–186. [Google Scholar]
- 5.Sacchi CT, Lemos AP, Whitney AM, Solari CA, Brandt ME, Melles CE, Frasch CE, Mayer LW. Correlation between serological and sequencing analyses of the PorB outer membrane protein in the Neisseria meningitidis serotyping system. Clin. Diagn. Lab. Immunol. 1998;5:348–354. doi: 10.1128/cdli.5.3.348-354.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacchi CT, Whitney AM, Popovic T, Beall DS, Reeves MW, Plikaytis BD, Rosenstein NE, Perkins BA, Tondella ML, Mayer LW. Diversity and prevalence of PorA types in Neisseria meningitidis serogroup B in the United States, 1992–1998. J. Infect. Dis. 2000;182:1169–1176. doi: 10.1086/315833. [DOI] [PubMed] [Google Scholar]
- 7.Thompson EA, Feavers IM, Maiden MC. Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology. 2003;149:1849–1858. doi: 10.1099/mic.0.26131-0. [DOI] [PubMed] [Google Scholar]
- 8.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl Acad. Sci. USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright MC, Spratt BG. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- 10.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 11.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 12.Gordon D, Desmarais C, Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jolley KA, Chan MS, Maiden MC. mlstdbNet – distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Platt S, Pichon B, George R, Green J. A bioinformatics pipeline for high-throughput microbial multilocus sequence typing (MLST) analyses. Clin. Microbiol. Infect. 2006;12:1144–1146. doi: 10.1111/j.1469-0691.2006.01541.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.