Abstract
MicroRNAs (miRNAs) are short RNAs that act as guides for the degradation and translational repression of protein-coding mRNAs. A large body of work showed that miRNAs are involved in the regulation of a broad range of biological functions, from development to cardiac and immune system function, to metabolism, to cancer. For most of the over 500 miRNAs that are encoded in the human genome the functions still remain to be uncovered. Identifying miRNAs whose expression changes between cell types or between normal and pathological conditions is an important step towards characterizing their function as is the prediction of mRNAs that could be targeted by these miRNAs. To provide the community the possibility of exploring interactively miRNA expression patterns and the candidate targets of miRNAs in an integrated environment, we developed the MirZ web server, which is accessible at www.mirz.unibas.ch. The server provides experimental and computational biologists with statistical analysis and data mining tools operating on up-to-date databases of sequencing-based miRNA expression profiles and of predicted miRNA target sites in species ranging from Caenorhabditis elegans to Homo sapiens.
INTRODUCTION
MicroRNAs (miRNAs) are a continuously growing class of small RNAs that act as guides in the translational silencing and degradation of target mRNAs (1). Many miRNAs are conserved over large evolutionary distances such as between human and worm (2). Fundamental biological processes such as development (3–6), metabolism (7–9), cardiac (10) and immune system function (11) have been shown to be regulated by miRNAs, and aberrant miRNA expression has been associated with cancers (12,13).
There are various approaches to miRNA expression profiling, one of which is small RNA sequencing. Classical cloning and sequencing of size-separated small RNAs have been used to generate a large atlas of miRNA expression profiles (14), and this approach can be scaled up considerably through deep sequencing technologies (15). Microarray-based expression profiling is also a popular approach, which has been used for instance to characterize the miRNA expression cancer samples (12). In contrast to sequencing, microarray-based profiling does not allow identification of novel miRNAs.
Numerous approaches have also been proposed for miRNA target prediction. Because the 5′-end of miRNAs (known as ‘seed’) appears to be important for target recognition, a number of tools focus on the evolutionary conservation of miRNA seed-complementary regions in 3′-UTRs (16–19). Other approaches emphasize the energy of hybridization between miRNA and target (20–22), the expected anti-correlation between the expression level of miRNAs and their mRNA targets (23,24), the properties of the environment of the miRNA target site (25,26) or combine various features of the miRNA target site itself (27,28).
Studies in both native expression (29) as well as transfection-induced miRNA overexpression situations (30,31) indicate that within a given tissue, the miRNAs that are most strongly expressed have the largest impact on mRNA targets. For this reason, deciphering the miRNA-dependent post-transcriptional regulatory layer in a given tissue or cell type needs to start from the miRNA expression profile of that tissue or cell type. Conversely, it is very common that one identifies differences in miRNA expression between cells at various stages of differentiation or between normal and malignant cells, and the natural question is what mRNAs are most likely to be affected by the change in miRNA expression. To address these types of questions, we developed MirZ (www.mirz.unibas.ch), a web service that integrates two resources that we developed in the context of previous research projects: the smiRNAdb miRNA expression atlas (14), and the ElMMo miRNA target prediction algorithm (18).
MATERIALS AND METHODS
The smiRNAdb miRNA expression atlas
smiRNAdb (14) is a unique, web-accessible and widely used resource of miRNA profiles determined by sequencing from hundreds of Homo sapiens, Mus musculus and Rattus norvegicus samples. The web interface of smiRNAdb features an extended repertoire of on-line analyses such as visualization and hierarchical clustering of miRNA expression profiles, principal component analysis, comparison of miRNA expression between two (sets of) samples with the aim of identifying the miRNAs whose expression differs most between the samples. We used the Brenda tissue ontology (32,33) as a guide in organizing the samples such that the user can readily identify related cell lineages or normal and pathological samples derived from a given tissue type. Our tissue hierarchy has four levels: the organ/system (e.g. hematopoietic system), subsystem (e.g. lymphoid lineage), cell type (e.g. B cell) and further cell type classification (e.g. B lymphocyte). MiRNAs themselves can be analyzed independently, grouped by their 2–7 subsequence, or grouped in precursor clusters. Two miRNAs are placed in the same precursor cluster if their loci are within 50 kb of each other in the genome, or if they share a mature form.
As an example, one may be interested in comparing miRNA expression between effector and naive human CD4+ T lymphocytes. SmiRNAdb features a ‘Sample comparison’ tool, which was specifically designed for the pairwise comparison of miRNA (sets of) samples. The user would select to compare the sample named ‘hsa_T-cell-CD4-effector’ to the sample named ‘hsa_T-cell-CD4-naive’. Because the naive CD4+ T cell sample and the effector CD4+ T cell sample differ widely in the total number of sequenced miRNAs (1374 versus 89), the precision of the miRNA frequency estimates in the two samples will also be very different. This situation is common in sequencing-based datasets making the identification of miRNAs whose expression is significantly different a non-trivial problem. At the heart of the tools offered by smiRNAdb, however, is a Bayesian model for computing the posterior probability that the frequency of a miRNA in the total miRNA population differs between two (sets of) samples. We compute this probability assuming a binomial sampling model and integrating over the unknown miRNA frequencies in the samples. This approach—described in details in Berninger et al. (34)—takes into account both the variability between sample sizes and the absolute miRNA counts.
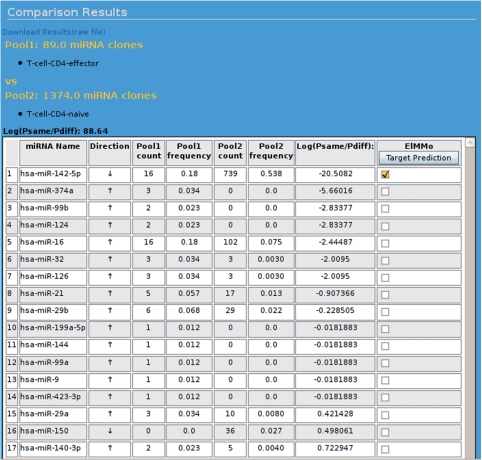
Figure 1 shows the results of comparing the miRNA expression profiles of naive versus effector CD4+ cells. The names and sizes of the samples being compared are shown at the top of the page, followed by the log-likelihood ratio log(Psame/Pdiff) of two models, one which assumes that the frequencies of miRNAs are the same and one that assumes that they can be different between the samples. The log-likelihood ratio takes positive values when the miRNA frequencies are similar and negative values when they are different. In this case, the log-likelihood ratio is positive, indicating that overall, the frequencies of miRNAs in these samples are more likely to have been the same. The list of miRNAs ranked from most dissimilar to most similar expression follows. Each row contains the name of an miRNA, the direction of regulation (up or down), the cloning counts and frequencies in both samples and provides a direct link to the predicted targets of the miRNA. The model indicates that with a 18% versus 54% cloning frequency, and despite the small size of the effector CD4+ T-cell sample, miR-142-5p is very likely to be downregulated in effector cells. Again, this can be inferred from the negative value of log(Psame/Pdiff) for miR-142-5p. From this page, the user can select one or several miRNAs that came out differentially expressed and can browse the list of predicted targets (Figure 2). In the case of miR-142-5p, the top 10 predicted targets include four transcription factors (AFF4, ONECUT2, ZFPM2 and ZNF148), and a kinase (PRPF4B) involved in pre-RNA splicing. These genes could provide a starting point for experimental studies on the function of miR-142-5p in T lymphocytes.
Figure 1.
Screenshot of the web page showing the result from comparing miRNA expression of human CD4+ effector T cells with the CD4+ naive T cells. Details are provided in the text.
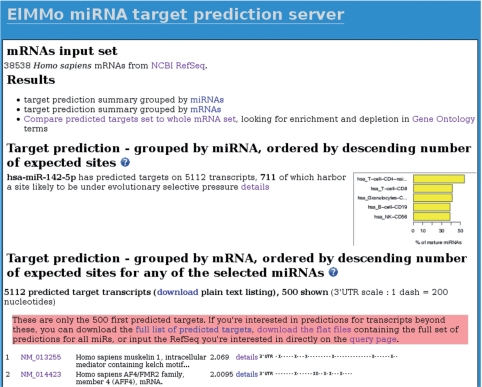
Figure 2.
Screenshot of the web page showing the ElMMo miRNA target predictions for miR-142-5p in all H. sapiens RefSeq mRNAs. The target predictions results are organized in two sections. The first section—located on the upper part of the web page—is miRNA-centric and features miRNA target predictions statistics as well as a figure showing the smiRNAdb tissues where the miRNA is mostly expressed. The second, mRNA-centric section is located on the lower part of the web page and provides a ranked list of mRNA predicted to be targeted by miR-142-5p.
Since the original release of smiRNAdb, we have implemented an additional tool for performing principal component analysis on the miRNA expression profiles, we added more possibilities for the user to download miRNA profile data for further processing and we started to incorporate other publicly available small RNA sequencing datasets from Danio rerio, Drosophila melanogaster and Caenorhabditis elegans. We reimplemented the software that was originally written in Perl CGI to use Java Server Faces technology and Apache/Tomcat. The computations are now performed on a computing cluster, with job distribution managed by the Sun Grid Engine queuing system. Finally, we enhanced the result screens of our on-line analysis tools with hyperlinks that directly take the user to the miRNA target predictions within the context of the smiRNAdb query, i.e. preserving the selected organism, miRNAs and tissue (if available). Refer to the web connectivity map in the Supplementary Material for an overview of the new links between smiRNAdb and ElMMo, as well as of the external ressources that we use in performing various analyses.
The ElMMo miRNA target prediction algorithm based on comparative genomic analysis
To be able to address the question of what mRNA is most likely affected by the change in expression of a miRNA, we coupled smiRNAdb to a PHP-based web interface to the ElMMo miRNA target predictions (18).
Returning to the example of the hsa-miR-142-5p miRNA that was highlighted in ‘The smiRNAdb miRNA expression atlas’ section, the web interface allows aside from browsing the predicted targets, a number of other queries. For instance, given an organism (H. sapiens in this example), the user can choose to scan for predicted miRNA target sites not only the default set of transcripts, which is all known RefSeq (35) mRNAs in the chosen organism, but also subsets of transcripts. The SymAtlas project (36) of the Genomics Institute of the Novartis Research Foundation (GNF) generated microarray-based mRNA expression profiles for a wide range of tissues. These profiles are incorporated in MirZ, giving the user the possibility to restrict target prediction to mRNAs that are expressed in a given cell type. The web interface further allows to scan an arbitrary number of mRNAs for up to 20 miRNAs simultaneously. Alternatively, the user can limit the number of mRNAs to scan to 20 mRNAs and then retrieve predicted target sites in these mRNAs for an arbitrary number of miRNAs.
MiRNAs exert their effector function through ribonucleoprotein complexes (miRNP) that contain, aside from the guiding miRNA a member of the Argonaute family of proteins. The determinants of productive miRNA–target site interactions are not entirely known, but a large body of work (16,37–41) established that perfect complementarity of the 7–8 ns from the 5′-end of the miRNA—the so-called miRNA ‘seed’—is critical for target recognition. Although miRNA target sites that do not satisfy this constraint have been described, at the genome-wide level the accuracy of predicting such sites is low (16,18). Other than perfect seed complementarity, the location of the putative target site within the 3′-UTR (18,25,42), structural accessibility (21,22,43), the nucleotide composition in its vicinity (25,26) and the complementarity of specific positions in the miRNA 3′-end to the target site (25) have all been reported to improve the accuracy of miRNA target prediction, yet the relative importance of these features remains unknown. The ElMMo miRNA target prediction method that we developed is based on a Bayesian model that only uses comparative genomics information. Yet it has as high an accuracy as other widely used target prediction programs that incorporate additional constraints, and measures of predictive performance on a set of experimentally validated miRNA targets in D. melanogaster can be found in the article describing the ElMMo method (18). Importantly, our model does not have any free parameters, and can easily accommodate additional species whose genome sequence becomes available.
Going back to our example, Figure 2 shows the ElMMo predictions for miR-142-5p in H. sapiens. This result screen is organized in two sections: (i) a miRNA-centric summary featuring per-miRNA target prediction statistics and a figure showing the smiRNAdb tissues where the selected miRNAs are mostly expressed, and (ii) a mRNA-centric summary that ranks all mRNAs predicted to be targeted by the selected miRNAs. In this later section, mRNAs are ordered by decreasing expected number of miRNA target sites under selective pressure, defined as the sum of all target site posterior probabilities for the selected miRNAs. The location of the putative target sites in the 3′-UTR is also indicated.
From the result screen, the user has the possibility to zoom onto a specific transcript to visualize the multiple genome alignments in the regions of the predicted target sites, and to find additional information about the targeted mRNAs from the Genbank database of the National Center for Biomedical Information (NCBI). Our web service also offers the possibility to run a Gene Ontology (GO) analysis searching for GO terms that are significantly over- or under-represented in the predicted miRNA targets through a modified version of the GeneMerge software (44). For instance, in the case of miR-142-5p, the most significantly enriched Biological Process GO term is ‘regulation of transcription, DNA-dependent’ (hypergeometric P-value <10−10, after Bonferroni multiple testing correction), followed by two ‘muscarinic acetylcholine receptor’-associated GO terms (P < 10−10). The muscarinic acetylcholine receptor has been shown to be involved in autocrine control of cell proliferation, including the proliferation of immune cells (45). This type of analyses could thus provide experimental scientists with clues to the function of miR-142-5p in the naive CD4+ T cells.
The current release of ElMMo features miRNA target predictions for H. sapiens, M. musculus, R. norvegicus, D. rerio, C. elegans and D. melanogaster. Of these, the M. musculus and R. norvegicus predictions were not present in our initial publication (18). Furthermore, for the remaining organisms, the current predictions are based on the genome sequences of a larger set of species, because more fully sequenced genomes became available since 2007. We further based our predictions on the most recent mRNA sequences and 3′-UTR annotations provided by the RefSeq database (35). Concerning the microarray profiles that the user can use to guide miRNA target discovery in specific tissues and aside from the H. sapiens profiles that were used in our original ElMMo release (18), we incorporated similar mRNA expression profiles for M. musculus and R. norvegicus. Finally, the ElMMo web interface now informs the user about the smiRNAdb samples in which the selected miRNAs are most strongly expressed.
Experimental data
The miRNA sequences that were used for miRNA sample annotation and for miRNA target prediction were obtained from the miRBase release 12.0 (46). For the miRNA profiles, MirZ includes a total of 297 samples: 173 for H. sapiens (14), 88 for M. musculus (14), 16 for R. norvegicus (14), 10 for D. melanogaster (47), 9 for D. rerio (48) and 1 for C. elegans (49).
For miRNA target predictions, we used the most recent genome assemblies available at the University of California Santa Cruz (UCSC) (50): hg18 for H. sapiens, mm9 for M. musculus, rn4 for R. norvegicus, danRer5 for D. rerio, ce6 for C. elegans and dm3 for D. melanogaster. We further used the following UCSC genome assemblies in the pairwise genome alignments: panTro2, rheMac2, mm9, rn4, canFam2, monDom4, bosTau4 and galGal3 for H. sapiens; panTro2, rheMac2, hg18, rn4, canFam2, monDom4, bosTau4 and galGal3 for M. musculus; panTro2, rheMac2, hg18, mm9, canFam2, monDom4, bosTau3 and galGal3 for R. norvegicus; tetNig1, fr2 and oryLat2 for D. rerio; caeJap1, caePb2, caeRem3, cb3 and priPac1 for C. elegans; and dp4, droAna3, droEre2, droGri2, droMoj3, droPer1, droSec1, droSim1, droVir3, droWil1 and droYak2 for D. melanogaster. mRNAs for all organisms were downloaded from the RefSeq database on 21 January 2009.
The links between sequence entities in various databases were made by mapping them all to the Gene database of NCBI (35). MiRNA expression profiles, microarray mRNA profiles and miRNA target predictions are stored as relational databases managed by a PostgreSQL server (www.postgresql.org).
CONCLUSION AND FUTURE DIRECTIONS
Using a concrete example comparing effector to naive CD4+ T-cells, we showed how MirZ can help isolating miRNAs that may be involved in a given biological function, and then provide clues into which molecular pathways may be controlled by these miRNAs to achieve their biological function. The integration of miRNA expression profiles with genome-wide miRNA target prediction combined with the tools we implemented—a Bayesian model for sample comparison, multivariate exploratory statistics, GO-term enrichment analysis—makes MirZ a powerful tool for studying miRNA-based regulation.
Since its publication, the miRNA expression atlas has been a valuable resource to the research community, and with the more general availability of deep sequencing technologies, more miRNA expression datasets are expected to emerge. Being able to explore and compare these datasets in a unified framework is highly desirable, and we plan to further support such analyses by updating MirZ as new datasets become available. Particularly for D. melanogaster, we currently only incorporate small-sized samples, and for C. elegans a whole-worm sample.
The target prediction methods also continue to evolve. In particular, additional determinants of miRNA targeting specificity must exist because not all transcripts that contain miRNA seed matches respond in a given experiment, but what these determinants are is still an open question (25,26). If a significantly better target prediction method emerges, this could be incorporated in our server.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Fund (grant #3100A0-114001 to M.Z.); Swiss Institute of Bioinformatics. Funding for open access charge: Swiss Institute of Bioinformatics.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the contribution of Robin Vobruba and Philip Handschin from Fachhochschule Nordwestschweiz (Brugg, Switzerland) to the re-implementation of smiRNAdb in Java; International Chicken Genome Sequencing Consortium (51) for the Gallus gallus genome; Chimpanzee Genome Sequencing Consortium for the Pan troglodytes genome; Baylor College of Medicine Human Genome Sequencing Center and the Rhesus Macaque Genome Sequencing Consortium for the Macaca mulatta genome; Mouse Genome Sequencing Consortium for the M. musculus genome (52); Rat Genome Project at the Baylor College of Medicine Human Genome Sequencing Center for the R. norvegicus genome (53,54); Dog Genome Sequencing Project for the Canis familiaris genome (55); Broad Institute for the Monodelphis domestica (opossum) genome; Baylor College of Medicine Human Genome Sequencing Center for the Bos taurus and Drosophila pseudoobscura genomes (56); Genoscope for the Tetraodon nigroviridis genome; Morishita laboratory for the Oryzias latipes (Medaka) genome (57); WormBase for the C. elegans genome (58); Agencourt Bioscience Corporation for the D. ananassae and D. erecta genomes; and the Drosophila 12 Genomes Consortium for all other Drosophila genomes (59). The Takifugu rubripes genome was provided freely by the Fugu Genome Consortium for use in this publication only. The C. brenneri, C. briggsae, C. japonica, C. remanei and P. pacificus genomes were produced by the Genome Sequencing Center at Washington University School of Medicine in St Louis and can be obtained from ftp://genome.wustl.edu/pub/organism/Invertebrates/.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Mller P, et al. Conservation of the sequence and temporal expression of let−7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 5.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 6.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR–430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 7.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir–14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 8.Poy MN, Eliasson L, Krützfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet–specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 9.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle–specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 11.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR–150 controls B cell differentiation by targeting the transcription factor c–Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, Alvarez–Saavedra E, Lamb J, Peck D, Sweet–Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.He L, Thomson JM, Hemann MT, Hernando–Monge E, Mu D, Goodson S, Powers S, Cordon–Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small rna library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 16.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad–Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 22.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 23.Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, Hughes TR, Blencowe BJ, Frey BJ, Morris QD. Using expression profiling data to identify human microRNA targets. Nat. Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 24.Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S, Cutillo L, Ballabio A, Banfi S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009;19:481–490. doi: 10.1101/gr.084129.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimson A, Farh KK, Johnston WK, Garrett–Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 28.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 29.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute–containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim LP, Lau NC, Garrett–Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 31.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. Transcripts targeted by the microRNA–16 family cooperatively regulate cell cycle progression. Mol. Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. Brenda, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berninger P, Gaidatzis D, van Nimwegen E, Zavolan M. Computational analysis of small RNA cloning data. Methods. 2008;44:13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post–transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 38.Lewis BP, Shih IH, Jones–Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 39.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev. Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majoros WH, Ohler U. Spatial preferences of microRNA targets in 3′ untranslated regions. BMC Genomics. 2007;8:152. doi: 10.1186/1471-2164-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL. The impact of target site accessibility on the design of effective siRNAs. Nature Biotech. 2008;26:578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- 44.Castillo-Davis CI, Hartl DL. Genemerge – post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- 45.Eglen RM. Muscarinic receptor subtypes in neuronal and non-neuronal cholinergic function. Auton. Autacoid Pharmacol. 2006;26:219–233. doi: 10.1111/j.1474-8673.2006.00368.x. [DOI] [PubMed] [Google Scholar]
- 46.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aravin AA, Lagos–Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 48.Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 50.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC genome browser database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;423:7018. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 52.Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 53.Havlak P, Chen R, Durbin KJ, Egan A, Yanru R, Song X.-Z, Weinstock GM, Gibbs RA. The atlas genome assembly system. Genome Res. 2004;14:721–732. doi: 10.1101/gr.2264004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rat Genome Sequencing Project Consortium. Genome sequence of the brown norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 55.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas E.J. 3rd, Zody MC, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 56.The Bovine Genome Sequencing and Analysis Consortium; Elsik CG, Tellam RL, Worley KC. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;446:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 58.Bieri T, Blasiar D, Ozersky P, Antoshechkin I, Bastiani C, Canaran P, Chan J, Chen N, Chen WJ, Davis P, et al. WormBase: new content and better access. Nucleic Acids Res. 2007;35:D506–D510. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.