Abstract
We have developed a set of online tools for measuring the semantic similarities of Gene Ontology (GO) terms and the functional similarities of gene products, and for further discovering biomedical knowledge from the GO database. The tools have been used for about 6.9 million times by 417 institutions from 43 countries since October 2006. The online tools are available at: http://bioinformatics.clemson.edu/G-SESAME.
INTRODUCTION
We provide and maintain a set of online tools to measure the semantic similarities of Gene Ontology (GO) terms and the functional similarities of gene products, and to discover biomedical knowledge through GO database. These tools are developed based on methods and algorithms proposed in our G-SESAME article (1) using MySQL 5.0.45 and PHP 5.1.6 and hosted by an Apache Web server (version 2.2.3) running on a Linux operating system (CentOS 5).
MOTIVATIONS
Although the GO project (2) has provided us the organized GO terms, one challenge is to accurately measure the semantic similarities of two GO terms, from which we can determine the functional similarities of genes. Some methods, such as Resnik's, Jiang's, Lin's (3–5) and their variants (6,7), have been used to measure the semantic similarities of GO terms. However, their accuracies for measuring GO terms were the main issues that we addressed in our previous article (1) as these methods were originally proposed for other specific taxonomies and were adapted to measure the semantic similarities of GO terms (8).
Background
The GO project was originally constructed and merged by researchers studying the genomes of three different model organisms: Drosophila melanogaster (fruity), Mus musculus (mouse) and Saccharomyces cerevisiae. A few methods have been proposed to compare the similarities of GO terms. In our previous article (1), we addressed the disadvantages of three methods (3–5). Besides their individual drawbacks, a common problem with existing methods is that they depend on the gene annotation statistics to measure the semantic similarities of GO terms. Hence, people may get different semantic similarity values for the same two GO terms if they use different gene annotation data sets (lexical corpus). This conflicts with the goal of having a set of controlled vocabularies (an ontology) for biological terms. A term in one ontology should have a fixed semantic meaning when it is used to annotate genes in a specific species.
The semantic difference of two GO terms is determined not only by their distance, but also by their locations in the whole directed acyclic graph (DAG). If two terms sharing the same parent are near the root of the ontology (terms are more general), they should have larger semantic differences than two terms having the same parent and being far away from the root of the ontology because the latter are more specific terms. However, using Jiang's or Lin's method, if two gene products are well annotated near the root of the ontology (shallow annotation), their semantic similarities will always be measured as very high (close to 1) and their semantic distance will always be computed as close to nil, thus providing a misleading result. The effect of shallow annotation is a serious drawback of both Jiang's and Lin's methods. Therefore, it is desirable to determine the semantic similarities of GO terms based only on the structure and annotation specification of GO ontologies. Unfortunately, most existing ontology-structure-based methods (8,9) also have their drawbacks in that they determine the semantic similarities of two GO terms either based on their distances to the closest common ancestor term or based on the number of their common ancestor terms. According to the true path rule, when calculating a GO term's semantic value, it must include the biological meanings of all its ancestor terms. Therefore, when measuring the semantic similarities of GO terms, we must consider not only the number of the common ancestor terms but also the locations of these ancestor terms related to the two specific terms in the GO graph.
Semantic similarities of two GO terms
According to our above analysis, we proposed a method to measure the semantic similarities of two GO terms in (1). We first decode the semantics of a GO term into a numeric value. Since the semantics (biological meanings) of a GO term are determined by its location in the entire GO hierarchy and its semantic relations with all of its ancestor terms, we use the DAG which is a subgraph of an ontology starting from this GO term and ending at any of the root term (biological process, cellular component, or molecular function) to represent this term. The detailed method has been introduced in Wang's article (1).
Our semantic similarities measurement algorithm has two advantages. First, it relies only on the relationships of the GO terms within a specific ontology (biological process, cellular component, or molecular function) to determine their semantic similarities. Therefore, it provides a consistent measurement for the semantic similarities between two GO terms, independent of the annotation statistics. Second, our algorithm is designed to decode the human perception of the semantic relationships between child and parent terms. Thus, the semantic similarities of GO terms obtained by our algorithm can reflect the closeness of their biological meanings in human perspectives.
In our previous study (1), we used many cases to evaluate the ‘is-a’ and ‘part-of’ values. We found that the gene clustering results by choosing 0.8 as the contribution factor for ‘is-a’ relations would be most consistent with the manual classification. In (8), the authors compared the covariance of the semantic similarities between GO terms in all three ontologies with the results of the BLAST. The covariance is not significantly impacted by using ‘part-of’ relations or not. We also know that, compared with the number of ‘part-of’ links, the number of ‘is-a’ links dominates the whole corpus in the GO database. Therefore, the semantic value of ‘is-a’ links should be bigger than that of ‘part-of’. Based on the evaluation of different values, we suggest to use the value 0.8 for ‘is-a’ and 0.6 for ‘part-of’.
Semantic similarities of two genes
Given the semantic similarities of two GO terms, a method to compare the similarities of two genes which are annotated by two sets of GO terms was also proposed by us in (1). When we compare two sets of GO terms, we let each pair of GO terms with higher semantic similarities dominate the relationship while neglecting other insignificant values. All the assumptions are based on the monotonic properties of the information content (IC) of a term with those of its ancestors and descendants. Each term in one GO term set should pair up its counterpart in the other set with close ‘distance’. The distance can be the one defined in Jiang's method; however, it is also feasible to use other definitions which should be in compliance with their own monotonic properties of IC. For example, in our previous article (1), we give our own distance definition.
Evaluation of our method and Resnik's
We use the gene annotation and classification information in pathways at the Saccharomyces genome database (SGD) as the reference for our evaluation.
There are 152 biological pathways in the SGD database. Most of these pathways contain at least three genes annotated by both GO molecular function terms and EC numbers (10). These genes are also manually clustered by their molecular functions. We compared the results of our method with Resnik's method on 111 pathways listed at SGD, which are well accepted pathways and the related genes are completely annotated in the GO database. From the pairs of clustering patterns, it is easy to tell that our method generated more similar pathways to those in SGD. The detailed comparisons in all cases between our method and Resnik's can be found in the supplemental materials of (1). The summary of evaluation results are shown in Table 1.
Table 1.
Comparison results of our method and Resnik's based on SGD pathways
| Cases | Numbers |
|---|---|
| Total of pathways in the SGD website | 152 |
| Number of pathways having at least three | 111 |
| Number of pathways used in evaluation | 111 |
| Evaluations showing our method is better | 66 |
| Evaluations showing that both methods are equal | 45 |
| Evaluations showing that our method is worse | 0 |
ONLINE TOOLS
The following sections introduce the set of our online tools, which include the tool for measuring the semantic similarities of two and multiple GO terms, the tool for the semantic comparison of two gene products from two different species, and multiple gene comparison and clustering tools. All the interfaces of our tools are consistent and user friendly. Users need to specify the values of ‘is-a’ and ‘part-of’ relations for all the tools. Our tools can run in an interactive mode or in a batch mode.
Tool for measuring the semantic similarities of two GO terms
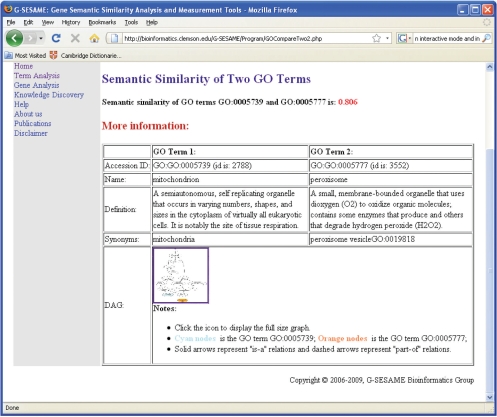
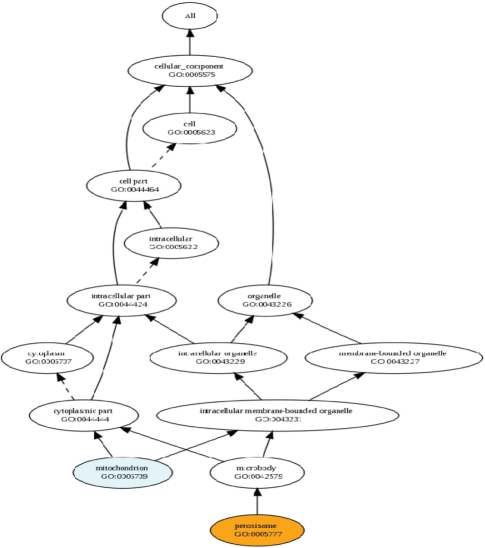
This tool is the simplest tool in our family of tools, with which users need to provide two GO terms and the ‘is-a’ and ‘part-of’ values. After receiving two GO terms online and the related ‘is-a’ and ‘part-of’ values, the program first searches the GO database to check the existence of the two GO terms. If both of the two GO terms exist, it retrieves the ancestors of these two GO terms. Figure 1 shows the output of comparing semantic similarities of two GO terms, GO: 0005739 and GO: 0005777. The DAGs of these two GO terms which are shown in Figure 2 are displayed as a hyperlink in the output webpage. In the DAG, solid arrows represent the ‘is-a’ relation and dashed arrows represent the ‘part-of’ relation.
Figure 1.
Output of semantic comparison of two GO terms, GO:0005739 and GO:0005777.
Figure 2.
DAGs of GO:0005739 and GO:0005777.
Tool for measuring the semantic similarities of two GO term sets
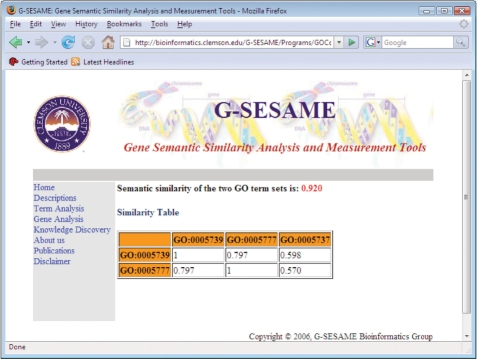
Based on the two GO term similarities comparison function, it is easy to implement the tool to compare the semantic similarities of two GO term sets. This tool allows users to upload two files which contain multiple GO terms in each file. Figure 3 shows the output of the comparison of two GO term sets.
Figure 3.
Output of comparison of two sets of GO terms. One set contains GO:0005739 and GO:0005777; the other one set contains GO:0005739, GO:0005777 and GO:0005737.
Tool for measuring the semantic similarities of two genes from two species
Based on the semantic correlation of GO terms used to annotate genes and gene products, we implemented the gene functional similarities comparison tool which can be used to compare the functional similarities of two genes. One unique feature of this tool is that it can compare two different genes from two different species. Users can specify one of the ontologies, two different species, and other criteria, such as data bases and evidence code. If multiple genes with the same symbols exist in the database, users have to choose one gene among these different species.
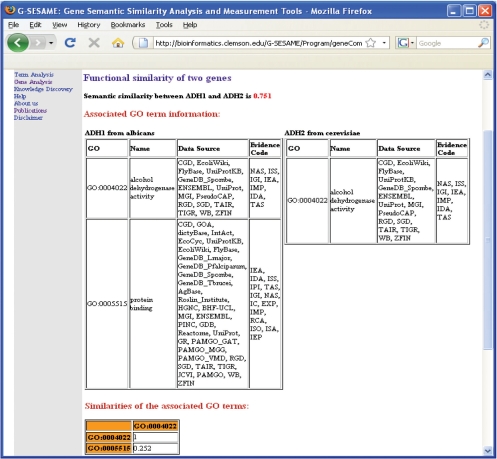
Figure 4 illustrates the output of measuring the semantic similarity of two genes, adh1 and adh4, from two different species. Not only the similarity value of these two genes is displayed at the top of the output webpage, but also the detail information of GO terms which annotate these two genes and other information, such as data sources and evidence codes, are displayed in two tables at the center of the page. The similarity table related with each pair of GO terms is also displayed at the bottom of the web page.
Figure 4.
Output of measuring the semantic similarity of two genes, adh1 and adh4, from two different species.
Tool for multiple genes comparison and gene clustering
Based on the gene functional similarity measurement, we implemented the multiple gene comparison and clustering tool for gene functionality analyses and knowledge discovery using our method, Jiang's, Lin's and Resnik's methods.
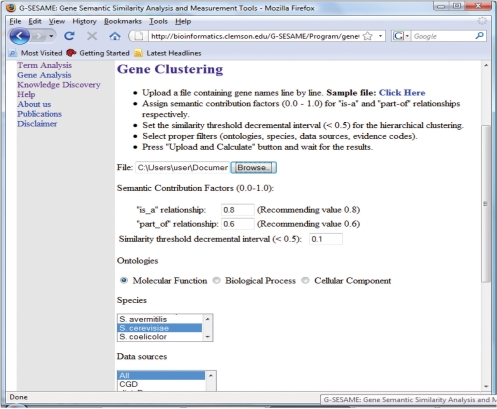
This tool needs to utilize a file which contains gene symbols with one symbol in each line. Users can specify different ontologies, species and other criteria. The interface of the tool is shown in Figure 5.
Figure 5.
Interface of the gene clustering tool.
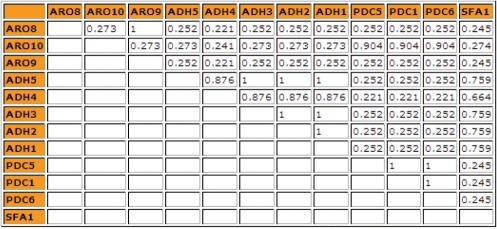
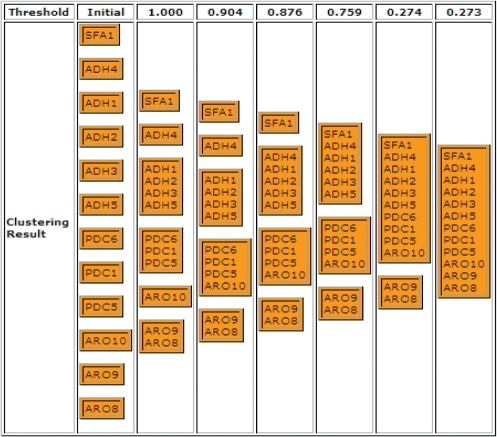
After users submit the query online, the tool not only outputs the similarities of these genes, but also displays the clustering results of these genes. It is easy to read the different patterns from these clustering results. Figures 6 and 7 show the similarity values and clustering results of genes in tryptophan-degradation pathway obtained by the tool based on Resnik's method.
Figure 6.
Semantic similarity table of genes in tryptophan degradation pathway obtained by Resnik's method.
Figure 7.
Clustering results of genes in tryptophan degradation pathway based on Resnik's method.
Using our tools in a batch mode
Our tools can be used not only in an interactive mode, but they are also designed and can be used in a batch mode for massive data analysis and data mining via the public Application Programming Interfaces (APIs). All our programs are listed at http://bioinformatics.clemson.edu/G-SESAME/Program/and all of them are well organized. We provide the public APIs, so it is very easy to let the third parties’ programs call the public APIs via the standard Hyper Text Transfer Protocol (HTTP).
Users can use their preferred languages to call our public APIs when they need to run our tools in a batch mode. Their programs need to provide the necessary query information which is needed by our tools. After sending the query information to our tools via the HTTP protocol, their programs will receive the results sent back by our tools and further perform their analysis. The details of how to use our tools in both modes have been described on our web pages.
Our tools have been optimized for efficiency and robustness. They have been used extensively and intensively by many institutions.
CONCLUSION AND FUTURE STUDIES
In this article, after introducing the background of the GO project, we introduce our solution for how to compare two different GO terms and genes. We then introduce a set of online tools based on our methods proposed in (1). These tools include the GO term comparison tool, the GO term set comparison tool, the comparison tool of two genes from two different species, the multiple gene comparison tool, and the clustering tool. We not only provide these online tools using our method, but also provide the clustering tools using Resnik's, Jiang's and Lin's methods to satisfy different user requirements.
All these tools can be used interactively or in a batch mode. They have been maintained and optimized by our team. As shown by our web log records, these tools have been widely used by researchers in biomedical research community.
Currently, we find that most of our knowledge discovery tools are related with one species scope. However, we believe that we need to do more research for multiple gene products among two or more species. Besides, due to the intensive usage of our tools, we may consider the feasibility to parallelize the implementation.
FUNDING
Funding for open access charge: National Institutes of Health grant 1R15CA131808-01.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang JZ, Du Z, Payattakool R, Yu PS, Chen C.-F. A new method to measure the semantic similarity of go terms. Bioinformatics. 2007;23:1274–1281. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
- 2.Consortium GO. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Resnik P. Semantic similarity in a taxonomy: an information-based measure and its application to problems of ambiguity in natural language. J. Artif. Int. Res. 1999;11:95–130. [Google Scholar]
- 4.Lin D. An information-theoretic definition of similarity, semantic similarity based on corpus statistics and lexical taxonomy. In the Proceedings of the 15th International Conference on Machine Learning; Morgan Kaufmann, San Francisco, CA. 1998. pp. 296–304. [Google Scholar]
- 5.Jiang JJ, Conrath DW. Semantic similarity based on corpus statistics and lexical taxonomy. In Proceedings of the 10th International Conference on Research on Computa-tional Linguistics (ROCLING X); Taiwan. 1997. [Google Scholar]
- 6.Couto FM, Silva MJ, Coutinho P. In DI/FCUL TR 03-29. University of Lisbon: Department of Informatics; 2003. Implementation of a functional semantic similarity measure between gene-products. [Google Scholar]
- 7.Lee SG, Hur JU, Kim YS. A graph-theoretic modeling on go space for biological interpretation of gene clusters. Bioinformatics. 2004;20:381–388. doi: 10.1093/bioinformatics/btg420. [DOI] [PubMed] [Google Scholar]
- 8.Lord PW, Stevens RD, Brass A, Goble CA. Investigating semantic similarity measures across the gene ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19:1275–1283. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- 9.Sevilla JL, Segura V, Podhorski A, Guruceaga E, Mato JM, Martinez-Cruz LA, Corrales FJ, Rubio A. Correlation between gene expression and go semantic similarity. IEEE/ACM Transact. Comput. Biol. Bioinformatics. 2005;2:330–338. doi: 10.1109/TCBB.2005.50. [DOI] [PubMed] [Google Scholar]
- 10.Ball CA, Dolinski K, Dwight SS, Harris MA, Issel-Tarver L, Kasarskis A, Scafe CR, Sherlock G, Binkley G, Jin H, et al. Integrating functional genomic information into the sac-charomyces genome database. Nucleic Acids Res. 2000;28:77–80. doi: 10.1093/nar/28.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]