Abstract
Chemical synthesis of custom DNA made to order calls for software streamlining the design of synthetic DNA sequences. GenoCAD™ (www.genocad.org) is a free web-based application to design protein expression vectors, artificial gene networks and other genetic constructs composed of multiple functional blocks called genetic parts. By capturing design strategies in grammatical models of DNA sequences, GenoCAD guides the user through the design process. By successively clicking on icons representing structural features or actual genetic parts, complex constructs composed of dozens of functional blocks can be designed in a matter of minutes. GenoCAD automatically derives the construct sequence from its comprehensive libraries of genetic parts. Upon completion of the design process, users can download the sequence for synthesis or further analysis. Users who elect to create a personal account on the system can customize their workspace by creating their own parts libraries, adding new parts to the libraries, or reusing designs to quickly generate sets of related constructs.
INTRODUCTION
In order to fully reap the potential benefits of de novo chemical gene synthesis (1) it has become necessary to develop tools and methodologies to streamline the design of custom DNA sequences (2). Protein expression for structural studies (3), functional genomics (4,5), metabolic engineering (6,7), or gene expression studies (8–11) are only some of the numerous possible applications of this emerging technology. Beyond small scale genetic constructs encompassing no more than a few interacting genes, it becomes possible to reengineer viral (12–15), bacterial (16), and even eukaryotic (17) genomes. While the number of users of this technology increases, so does the need to streamline the design of synthetic DNA sequences. GenoCAD is a web-based application filling this need by providing users with an integrated graphical development environment that no other software provides.
GenoCAD's design philosophy derives from the notion of genetic parts, which was first articulated to analyze genomics data (18). Thinking of genetic systems as composed of parts, each with its own function and characteristics, is akin to the way parts are described and used in various engineering fields. Designing complex systems through a bottom up integration of components is a dominant paradigm in engineering. It was therefore natural that engineers approaching DNA as an engineering substrate, rather than a natural macromolecule, used the notion of biological parts as building blocks (19,20). For instance, promoters, ribosome-binding sites (RBS), genes and terminators are all categories of parts that are needed for designing complex prokaryotic genetic constructs such as switches (21–23) and oscillators (23–25). One could argue that systematic efforts to decompose biological sequences into functional modules that can be recombined to meet user-defined specifications is one of the most distinctive features of synthetic biology compared to more traditional uses of recombinant DNA technologies (2,26–28).
GenoCAD facilitates the design of artificial DNA sequences in three ways. First, GenoCAD includes a flexible system to manage libraries of public and user-defined genetic parts. Second, GenoCAD relies on formal design strategies to guide both novice and experienced users in the design of structurally valid constructs for various biological applications. Finally, GenoCAD's sophisticated data model enables individual users and research groups to customize their workspace to their specific needs.
FLEXIBLE MANAGEMENT OF GENETIC PARTS LIBRARIES
Nothing better attests the benefits of a parts-based approach to the design of genetic constructs than the success of the Registry of Standard Biological Parts (www.partsregistry.org). By defining the BioBrick™ standard allowing the composition of parts and implementing mechanisms to share parts, the Registry has been critical in fostering the development of a vibrant synthetic biology community (1,19,20,29). We recently analyzed the content of the Registry database and the associated collection of clones to better understand how the successes and limitations of this pioneering experiment could guide the development of a second generation of registries of biological parts (29). GenoCAD attempts to refine some of the concepts upon which the Registry was developed.
By assuming that genetic designs can be synthesized, GenoCAD eliminates the need for standardizing the means by which parts are connected. It also eliminates the need to develop a collection of bacterial clones to manage the physical implementation of the parts. Our analysis also stressed the importance of basic parts used to generate new combinations of parts with specific functions. Ensuring the accuracy of the sequence and annotation of the basic parts is essential since inaccuracies at this level may affect numerous designs. As a result of this observation, GenoCAD parts libraries are exclusively composed of basic parts while sequences composed of multiple parts are called designs. The libraries of parts available to all GenoCAD users are limited to sequences used in peer-reviewed publications or commercial vectors. Parts are curated by a small number of experts according to a process that will be described in a future publication.
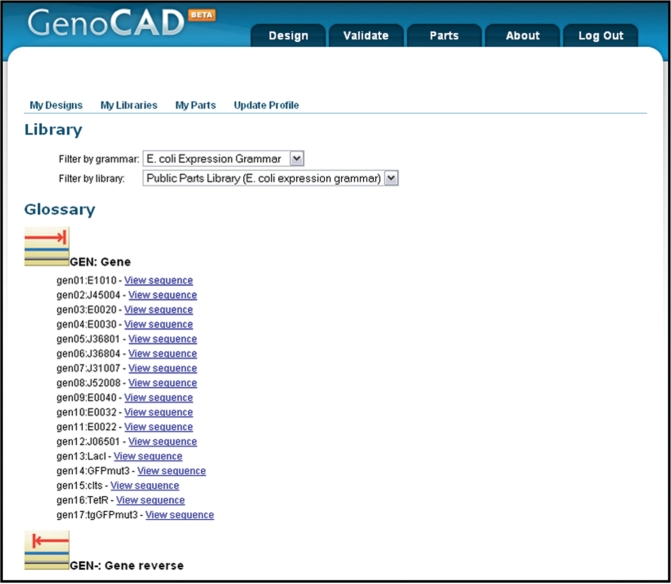
Categorizing parts into functional groups has also proved challenging as the number and diversity of parts increases. It would be, for instance, questionable to record bacterial and eukaryotic promoters in the same group. Developing a more granular categorization system may lead to an exponential growth of categories that would prove cumbersome to navigate. GenoCAD overcomes this challenge by relying on the notion of grammar (30). A grammar is composed of rules describing the structure of DNA sequences. One of the rules of the Escherichia coli Expression Grammar is CAS→PRO CIS TER which reads: an expression cassette is composed of a promoter, a cistron and a terminator. Another rule of the same grammar is CIS→RBS GEN (a cistron is composed of a RBS and a gene). The two rules can be used successively to create a basic expression cassette PRO RBS GEN TER. Different grammars can be developed for different applications and each grammar has its own parts categorization hierarchy. This approach ensures, for instance, that parts suitable for designing constructs for specific organisms like E. coli or yeast can easily be identified. It also enables the development of grammars for specific applications like protein production, homologous recombination in yeast, etc. Instead of attempting to develop a universal parts categorization system, GenoCAD provides a generic framework for the development of smaller more manageable application-specific parts-libraries. The ‘Parts’ tab of the GenoCAD user interface provides a parts library browser (Figure 1).
Figure 1.
The GenoCAD parts library browser. Parts are associated with individual libraries, each of which is associated with a specific grammar. Users select which parts library they view through choice of a grammar and specific library in drop down boxes on the page. The part category ‘Gene’ is displayed in this figure along with the icon that represents genes in the designs. By clicking on the icon, the list of genes expands, allowing the user to see the available choices in the library. Selecting the link to ‘View Sequence’ for any part opens a small window containing the sequence of the individual part.
POINT-AND-CLICK DESIGN OF GENETIC CONSTRUCTS
In addition to providing a hierarchy of categories, grammars include sets of rewriting rules that formalize design strategies for various types of genetic constructs (2,30). The design feature of GenoCAD embeds the grammars in a graphical user interface that leads users through the design workflow formalized in the grammar. Grammars usually prompt users to begin by choosing high-level structures of their system and systematically decomposing them into individual part categories. The last step of the design process consists in selecting actual parts corresponding to specific DNA sequences (Figure 2). When starting from one of the public design templates, users can quickly design constructs by simply selecting parts in a parts library instead of going through the entire design process described below.
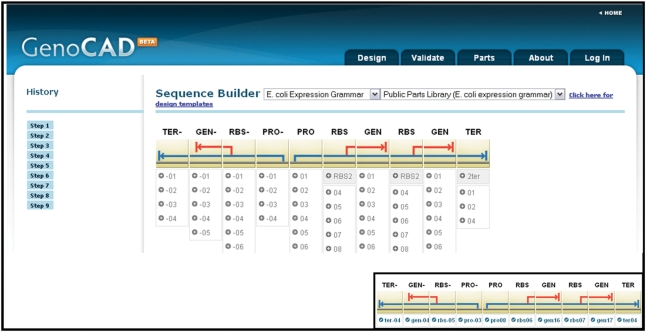
Figure 2.
The design interface showing the structure of a genetic toggle switch. The interface has drop down boxes at the top to select the grammar and parts library that will be used in the design. The history panel allows users to select one of the steps in the design process and see the structure of the design at that step. Users are permitted to go back to any step and redesign from that point. The design is presented in the main panel of the page, and icons for each part and the abbreviated parts categories are shown at the top of the design. Choices for each part are shown underneath the part icon. The inset shows the final design for this construct after specific choices (terminals) are selected for each part category.
Here, we use the design of a bistable genetic switch to illustrate GenoCAD's workflow (22). Selection of a grammar and an associated parts library (Figure 2) is the first step of this process. By selecting the grammar the user defines the type of construct that is possible, and by selecting the library they define the set of parts available to complete the design. Determining the high level structure of a functional genetic system de novo could potentially be confusing to users that do not have an intimate knowledge of the role of each part in the regulation of gene expression. However, each grammar behind GenoCAD provides the design strategy for specific types of constructs. GenoCAD's default grammar, ‘E. coli-Expression Grammar’, is suitable for the design of prokaryotic expression constructs. Additional grammars will be added for other applications.
The toggle switch construct was designed in nine steps as shown in Figure 2. The history pane (left side) allows users to review their work at any stage of the design process by clicking on that step. Users may click in reverse or in forward steps, and they are able to redesign from any point if they wish. Figure 3 shows a slightly condensed version of the design process of the toggle switch construct. Displayed below each part of a design are the options available to the user to transform the design. Choices in gray correspond to transformations that affect the design structure while options in white correspond to the selection of a specific part. Structural transformations are either part categories for which a specific part selection can be made or a higher level feature that must be decomposed to features lower in the abstraction hierarchy before the design can be finalized. An example of a high level part is the cistron (CIS in Figure 3), which is transformed into a RBS and gene (GEN). For each part category users have a choice of one or more specific sequences such as a specific RBS or gene. A mouse-over feature in the interface provides more information about the available choices. For example, the name of choice 04 for the promoter (PRO) category is displayed through this mechanism.
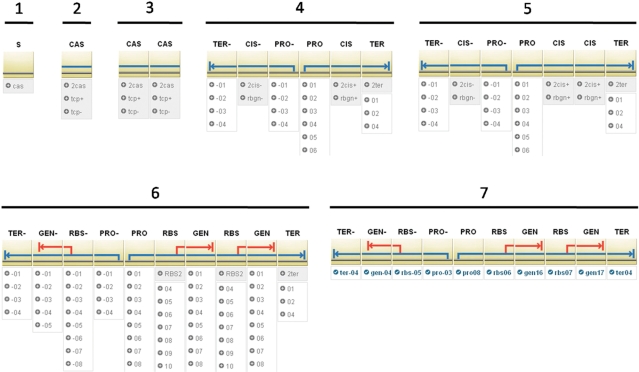
Figure 3.
A progression through the design of a bistable toggle switch. The starting symbol, S, (1) is where each design begins, and it is transformed into a transcription cassette, CAS, (2). Since the toggle switch contains two transcription cassettes, the single cassette is doubled (3). The design we are following has the cassettes oriented in opposite directions, and we achieve this by transforming the left cassette to the tpc- option, and the right to the tcp+ option (4), which contain a promoter, cistron, and terminator, but in opposite orientations. The right cassette is meant to express a transcriptional repressor and reporter gene in a bicistronic manner, so the cistron is doubled by selecting the 2cis+ option (5). Each cistron is then decomposed to a RBS and gene (6), with the RBS and gene in the reverse orientation in the left cassette. Selection of the specific promoters, RBSs, genes, and terminators produces a final construct that is associated with a DNA sequence (7).
A design is complete when specific sequences have been selected for each of its structural features. It is then possible to click on the ‘Download Sequence’ button to export the construct sequence as a text file that can be imported into software to design oligos for gene synthesis (31–34). Alternatively the sequence synthesis can be ordered from a fast growing number of vendors providing contract gene synthesis services (1).
CUSTOM USER WORKSPACE
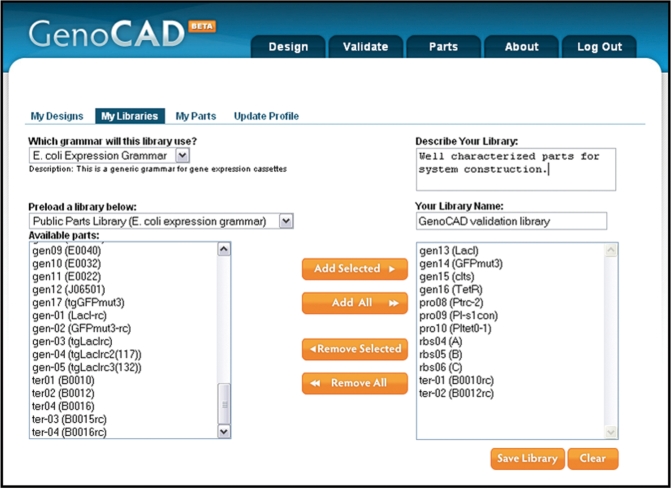
Parts available to all users in the public parts library have been derived from peer-reviewed publications and the documentation of commercial vectors. Registered users are able to create both their own parts library and their own parts. To create a new parts library, the user selects the ‘My Libraries’ link on the ‘Design’ or ‘Parts’ tab. When the user selects the link to create a new library they are presented with a user-friendly interface to do so (Figure 4). Since parts libraries are associated with specific grammars, the user must select the grammar to which the new library will be linked. Parts can then be copied from other libraries linked to the same grammar by first preloading that library at the lower left of the interface. Parts can be copied into and removed from the new library with the ‘add’ and ‘remove’ buttons. Once a user has created their own library, they are then able to create new parts and associate them with that library. In Figure 5, the part creation page allows the user to specify a grammar, select which part category it belongs to, and enter the part name, sequence and description. One or more user libraries associated with the grammar must be selected at the bottom of the page before the part can be saved into specific libraries.
Figure 4.
Creating a custom parts library. Users who create an account at the website are able to create their own parts libraries, and are then able to add custom parts to these libraries. Through the library creation interface, users select the grammar that their library will belong to, provide a name for their library, and can enter a description. Parts can be added to a new library from other libraries of the same grammar, which are loaded in the lower left box on the web page. All parts or a select subset of parts, from the existing library can be copied into or removed from the new library using the orange ‘add’ and ‘remove’ buttons.
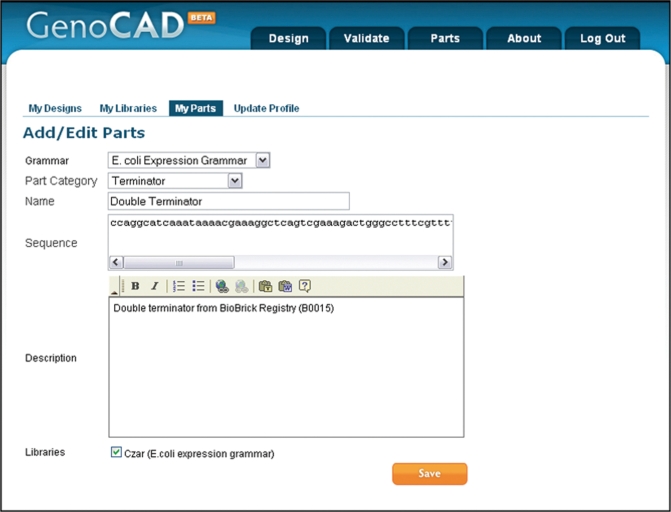
Figure 5.
Interface to add a new part. Users that have created a custom library are able to add and save parts that can be used in their designs. The categories of parts permissible in designs are defined in the grammar, so the grammar must be chosen first through a drop down menu. A second drop down, then allows users to choose which part category the new part will belong to; in this case a new terminator is being created. The sequence and description of the part are entered in text boxes, and the library(s) to which the part will be added must be checked.
Users can also save their designs in their workspace. Designs can be saved and named at any stage of the design process. The ‘My Designs’ page provides links to delete and clone (i.e. copy) previously saved designs. By saving a design prior to selecting parts, users can quickly clone a design template into multiple variants without having to go through the entire design process for each of them.
IMPLEMENTATION AND DATA MODEL
The GenoCAD website is written in a combination of PHP and JavaScript and runs on an Apache server. The MySQL database is on a different server, and both servers use the Linux operating system. The validation page relies on a custom parser developed in C++.
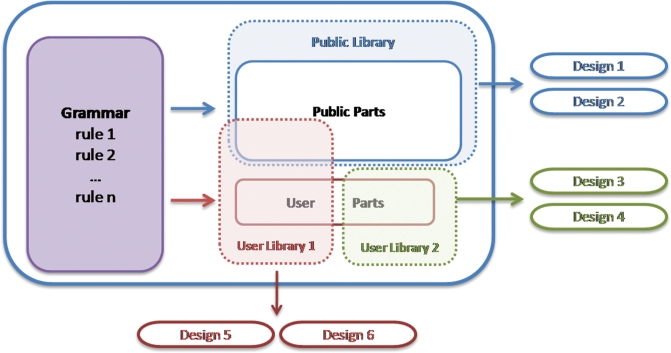
The data model for GenoCAD is summarized on Figure 6. Each design is associated with a specific parts library which, in turn, is linked to a specific grammar. Multiple public and user-defined libraries can be associated with each grammar and multiple designs can be associated with a specific library. Parts defined by users need to be associated with one of the user's parts libraries.
Figure 6.
GenoCAD data model. Each grammar encompasses a set of rules by which constructs can be designed. The grammar also defines the categories of parts that are available to design the constructs. For each grammar there is a collection of public parts (solid, blue rectangle), which constitute a publicly available parts library (dashed, blue rectangle). ‘User Libraries’ can be created from any subset of the public parts, and this library can be supplemented with user-created parts (solid, red rectangle). Two user libraries (dashed, red and dashed, green rectangles) are shown here that contain different subsets of public and user-created parts. User library 2 contains all user-created parts. When a design is created, all the parts to complete the design must be contained within a single library.
This simple data model has several limitations. Since many parts such as coding sequences can be used in different organisms, it would be desirable to replace the current hierarchical data model with a more refined model allowing the same part to be used in multiple libraries and grammars. It would also be desirable to define parts corresponding to coding regions by their amino-acid sequences instead of being limited to a DNA sequence with codons that are optimal for expression in a specific organism.
Additional grammars will be defined for the use of specific applications or for applications relevant to specific organisms in collaboration with organism or domain experts. Defining parts categories and design strategies suitable for a particular application will require a dialogue between biologists and computer scientists having experience in grammar development. Once agreed upon, grammars can easily be recorded in the MySQL database. Even though it would be attractive to guide the user in the definition of new grammars, the complexity of this process makes it unlikely that it will be possible to develop a grammar-building wizard in the foreseeable future.
SUMMARY AND FUTURE WORK
Like the stand-alone Gene Designer (34) or the web-based Registry of Standard Biological Parts, GenoCAD allows users to quickly design new genetic constructs by combining sequences corresponding to various functional elements known as parts. Unlike its predecessors though, GenoCAD guides the user through a design workflow corresponding to previously agreed upon design principles captured in grammars. Since it relies on linguistic models of DNA sequences (35), GenoCAD is a tool to help users write in the language of DNA sequences.
GenoCAD is a work in progress. The sequence validation tool makes it possible to test whether a sequence developed outside of GenoCAD is consistent with a specific grammar and parts library. This tool is still rudimentary since it simply provides pass/fail information. Eventually, more sophisticated error messages will be generated to help user troubleshoot their sequence. As the GenoCAD user base grows, GenoCAD will support workgroups by allowing them to share parts, libraries and grammars.
The next major improvement to GenoCAD will include tools to predict the design behavior. By augmenting GenoCAD data model it is possible to compile a design DNA sequence into a SBML file (36) that can be simulated by one of the numerous applications that supports this standard (37). GenoCAD will then join a growing number of applications experimenting with mechanisms to derive the gene network model encoded in genetic constructs composed of standard biological parts (2,28,38,39). GenoCAD will also integrate tools to track the synthesis and assembly of designs generated in GenoCAD. Optimizing the DNA fabrication process based on the strategies used to design a series of constructs would be extremely valuable.
While the GenoCAD web site is stable and has been in operation since 2007, the experimental validation of the concepts upon which it has been developed is still ongoing. As a reminder of the necessity to test designs in the lab, we will consider GenoCAD in beta test until extensive characterization of GenoCAD-designed systems has been described in peer-reviewed publications.
FUNDING
Virginia Commonwealth Research Initiative; fellowship from the Virginia Tech Genetics, Bioinformatics and Computational Biology graduate program awarded to YC. Funding for open access charge: Virginia Bioinformatics Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Czar MJ, Anderson JC, Bader JS, Peccoud J. Gene synthesis demystified. Trends Biotechnol. 2009;27:63–72. doi: 10.1016/j.tibtech.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Goler JA, Bramlett BW, Peccoud J. Genetic design: rising above the sequence. Trends Biotechnol. 2008;26:538–544. doi: 10.1016/j.tibtech.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, et al. Protein production and purification. Nat. Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 5.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 6.Keasling JD. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 2008;3:64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- 7.Keasling JD, Chou H. Metabolic engineering delivers next-generation biofuels. Nat. Biotechnol. 2008;26:298–299. doi: 10.1038/nbt0308-298. [DOI] [PubMed] [Google Scholar]
- 8.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 10.Murphy KF, Balazsi G, Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc. Natl Acad. Sci. USA. 2007;104:12726–12731. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox RS, 3rd, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 13.Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100025. 2005 0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 16.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 17.Dymond JS, Scheifele LZ, Richardson S, Lee P, Chandrasegaran S, Bader JS, Boeke JD. Teaching synthetic biology, bioinformatics and engineering to undergraduates: the interdisciplinary build-a-genome course. Genetics. 2009;181:13–21. doi: 10.1534/genetics.108.096784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bains W. The parts list of life. Nat. Biotechnol. 2001;19:401–402. doi: 10.1038/88044. [DOI] [PubMed] [Google Scholar]
- 19.Baker D, Church G, Collins J, Endy D, Jacobson J, Keasling J, Modrich P, Smolke C, Weiss R. Engineering life: building a fab for biology. Sci. Am. 2006;294:44–51. doi: 10.1038/scientificamerican0606-44. [DOI] [PubMed] [Google Scholar]
- 20.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 21.Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 22.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 23.Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- 24.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 25.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ball P. Synthetic biology: designs for life. Nature. 2007 doi: 10.1038/448032a. [DOI] [PubMed] [Google Scholar]
- 27.Drubin DA, Way JC, Silver PA. Designing biological systems. Genes Dev. 2007;21:242–254. doi: 10.1101/gad.1507207. [DOI] [PubMed] [Google Scholar]
- 28.Marchisio MA, Stelling J. Computational design of synthetic gene circuits with composable parts. Bioinformatics. 2008;24:1903–1910. doi: 10.1093/bioinformatics/btn330. [DOI] [PubMed] [Google Scholar]
- 29.Peccoud J, Blauvelt MF, Cai Y, Cooper KL, Crasta O, DeLalla EC, Evans C, Folkerts O, Lyons BM, Mane SP, et al. Targeted Development of Registries of Biological Parts. PLoS ONE. 2008;3:e2671. doi: 10.1371/journal.pone.0002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai Y, Hartnett B, Gustafsson C, Peccoud J. A syntactic model to design and verify synthetic genetic constructs derived from standard biological parts. Bioinformatics. 2007;23:2760–2767. doi: 10.1093/bioinformatics/btm446. [DOI] [PubMed] [Google Scholar]
- 31.Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayaraj S, Reid R, Santi DV. GeMS: an advanced software package for designing synthetic genes. Nucleic Acids Res. 2005;33:3011–3016. doi: 10.1093/nar/gki614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson SM, Wheelan SJ, Yarrington RM, Boeke JD. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 2006;16:550–556. doi: 10.1101/gr.4431306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics. 2006;7:285. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Searls DB. The language of genes. Nature. 2002;420:211–217. doi: 10.1038/nature01255. [DOI] [PubMed] [Google Scholar]
- 36.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 37.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. COPASI – a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 38.Hill AD, Tomshine JR, Weeding EM, Sotiropoulos V, Kaznessis YN. SynBioSS: the synthetic biology modeling suite. Bioinformatics. 2008;24:2551–2553. doi: 10.1093/bioinformatics/btn468. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigo G, Carrera J, Jaramillo A. Asmparts: assembly of biological model parts. Syst. Synth. Biol. 2007;1:167–170. doi: 10.1007/s11693-008-9013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]