Abstract
Ortholog detection is essential in functional annotation of genomes, with applications to phylogenetic tree construction, prediction of protein–protein interaction and other bioinformatics tasks. We present here the PHOG web server employing a novel algorithm to identify orthologs based on phylogenetic analysis. Results on a benchmark dataset from the TreeFam-A manually curated orthology database show that PHOG provides a combination of high recall and precision competitive with both InParanoid and OrthoMCL, and allows users to target different taxonomic distances and precision levels through the use of tree-distance thresholds. For instance, OrthoMCL-DB achieved 76% recall and 66% precision on this dataset; at a slightly higher precision (68%) PHOG achieves 10% higher recall (86%). InParanoid achieved 87% recall at 24% precision on this dataset, while a PHOG variant designed for high recall achieves 88% recall at 61% precision, increasing precision by 37% over InParanoid. PHOG is based on pre-computed trees in the PhyloFacts resource, and contains over 366 K orthology groups with a minimum of three species. Predicted orthologs are linked to GO annotations, pathway information and biological literature. The PHOG web server is available at http://phylofacts.berkeley.edu/orthologs/.
INTRODUCTION
Gene families evolve diverse functions via gene duplication, domain architecture rearrangements, mutations at key positions and other evolutionary processes (1,2). Since orthologs (related by speciation events from a common ancestor) are more likely to maintain the same function than paralogs (related by duplication) (3), orthology identification is a key first step in protein function prediction and functional annotation of genomes (4,5), and has additional applications to species tree estimation (6), and prediction of protein–protein interaction (7).
Although orthology is an evolutionary term and thus ideally determined using phylogenetic analysis (8), the computational cost of phylogenetic reconstruction has spurred the development of more computationally efficient approaches. Chief among these are methods relying on pairwise sequence comparison between genomes [e.g. InParanoid (9), OrthoMCL (10), COG (11) and eggNOG (12)]. Nevertheless, an increasing number of web servers have been developed that provide orthology predictions based on phylogenetic analysis [e.g. TreeFam (13), HOGENOM (14), PhylomeDB (15) and Ensembl-Compara (16)]. Fundamental differences between these resources lie in the taxonomic range of species considered, whether orthology predictions are restricted to fully sequenced genomes, whether trees are reconciled and/or manually curated, the inclusion of auxiliary data such as synteny and gene order information, and modes of access.
We present the Berkeley PhyloFacts Orthology Group (PHOG) web server using a novel phylogenetic approach to identify orthologs without the computational cost of species tree-gene tree reconciliation. Berkeley PHOG makes use of over 57 000 phylogenetic trees in the PhyloFacts resource. PhyloFacts is an encyclopedia of protein superfamilies including sequences from both whole genomes and only partly sequenced genomes across the Tree of Life (17,18); it is designed to reduce the systematic errors associated with homology-based function prediction (1,19,20) by providing a system for phylogenomic inference of gene function (2,4). Different variants of the PHOG algorithm are provided to allow users to target a selected recall or precision depending on individual preferences. Results on a benchmark dataset from the TreeFam-A manually curated orthology database show that Berkeley PHOG provides a combination of high recall and precision that is competitive with both OrthoMCL and InParanoid. PHOG can also be tuned for specified taxonomic distances using a tree-distance threshold. For instance, a mouse-specific threshold achieves 95% recall at 91% precision on mouse orthology detection, while OrthoMCL-DB achieves 83% recall at 77% precision and InParanoid achieves 96% recall but only 21% precision.
THE PHOG ALGORITHM
The Berkeley PHOG server provides PhyloFacts Orthology Groups targeting different evolutionary distances and precision levels. At one extreme, we aim to provide highly specific clusters of ‘super-orthologs’—sequences related strictly by speciation, i.e. every node on the path in the tree joining the sequences represents a speciation event (21). Super-orthology is thus a more restrictive definition than the standard definition of orthology, which requires only that the most recent common ancestor of the pair must represent a speciation event. Note that while the standard definition of orthology is not transitive (22), super-orthology is. At the other extreme, we aim at maximizing recall while still maintaining high precision. Intermediate levels balancing recall and precision for different taxonomic distances are also provided.
The PHOG algorithm takes as input a phylogenetic tree (typically, a multi-gene tree containing sequences from many species). For each sequence (leaf), we identify the closest sequence in each other species by tree distance (i.e. the sum over the edge lengths). Orthologs can then be defined as sequences from different species that are each other's reciprocal nearest neighbor (RNN) in the tree. A maximal subtree that contains only RNN orthologs, having at most one representative of each species (allowing subtrees containing members exclusively from one species as either very recent inparalogs or redundant variants of the same gene in a sequence database), can be inferred to form a super-orthology group. (We explain in the ‘Future work’ section some of the reasons why this inference may not always be correct.) This version of the PHOG algorithm is called PHOG-S, for PHOG super-orthologs. A second variant of PHOG is designed to approximate the standard definition of orthology, termed PHOG-O, for PHOG-Orthologs. Finally, we provide a tree-distance thresholded version of the PHOG algorithm, termed PHOG-T, allowing super-orthologous subtrees identified by PHOG-S to expand to include additional sequences based on a tree-distance criterion. Details of these methods are available at http://phylofacts.berkeley.edu/orthologs/supplement/v1/.
In predicting orthologs, the PHOG algorithm makes use only of information about the species of origin, the tree topology and the tree distances. As is the case for all orthology prediction methods that are similarly restricted, there are limitations to this approach. For instance, synteny and gene order relationships can provide important information to disambiguate between true orthologs and paralogs. As in most areas of bioinformatics, there is an inherent trade-off between computational efficiency and accuracy. By not requiring genomic locus information, PHOG can analyze sequences for which whole genomes are unavailable; however, the inclusion of genomic information (where available) will obviously result in increased performance accuracy. We review these issues and our plans in the ‘Future work’ section.
EVALUATING PHOG PERFORMANCE
To assess the accuracy of PHOG, we used a set of human sequences and their predicted orthologs in three model organisms—Mus musculus (mouse), Danio rerio (zebrafish) and Drosophila melanogaster (fruit fly)—from the TreeFam-A resource (13) as a gold standard benchmark. TreeFam-A uses a sophisticated ortholog-identification protocol (including tree reconciliation and manual curation) providing for a high-accuracy dataset. Mouse, zebrafish and fruit fly were selected since they had been targeted for analysis by both OrthoMCL-DB and InParanoid and represented a range of evolutionary distances. We chose 100 human sequences from TreeFam-A meeting the following requirements. First, TreeFam-A had to include orthologs for at least two of the targeted species. Second, to ensure that results of these experiments would generalize to new data, we filtered candidate sequences to eliminate homologs (accomplished by removing sequences having a BLAST E-value <1 or sharing a common PFAM domain). For each human sequence, we retrieved orthologs identified by InParanoid and OrthoMCL-DB in mouse, zebrafish and fruit fly. We analyzed phylogenetic trees in PhyloFacts containing the human sequences to predict orthologs for the different PHOG variants, and produced a full recall-precision curve for PHOG-T through the use of different thresholds. Results of the SCI-PHY functional subfamily identification algorithm (23) are included for comparison (we treated all members of a SCI-PHY subfamily as predicted orthologs).
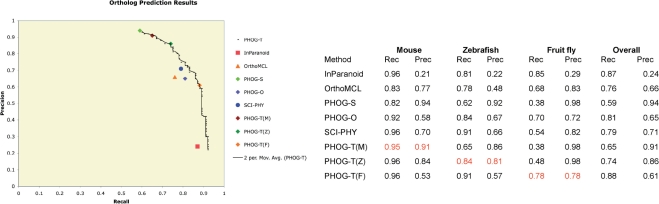
The results, shown in Figure 1, demonstrate the classic trade-off between precision and recall. PHOG-S (super-orthology prediction) has the best overall precision (94%) but the lowest recall (59%). InParanoid achieves very high recall (87%), but at a cost of quite low precision (24%). OrthoMCL-DB has significantly higher precision than InParanoid but lower overall recall (76% recall and 66% precision). PHOG-O performance is very close to OrthoMCL-DB, with a modest increase in recall and a tiny drop in precision (81% recall and 65% precision). SCI-PHY, which is not designed for orthology detection per se and makes no use of species information, has surprisingly good performance on this dataset (79% recall and 71% precision) with slightly better recall and precision than OrthoMCL-DB.
Figure 1.
Results of orthology prediction methods assessed on a benchmark dataset from the TreeFam-A resource. Performance was evaluated on 100 human proteins selected from the TreeFam-A manually curated orthology database, with orthologs to each human protein from mouse, zebrafish and fruit fly. Methods evaluated include several PHOG variants, OrthoMCL-DB, InParanoid and SCI-PHY. PHOG-S represents super-orthology predictions, PHOG-O represents standard orthology predictions and PHOG-T represents the tree-distance thresholded variants. PHOG-T variants PHOG-T(M), PHOG-T(Z) and PHOG-T(F) correspond to tree-distance thresholds selected for optimal performance on this dataset for mouse, zebrafish and fruit fly, respectively. Tree distance thresholds were 0.09375 (mouse), 0.296875 (zebrafish) and 0.9375 (fruit fly). SCI-PHY uses hierarchical clustering and encoding cost measures to define functional subtypes and is included for comparison. Recall measures the fraction of TreeFam-A orthologs detected by a method. Precision measures the fraction of a method's predicted orthologs that are included in TreeFam-A. A True Positive (TP) is an orthology pair included in TreeFam-A that is also predicted by a method, a False Positive (FP) is an orthology pair predicted by a method that is not included in TreeFam-A and a False Negative (FN) is a TreeFam-A ortholog that is missed by a method. Left: recall-precision curves over the entire dataset. Right: table of results for each method for individual species as well as over the entire dataset. Values in red highlight the recall and precision for species-specific threshold selections.
PHOG-T provides the best performance overall, by allowing the user to control the recall-precision tradeoff. For instance, while OrthoMCL-DB achieves 76% recall and 66% precision on this dataset, PHOG-T achieves 86% recall at 68% precision, making an improvement of 10% in recall at a slightly higher precision. At a recall of 77%, PHOG-T achieves 80% precision—an improvement of 14% in precision over OrthoMCL. Similarly, InParanoid achieves 87% recall at 24% precision; at a recall of 88% PHOG-T achieves 61% precision—37% higher precision. PHOG-T also allows users to target ortholog detection for specific taxonomic distances. For example, a PHOG-T threshold of 0.296875 provides 84% recall and 81% precision on zebrafish orthology prediction. By contrast, InParanoid attains almost the same recall as PHOG-T (81%) on the zebrafish orthologs but has much lower precision (22%), and OrthoMCL achieves 78% recall and 48% precision. When evaluated on mouse ortholog prediction, PHOG-T attains 95% recall at 91% precision, while OrthoMCL-DB achieves 83% recall at 77% precision and InParanoid achieves 96% recall but only 21% precision.
Details of these experiments, including species-specific results for each method, are available at http://phylofacts.berkeley.edu/orthologs/supplement/v1/.
PHOG WEB SERVER
The PHOG web server is available at http://phylofacts.berkeley.edu/orthologs/.
INPUT
We enable two ways of accessing PHOG orthologs: by sequence identifier and by FASTA sequence input. Allowed sequence identifier inputs (for protein sequences only) include UniProt accessions or IDs and GenBank IDs (but not accessions).
OUTPUT
Results based on input sequence accessions
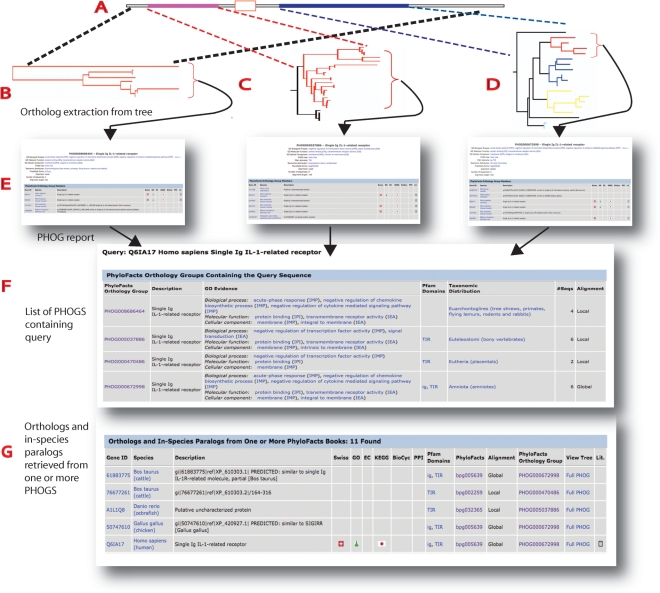
We present two tables, as shown in Figure 2. The first presents a list of all the PHOGs containing the sequence, along with summary data including taxonomic distribution, number of members, and Gene Ontology annotations and evidence codes. The second table displays a table of all orthologs found over all PHOGs containing the query, listed in alphabetic order by taxonomic origin. Links to data relevant to the function of these predicted orthologs are provided, including KEGG, BioCyc, SwissProt and protein–protein interaction (PPI) data. The PPI data links to a table of interacting partners, which are then linked to their orthologs, enabling the user to predict interacting partners via interolog analysis. Users can also follow a link to the complete PhyloFacts family ‘book’ containing a multiple sequence alignment, phylogenetic tree, homologous PDB structures, GO annotations and evidence codes, predicted functional residues, predicted subfamilies, hidden Markov models for the family and subfamilies, biological literature, and links to external resources. From this page users can click a button to access the downloads page, from which they can download the alignment, the tree, hidden Markov models and other information associated with the family. The ‘Alignment’ column indicates whether the phylogenetic tree is based on global homology [a common domain architecture, clustered using the FlowerPower algorithm (24)] or local homology. A link to a ‘PHOG Report’ displaying additional data on each orthology group is displayed under the ‘PhyloFacts Orthology Group’ column. Cross-references provided by UniProt are used to provide many of these links. The ‘View Tree’ column provides a link to the tree providing the basis for the predicted orthology. We provide two tree views—one for the subtree corresponding to the predicted orthology, and one for the full tree (i.e. including out-paralogs). Data for orthologs can also be downloaded in CSV format.
Figure 2.
PhyloFacts ortholog identification pipeline. The input is a protein sequence, in either FASTA format (for BLAST search) or by accession. Results of a sequence accession search are displayed in an Orthology Report including a table of all PHOGs containing the query (F) followed by a table displaying the sequences contained in these PHOGs (G). Links in the columns labelled PhyloFacts Orthology Group retrieve the corresponding PHOG report (E). BLAST results are displayed in an initial table of results (not shown); users would then select one of the sequences in the table, to retrieve the Orthology Report for their selected sequence. (A) Protein sequence query. In this example, the query sequence consists of two evolutionarily conserved domains—an N-terminal Ig domain (pink) followed by a transmembrane helix and and a C-terminal Toll Interleukin Receptor (TIR) domain (blue). (B–D) PhyloFacts trees containing the query sequence are identified, and orthologs are extracted from the orthology group for the sequence (indicated by red subtrees). In this example, the sequence is contained in three PhyloFacts trees. The tree shown in B corresponds to sequences sharing the same overall domain architecture (global homologs). The trees shown in C and D contain sequences that share local (partial) homology along a single domain; the tree in C contains sequences having an Ig domain and the tree in D contains sequences having a TIR domain. (Note that the taxonomic distributions of these PHOGs differ, corresponding to differences in orthology predictions across these domains.) (E) PHOG report—this report displays summary data for the PHOG, followed by a table listing all the orthologs in the PHOG including a link to the sequence database from which the member was drawn, the species of origin, description and links to external resources (e.g. SwissProt, KEGG and BioCyc). (F) List of PHOGs containing the query. This table contains summary data about each PHOG, including PFAM domains, GO annotations and evidence codes and taxonomic distribution. (G) Orthology report: all members of all PhyloFacts orthology groups containing the query are gathered and presented in a table. Note that some orthologs to the query will belong to more than one PHOG (i.e. containing both the ortholog and the query); the column ‘PhyloFacts Orthology Group’ provides a link to the most informative PHOG for each sequence as well as to the PhyloFacts book containing that PHOG. GO annotations and evidence codes, PFAM domains and links to external resources (e.g. SwissProt, KEGG, BioCyc and GO) are also provided. These data are also overlaid on the phylogenetic tree for the PHOG as well as for the family tree from which the PHOG was drawn, and can be viewed using the PhyloScope tree viewer.
Results based on FASTA sequence input
Results are returned in a table, ordered by BLAST score. Links are provided to the sequence database from which the sequence was drawn, orthologs to the sequence, the species, description, E-value, percent identity, and BLAST bit score. Users can click on any sequence in the table to view its PHOG orthologs (retrieving the table of results described in the previous section).
Statistics
PHOG currently contains 366 610 super-orthology groups (the most restrictive criterion for orthology) containing a minimum of three species, of which 141 170 contain at least one eukaryotic member, 242 907 contain at least one prokaryotic member and 17 467 contain both eukaryotes and prokaryotes. More than 155 K of these super-orthology groups contain at least one member in the SwissProt manually curated sequence database. Tables of orthologs to human and E. coli genes for selected species are available for download at http://phylofacts.berkeley.edu/orthologs/downloads/.
DISCUSSION
PhyloFacts orthology groups (PHOGs) are derived from analysis of phylogenetic trees of protein families in the PhyloFacts phylogenomic encyclopedia. Experiments on a benchmark dataset of mouse, zebrafish and fruit fly orthologs to human proteins from the TreeFam-A manually curated orthology database show that PHOG has performance competitive with both InParanoid and OrthoMCL-DB, while offering the user control over the specificity-recall tradeoff and providing versions targeting different taxonomic distances. The demonstrated precision of PhyloFacts orthology groups, validated against the reconciled and manually curated orthologs in the TreeFam-A resource, makes them a source of high-accuracy orthologs for inference of protein function in a phylogenomic context.
FUTURE WORK
The results reported here, as well as the thresholds determined for PHOG-T, are based on a small dataset of animal orthologs to human proteins drawn from the TreeFam-A resource. Note that although none of the TreeFam-A orthologs were used to train this method, the problem of small datasets and potential sample skew will need to be addressed in future work on significantly expanded datasets. In particular, tree-distance thresholds determined here are unlikely to be optimal for ortholog detection at greater evolutionary distances (e.g. human-yeast orthologs). Future experiments on other manually curated datasets will be performed to determine the optimal tree-distance thresholds for different species pairs. Note that the results presented here are based on Neighbor-Joining trees; we expect that Maximum Likelihood trees will produce better results, and will be switching to the use of ML trees in the future. As noted earlier, the current implementation of PHOG does not use gene neighborhood or synteny information. This can lead PHOG to make mistakes that would have been avoided had this information been included. For instance, after whole genome duplication, gene loss occurring independently in different lineages may lead to the retention of different copies in each lineage. The resulting paralogs would be mistakenly identified by PHOG as (super)-orthologs. We plan to incorporate gene neighborhood and synteny information to improve the accuracy of our predictions in the future. Other plans include expanding sequence identifiers recognized to include Ensembl identifiers and those of the main model organism databases, providing orthology confidence scores based on phylogenetic support (e.g. bootstrap, ML and Bayesian support), alignment overlap and other relevant criteria, and using tree reconciliation software (as computational resources permit) to label internal nodes of trees as duplication and speciation events.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
PECASE Award (grant number 0238311 to K.S.) from the National Science Foundation, National Institutes of Health ( R01HG02769 to K.S.) and Microbial Genome Sequencing Program of the National Science Foundation ( 0732065 to K.S.). Funding for open access charge: National Science Foundation; National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr João Carlos Setubal for testing the server and helpful comments, and the anonymous referees for their perceptive comments and suggestions for future extensions to PHOG.
REFERENCES
- 1.Sjölander K. Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics. 2004;20:170–179. doi: 10.1093/bioinformatics/bth021. [DOI] [PubMed] [Google Scholar]
- 2.Brown D, Sjölander K. Functional classification using phylogenomic inference. PLoS Comput. Biol. 2006;2:e77. doi: 10.1371/journal.pcbi.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 4.Eisen JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg I. Automated protein function prediction—the genomic challenge. Brief Bioinform. 2006;7:225–242. doi: 10.1093/bib/bbl004. [DOI] [PubMed] [Google Scholar]
- 6.Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- 7.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thornton JW, DeSalle R. Gene family evolution and homology: genomics meets phylogenetics. Ann. Rev. Genomics Hum. Genet. 2000;1:41–73. doi: 10.1146/annurev.genom.1.1.41. [DOI] [PubMed] [Google Scholar]
- 9.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 10.Chen F, Mackey AJ, Stoeckert C.J., Jr., Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruan J, Li H, Chen Z, Coghlan A, Coin LJ, Guo Y, Heriche JK, Hu Y, Kristiansen K, Li R, et al. TreeFam: 2008 update. Nucleic Acids Res. 2008;36:D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dufayard JF, Duret L, Penel S, Gouy M, Rechenmann F, Perriere G. Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- 15.Huerta-Cepas J, Bueno A, Dopazo J, Gabaldon T. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 2008;36:D491–D496. doi: 10.1093/nar/gkm899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birney E, Andrews D, Caccamo M, Chen Y, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, Cutts T, et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glanville JG, Kirshner D, Krishnamurthy N, Sjölander K. Berkeley Phylogenomics Group web servers: resources for structural phylogenomic analysis. Nucleic Acids Res. 2007;35:W27–W32. doi: 10.1093/nar/gkm325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnamurthy N, Brown DP, Kirshner D, Sjölander K. PhyloFacts: an online structural phylogenomic encyclopedia for protein functional and structural classification. Genome Biol. 2006;7:R83. doi: 10.1186/gb-2006-7-9-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brenner SE. Errors in genome annotation. Trends Genet. 1999;15:132–133. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- 20.Gilks WR, Audit B, De Angelis D, Tsoka S, Ouzounis CA. Modeling the percolation of annotation errors in a database of protein sequences. Bioinformatics. 2002;18:1641–1649. doi: 10.1093/bioinformatics/18.12.1641. [DOI] [PubMed] [Google Scholar]
- 21.Zmasek CM, Eddy SR. RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitch WM. Homology a personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- 23.Brown DP, Krishnamurthy N, Sjölander K. Automated protein subfamily identification and classification. PLoS Comput. Biol. 2007;3:e160. doi: 10.1371/journal.pcbi.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnamurthy N, Brown D, Sjölander K. FlowerPower: clustering proteins into domain architecture classes for phylogenomic inference of protein function. BMC Evol. Biol. 2007;7(Suppl 1):S12. doi: 10.1186/1471-2148-7-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.