Abstract
Current protein sequence databases employ different classification schemes that often provide conflicting annotations, especially for poorly characterized proteins. ProGMap (Protein Group Mappings, http://www.bioinformatics.nl/progmap) is a web-tool designed to help researchers and database annotators to assess the coherence of protein groups defined in various databases and thereby facilitate the annotation of newly sequenced proteins. ProGMap is based on a non-redundant dataset of over 6.6 million protein sequences which is mapped to 240 000 protein group descriptions collected from UniProt, RefSeq, Ensembl, COG, KOG, OrthoMCL-DB, HomoloGene, TRIBES and PIRSF. ProGMap combines the underlying classification schemes via a network of links constructed by a fast and fully automated mapping approach originally developed for document classification. The web interface enables queries to be made using sequence identifiers, gene symbols, protein functions or amino acid and nucleotide sequences. For the latter query type BLAST similarity search and QuickMatch identity search services have been incorporated, for finding sequences similar (or identical) to a query sequence. ProGMap is meant to help users of high throughput methodologies who deal with partially annotated genomic data.
INTRODUCTION
Functional annotation of new protein sequences is primarily a classification exercise that is based on searching several pre-classified protein or domain family databases (1,2). Current databases use a variety of classification schemes and methods, and therefore the resulting protein groups (e.g. families or orthologous groups) and functional annotations provided may vary from database to database (3,4). This problem is often encountered by users of high throughput methodologies especially when dealing with partially annotated genomes and poorly characterized proteins. Unifying and/or reclassifying the protein databases appears to be a plausible solution, however it also has major drawbacks. First, if properly done, this approach would require an effort equivalent to establishing and maintaining a new, curated protein database. Second, the individual classification schemes of the databases represent a very important added value which would go at least partly lost if we replace them with a new classification scheme. These problems led us to seek solutions that preserve all the information present in the underlying datasets and yet can be maintained in a largely automated fashion.
ProGMap is a single-entry web-tool that unifies the classification information of the current protein databases. Instead of creating a new classification scheme in which some of the expert knowledge used to construct the underlying databases would be inevitably lost, ProGMap combines the distinct classification schemes through constructing a network of links using a fast and fully automated hashing/mapping method originally developed for document classification (5). Briefly, this algorithm converts sequences into unique ‘message digests’ or ‘fingerprints’ which can then be used for mapping sequences (identifiers) from various database rapidly. The purpose of ProGMap is 3-fold: (i) to provide a direct insight into the relationships among the various data sets through a single entry point, (ii) to refine and improve upon existing protein classification (clustering) methodologies, and ultimately, (iii) to gain better understanding of the concepts used for grouping proteins. ProGMap consists of a non-redundant dataset of over 6.6 million protein sequences which are mapped to 240 000 protein and group descriptions collected from UniProt (6), RefSeq (7), Ensembl (8), COG and KOG (9), HomoloGene (10), OrthoMCL-DB (11), TRIBES (12) and PIRSF (13). Looking up a query sequence or a group name in ProGMap provides information whether or not the underlying databases are in agreement on a certain term, and it also gives a plausible indication on how the conflicting annotations and/or group assignments could be improved. Therefore ProGMap is an annotation tool designed not only for database annotators, but also for users of high throughput methodologies such as microarrays or proteomics.
METHODS
We used a centralized data warehouse approach implemented in a relational database (Oracle version 10.2g) to store protein-to-protein, protein-to-group and group-to-group mappings as well as functional descriptions of proteins and groups. Specifically, these descriptions and mappings can be best pictured as nodes and edges in ProgMap's network, respectively. This network-based architecture enables queries to be made, for example, with distinct protein identifiers without explicitly specifying their type. For instance, queries such as HBA_HUMAN, P69905, NP_000549, ENSP00000251595 and 3039 used by UniProt, Refseq, Ensembl and EntreGene databases, respectively, yield identical results as they point to the same node within the network. The data used to build ProGMap (Table 1) were extracted from the source databases using our local Sequence Retrieval Server (SRS) (14) as well as using modules written in Perl. Our goal is to keep the database up-to-date by following the regular updating schedule of the HomoloGene database (using only the odd-numbered releases).
Table 1.
Database members and supported identifiers in the ProGMap database
| Database | Supported identifiers | Notes | URL |
|---|---|---|---|
| UniProt | • Protein ID (e.g. HBA_HUMAN) primary/secondary protein ACCESSION (e.g. P69905, P01922) | ftp://ftp.ebi.ac.uk/pub/databases/uniprot/current_release/ knowledgebase/complete/ | |
| RefSeq (proteins) | • Protein ACCESSION (e.g. NP_000549) • Protein GI (e.g. 4504347) | ftp://ftp.ncbi.nih.gov/refseq/release/ | |
| Ensembl (proteins) | • Translation ID (e.g. ENSP00000251595) | ftp://ftp.ensembl.org/pub/ | |
| Ensembl Compara (families) | • Family ID (e.g. ENSF00000005499) | Protein families | |
| HomoloGene | • RefSeq protein ACCESSION (e.g. NP_000549) • Protein GI (e.g. 4504347) • Entrez GeneID (e.g. 3039) • Official gene symbol (e.g. HBA1) • Group ID (e.g. 469) | Orthologous clusters of 20 eukaryotic proteomes | ftp://ftp.ncbi.nih.gov/pub/HomoloGene/current/ |
| COG | • COG-specific protein ID (e.g. ampG) • Group ID (e.g. COG0477) | Orthologous clusters of 66 prokaryotic and eukaryotic (unicellular only) proteomes | ftp://ftp.ncbi.nih.gov/pub/COG/KOG/ |
| KOG | • KOG-specific protein ID (e.g. Hs4504345) • Group ID (e.g. KOG3378) | Orthologous clusters of seven eukaryotic proteomes | ftp://ftp.ncbi.nih.gov/pub/COG/KOG/ |
| OrthoMCL-DB | • DB-specific protein ID (e.g. hsa11326) • Group ID (e.g. OG1_7606) | Orthologous clusters of 87 proteomes (both eukaryotes and prokaryotes) | http://orthomcl.cbil.upenn.edu/OrthoMCL_DB_Data/ |
| TRIBES | • Tribes specific protein ID (e.g. MMUS-XXX-02-000372) • Group ID (e.g. TR-006821) | Protein families | http://cgg.ebi.ac.uk/services/tribes/tribe.sql.gz Website no longer supported. |
| PIRSF | • UniProt ACCESSION (e.g. P68871) • Group ID (e.g. PIRSF500045) | Protein families, subfamilies and superfamilies | ftp://ftp.pir.georgetown.edu/databases/pirsf/ |
First, we constructed a non-redundant set of over 6.6 million protein sequences and cross-referenced them using a fast and reliable hashing/mapping method implementing the MD4 algorithm (5). This algorithm was intended for digital signature applications such as for ‘compressing’ large files prior secure encryption. As the algorithm can take any string of characters and convert it into a unique 128-bit ‘message digest’ or ‘fingerprint’ in an efficient manner, we applied it for comparing protein sequences to each other as well as to group only sequences identical over the entire length into uniquely labeled ‘Protein Identity Groups’ (PIGs). Sequences which differ by a single (amino acid) residue give rise to different fingerprints (except for the N-terminal methionine which is disregarded) whereas sequences identical over the entire length share the same fingerprint. Each PIG corresponds to a unique protein sequence associated with various synonymous source databases’ protein identifiers (labels) and descriptions, therefore these are kept intact as present originally in the source databases. Importantly, the algorithm guarantees that no two distinct protein sequences produce identical message digests, and hence be members of the same PIG.
Once the initial mapping was completed, group-to-group mappings were established through the process of translating the group members’ identifiers into the unique keys and directly linking only those groups which shared at least one common member.
RESULTS
Network of protein group mappings
The large databases underlying ProGMap were integrated using a centralized (data warehouse) approach to enable fast response to user queries. Once the datasets were downloaded and formatted according to ProGMap's database scheme, mapping these onto each other and building the Oracle database (16.4 GB in total) took less than an hour on a database server with two Intel Xeon processors (4 GB RAM). This fully automated mapping procedure resulted in a complex network of groups interlinked by one-to-one, one-to-many and many-to-many relationships. The resulting network of links on this centralized system enables functional as well as evolutionary information to be retrieved for many proteins being studied in high throughput experiments.
Web interface
The ProGMap database is equipped with a web interface that enables queries to be performed on the entire data sets (provided by the member databases) from a single entry point. The results are presented in both numerical and graphical forms. The web interface consists of six pages: (i) the ‘About’ page provides some background information about ProGMap; (ii) the ‘Query’ page is the main entry point for submitting queries; (iii) the ‘BLAST’ page enables protein or nucleotide sequences to be compared to the non-redundant ProGMap dataset using the BLAST algorithm (15); (iv) the ‘Quick Match’ page is an interface to an exact protein sequence retrieval service which is much faster than a BLAST similarity search; (v) the ‘Statistics’ page summarizes the ProGMap's content in several tables and charts; and (vi) the ‘Help’ page. These pages were developed using Oracle's rapid application development environment (APEX version 3.1.1) which facilitates both easy maintenance and implementation of new features.
The main ‘Query’ page offers eight predefined queries (Q0-7) using ‘keywords’, ‘proteinID’, ‘groupID’ or combinations of thereof. Importantly, valid gene symbols and database-specific identifiers (Table 1) can be used for querying ProGMap without the need to convert these into a specific type prior to searching the databases owing to its network-based architecture. Moreover, users can use batch mode to upload more than one query item in a space-delimited file. Once the results of a query have been retrieved, these can be saved in a text file or inspected visually using built-in graphical web tools.
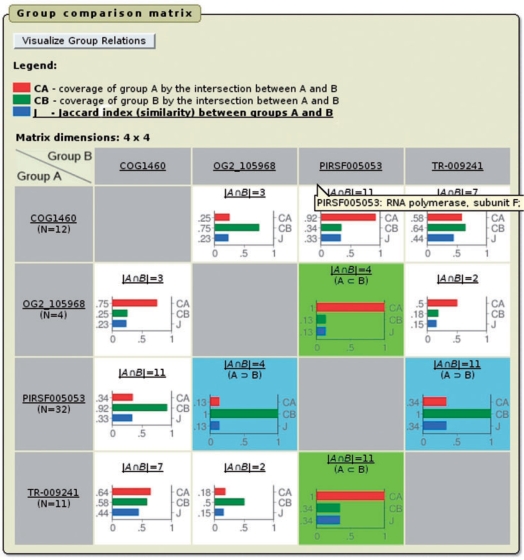
The ProGMap interface provides numeric and graphical tools for visualizing group-to-group relations. For example, the ‘Group comparison matrix’ (denoted as ‘matrix’ from here on) is available via the ‘Compare Protein Groups’ button (applicable to only some queries) in the upper left corner of the query results (Figure 1). Each cell in the matrix corresponds to a pairwise group comparison, and provides several measures shown in a bar chart and explained in the help message. This chart consists of three bars that indicate the extent of the overlap (coverage) of two groups A and B (denoted as CA and CB for groups A and B, respectively), as well as the similarity between them using the Jaccard index (denoted as J). This index equals to one for identical groups (that have all sequences in common) and equals to zero for non-overlapping groups (that do not share any common sequence). Additionally, the number of common members shared by two groups (intersection) and their set relations such as identity, superset and subset, are indicated in each non-empty cell.
Figure 1.
Comparing protein groups using the matrix comparison tool. Using an uncharacterized protein from M. jannaschii (RefSeq: NP_247002), ProGMap annotates this protein sequence as a ‘RNA polymerase subunit F’ on the basis of the manually curated PIRSF family (PIRSF005053). Although three other groups—wherein the protein is also found—do not provide plausible functional annotations (COG: COG1460; TRIBES: TR-009241; OrthoMCL-DB: OG2_105968), these, however, have more than one member in common as well as form either perfect (TR-009241 and OG2_105968) or nearly perfect subsets (COG1460) of the PIRSF family. The matrix comparison tool provides detailed information on set theoretic relations, per-group coverage (CA and CB, bars in red and green) and Jaccard index (J, bars in blue).
Another complementary visualization tool, which is available via the ‘Visualize Group Relations’ button in the upper left corner of the matrix (Figure 2), has been developed to gain a direct insight into the interlinked network of relations between protein groups. One can choose between three different network layouts, namely circle (default), spiral, or random, and adjust the representation of data to his/her own needs. The active nodes and edges (highlighted in red) are accompanied by hyperlinks to additional information about protein groups and relationships. The tools above have been developed using PL/SQL, scalable-vector graphics (SVG) and Javascript and have been extensively tested using the Firefox, Internet Explorer, and Opera web browsers.
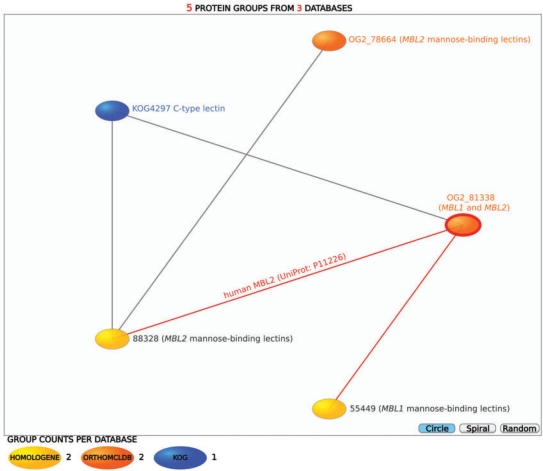
Figure 2.
Comparing protein groups using the network visualization tool. The relationships among five orthologous groups of mannose-binding lectins (KOG: KOG4297; OrthoMCL-DB: OG2_78664, OG2_81338; HomoloGene: 55449, 88328). Groups sharing at least one protein are connected with an edge. In this particular example, the HomoloGene database (yellow) divides the lectins precisely into the two orthologous groups described in the literature (16,17), whereas the other databases either combine them into one group (KOG, blue), or divide them differently (OrthoMCL, orange).
Examples
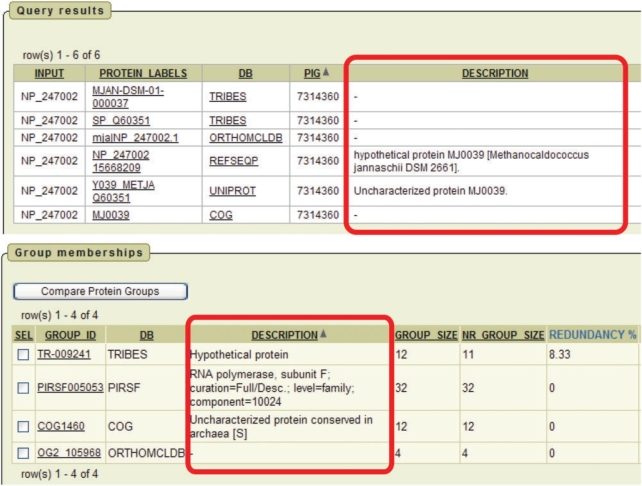
If a protein sequence is found in the databases underlying ProGMap, submitting the sequence ID (using the ‘Q6’ option) will return all synonymous sequence IDs of the protein in ProGMap, along with the functional annotations. One can then view the groups into which this protein is classified in the various databases. Figure 3 shows an example of an ID-based query using an uncharacterized protein of Methanococcus jannaschii that is referenced in some of the databases; however a table of parallel annotations shows that only the PIRSF group was manually curated and provides a plausible biological function for the query protein. In practice, a list of protein (gene) IDs or names obtained from microarray or proteomics experiments can be submitted for ID-based search in batch mode to retrieve the corresponding proteins’ annotations.
Figure 3.
Finding functional annotations with ProGMap. A hypothetical protein query is submitted to the BLAST server that shows significant similarities with an uncharacterized protein from M. jannaschii (RefSeq: NP_247002) (output not shown). By submitting this entry to ProGMap, all the synonymous protein identifiers along with protein descriptions and links to protein groups are retrieved from the underlying databases. Only one of the databases, PIRSF assigns this protein to a curated family annotated as ‘RNA polymerase subunit F’. The annotation of the PIRSF group indicates manual curation, which is an argument for accepting this tentative function. Although the group comparison view (Figure 1) shows that the databases are highly consistent with respect to this group (the groups are in nearly perfect agreement in all databases), the functional annotations are different for the groups compared.
If the protein ID is not found, there are two alternatives for submitting a query sequence: (i) searching for exact matches (no mismatches allowed) using the ‘Quick Match’ service, or (ii) use the BLAST algorithm to search for similar sequences in ProGMap. In this case, one can simply select the desired entries by clicking on the top list and submit them to ProGMap for functional annotation.
Some automatically inferred protein families contain conflicting annotations. For instance, the putative ENSF00000005499 family of Ensembl Compara is annotated as ‘heat shock homolog hsp20’. ProGMap allows one to compare this family to curated (reference) groups from other databases. The ‘Q2’ query returns four groups (without the query group), of which three are from PIRSF (PIRSF036514, PIRSF000228 and PIRSF002680) and one from TRIBES (TR-000776). The pairwise comparisons between these four groups and Ensembl's family show minimal overlaps, which include a small heat shock protein (hsp20), collagen and an NADH dehydrogenase subunit. In contrast, the TR-000776 family in TRIBES is functionally coherent when compared to manually curated PIRSF036514 (alpha-crystallin-related small heat-shock proteins) or KOG3591 (alpha crystallins), so the user has an option to choose.
Reliable orthology detection is crucial, amongst others, for functional annotation of uncharacterized proteins (3). ProGMap can also help in finding false positive orthology assignments (i.e. paralogs) in protein orthology databases. An example is the human mannose-binding lectin MBL2. Previous phylogenetic and functional studies showed that mannose-binding lectin proteins of vertebrates belong to two distinct orthologous groups (represented by MBL1 and MBL2 genes), which duplicated before the divergence of primates and rodents, as well as show tissue-specific gene expression (16,17). Due to loss of the MBL1 gene, humans retained only MBL2. Mannose-binding lectins can be retrieved by using the MBL1 and MBL2 gene symbols in the ‘Q7’ query of the ProGMap interface. This results in a list of 15 groups; orthology is explicitly shown only in five of the groups, OrthoMCL-DB (OG2_78664 and OG2_81338), HomoloGene (55449 and 88328) and KOG (KOG4297) so we compare these groups using the ‘Q4’ query. By examining these orthologous groups we find that only the HomoloGene database infers the orthologs of the mannose-binding proteins in agreement with the cited paper, i.e. the out-paralogous families being separated into two distinct groups (Figure 2). OrthoMCL-DB's assigns human MBL2 protein to the paralogous group OG2_81338 instead of the orthologous group OG2_78664. On the other hand, KOG4297 includes species at large phylogenetic distances.
Comparison with other tools
There are an increasing number of tools designed to interlink multiple databases and make the information available through single WWW entry points, among others MatchMiner (18), SOURCE (19), Harvester (20), iHOP (21), IDConverter (22), CARGO (23), YOGY (24) and HCOP (25). Some functionalities of ProGMap are also included in several other services. For example, some services including IDconverter enable queries to be made using synonymous names or IDs for various genes (proteins); however, the relevant biological information can only be retrieved for a limited number of well-annotated eukaryotic genomes including human and mouse. In contrast, ProGMap includes all currently known proteins i.e. it covers all the kingdoms of life. At present only ProGMap includes a sequence similarity and an identity search service. Text searches using multiple keywords, gene symbols or protein IDs/accessions are supported by several other web portals including IDConverter, MatchMiner, SOURCE, CARGO and HCOP, but in addition ProGMap allows full text queries to be combined using Boolean operators. Graphic presentation of query results is an integral part of ProGMap, CARGO, YOGY and iHOP. ProGMap is unique among these portals because it can directly compare protein groups in different databases, and thereby provide statistical support to annotation decisions.
CONCLUSIONS AND PERSPECTIVES
In this article we present ProGMap, a comprehensive mapping of the UniProt, RefSeq, Ensembl, COG, KOG, OrthoMCL-DB, HomoloGene, TRIBES and PIRSF databases that can be queried via a single interface (http://www.bioinformatics.nl/progmap). ProGMap is meant for users such as biologists and database annotators, who want to find the most probable functions for poorly characterized sequences, or want to assess the coherence between automatically inferred and expert curated protein families/orthologous groups. The ProGMap interface is freely accessible and presents the results both in numerical and graphical form. Future work includes the development of a web services-based interface suitable to link to high throughput pipelines.
FUNDING
Graduate School Experimental Plant Sciences (to A.K.); Dutch NutriGenomics Consortium (to K.L.); BioAssist programme of the Netherlands Bioinformatics Centre (to H.N.). Funding for open access charge: Wageningen University and Research Centre.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Hong Luo for expert help in the initial set-up of the Oracle database and Blaise Alako for stimulating discussions.
REFERENCES
- 1.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuzniar A, van Ham RCHJ, Pongor S, Leunissen JAM. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24:539–551. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Rost B. Domains, motifs and clusters in the protein universe. Curr. Opin. Chem. Biol. 2003;7:5–11. doi: 10.1016/s1367-5931(02)00003-0. [DOI] [PubMed] [Google Scholar]
- 5.Rivest R. The MD4 Message-Digest Algorithm. Cambridge (MA), United States: RFC 1320, MIT; 1992. [Google Scholar]
- 6.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2009;35:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tatusov RL, Fedorova MD, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya A, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright AJ, Kunin V, Ouzounis CA. Protein families and TRIBES in genome sequence space. Nucleic Acids Res. 2003;31:4632–4638. doi: 10.1093/nar/gkg495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu CW, Nikolskaya AN, Huang H, Yeh LL, Natale DA, Vinayaka CR, Hu Z, Mazumder R, Kumar S, Kourtesis P, et al. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etzold T, Argos P. SRS – an indexing and retrieval tool for flat file data libraries. Comput. Appl. Biosci. 1993;9:49–57. doi: 10.1093/bioinformatics/9.1.49. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sastry R, Wang JS, Brown DC, Ezekowitz RA, Tauber AI, Sastry KN. Characterization of murine mannose-binding protein genes Mbl1 and Mbl2 reveals features common to other collectin genes. Mamm Genome. 1995;6:103–110. doi: 10.1007/BF00303252. [DOI] [PubMed] [Google Scholar]
- 17.Phatsara C, Jennen DGJ, Ponsuksili S, Murani E, Tesfaye D, Schellander K, Wimmers K. Molecular genetic analysis of porcine mannose-binding lectin genes, MBL1 and MBL2, and their association with complement activity. Int. J. Immunogenet. 2007;34:55–63. doi: 10.1111/j.1744-313X.2007.00656.x. [DOI] [PubMed] [Google Scholar]
- 18.Bussey KJ, Kane D, Sunshine M, Narasimhan S, Nishizuka S, Reinhold WC, Zeeberg B, Ajay W, Weinstein JN. MatchMiner: a tool for batch navigation among gene and gene product identifiers. Genome Biol. 2003;4:R27. doi: 10.1186/gb-2003-4-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, Hernandez-Boussard T, Rees CA, Cherry JM, Botstein D, Brown PO, et al. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liebel U, Kindler B, Pepperkok R. ‘Harvester’: a fast meta search engine of human protein resources. Bioinformatics. 2004;20:1962–1963. doi: 10.1093/bioinformatics/bth146. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann R, Valencia A. A gene network for navigating the literature. Nat. Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- 22.Alibés A, Yankilevich P, Cañada A, Díaz-Uriarte R. IDconverter and IDCIlight: conversion and annotation of gene and protein IDs. BMC Bioinformatics. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cases I, Pisano DG, Andres E, Carro A, Fernandez JM, Gomez-Lope G, Rodriguez JM, Vera JF, Valencia A, Rojas AM. CARGO: a web portal to integrate customized biological information. Nucleic Acids Res. 2007;35:W16–W20. doi: 10.1093/nar/gkm280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penkett CJ, Morris JA, Wood V, Bähler J. YOGY: a web-based, integrated database to retrieve protein orthologs and associated Gene Ontology terms. Nucleic Acids Res. 2006;34:330–334. doi: 10.1093/nar/gkl311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eyre TA, Wright MW, Lush MJ, Bruford EA. HCOP: a searchable database of human orthology predictions. Brief. Bioinform. 2007;8:2–5. doi: 10.1093/bib/bbl030. [DOI] [PubMed] [Google Scholar]