Abstract
The MEME Suite web server provides a unified portal for online discovery and analysis of sequence motifs representing features such as DNA binding sites and protein interaction domains. The popular MEME motif discovery algorithm is now complemented by the GLAM2 algorithm which allows discovery of motifs containing gaps. Three sequence scanning algorithms—MAST, FIMO and GLAM2SCAN—allow scanning numerous DNA and protein sequence databases for motifs discovered by MEME and GLAM2. Transcription factor motifs (including those discovered using MEME) can be compared with motifs in many popular motif databases using the motif database scanning algorithm Tomtom. Transcription factor motifs can be further analyzed for putative function by association with Gene Ontology (GO) terms using the motif-GO term association tool GOMO. MEME output now contains sequence LOGOS for each discovered motif, as well as buttons to allow motifs to be conveniently submitted to the sequence and motif database scanning algorithms (MAST, FIMO and Tomtom), or to GOMO, for further analysis. GLAM2 output similarly contains buttons for further analysis using GLAM2SCAN and for rerunning GLAM2 with different parameters. All of the motif-based tools are now implemented as web services via Opal. Source code, binaries and a web server are freely available for noncommercial use at http://meme.nbcr.net.
INTRODUCTION
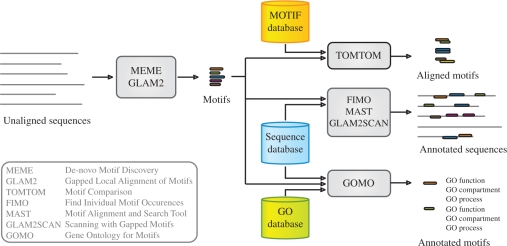
The MEME Suite is a software toolkit with a unified web server interface that enables users to perform four types of motif analysis: motif discovery, motif–motif database searching, motif-sequence database searching and assignment of function. It offers a significantly expanded set of programs for these tasks compared with the earlier web server (1). Figure 1 shows an overview of the MEME Suite. MEME (2) and GLAM2 (3) are tools for motif discovery, Tomtom (4) searches for similar motifs in databases of known motifs, FIMO, GLAM2SCAN (3) and MAST (5) search for occurrences of motifs in sequence databases, and GOMO (6) provides associations between motifs and GO terms. The components of the MEME Suite are implemented in ANSI C as command line tools. These are published as SOAP (Simple Object Access Protocol) web services using Opal (7) and the Tomcat Java servlet container. Opal provides job management services allowing the MEME Suite to queue multiple simultaneous requests.
Figure 1.
Overview of the MEME Suite tools.
MOTIF DISCOVERY
The MEME algorithm (2) has been widely used for the discovery of DNA and protein sequence motifs, and MEME continues to be the starting point for most analyses using the MEME Suite. Detailed protocols describing how to use MEME are available (8).
Some biosequence motifs exhibit insertions and deletions, but MEME cannot discover such motifs, because it does not allow gaps. To overcome this limitation, we have incorporated a recent algorithm for gapped motif discovery—GLAM2 (3)—into the MEME suite. Discovering gapped motifs is intrinsically more difficult than discovering ungapped motifs, because there are vastly more possible gapped motifs than ungapped motifs. Therefore, when trying to discover gapped motifs, we recommend performing a simpler gapless motif analysis as well.
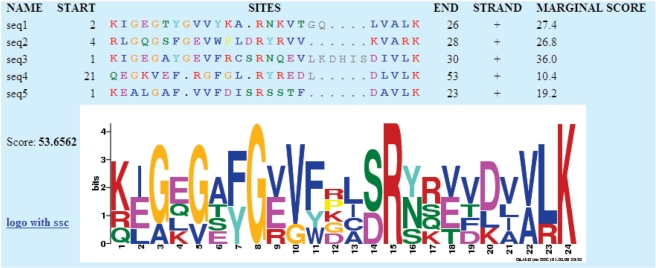
GLAM2 uses a particular ‘model’ of gapped motifs, which is illustrated in Figure 2. A motif has a certain number of aligned columns, indicated by colored letters in the figure. Aligned columns may exhibit deletions (indicated by dots), and residues may be inserted between them (gray letters). No attempt is made to align inserted (gray) residues with one another: GLAM2 assumes that their identity is unimportant. Inserted residues are also omitted from the LOGO.
Figure 2.
A sample GLAM2 gapped motif.
GLAM2 reports a score for each motif that it discovers, with higher scores indicating stronger motifs. GLAM2 also reports a score for each site, with higher scores indicating better matches to the overall motif.
Using of GLAM2 is similar to using MEME, with only a few differences. Unlike MEME, GLAM2 does not search for multiple distinct motifs. Instead, it performs replicates: it attempts to discover the strongest possible motif 10 times, and displays the results in order of score. If the top few results are similar, this may be regarded as successful replication. If not, GLAM2 can be rerun more thoroughly (but slowly) by increasing the ‘number of iterations’ parameter.
The gappiness of GLAM2 motifs can be controlled by four pseudocount options. Their relative values control GLAM2's aversion to gaps: increasing the no-deletion pseudocount relative to the deletion pseudocount makes it more averse to deletions, and likewise for the no-insertion and insertion pseudocounts. The absolute pseudocount values control GLAM2's preference for putting gaps together in the same positions: decreasing the deletion and no-deletion pseudocounts makes it more prone to gather deletions into a few columns, and likewise for the (no-)insertion pseudocounts. Note that the pseudocounts affect the score calculation, so scores are not comparable between motifs discovered with different pseudocount settings.
GLAM2 has options to set the maximum and minimum number of aligned columns, similar to MEME's maximum and minimum width options. It also has an option for the initial number of aligned columns: setting this can help it find an appropriate motif. GLAM2 has difficulty adjusting the motif width when there are many sequences, especially if they are short. It should be noted that both protein and DNA motifs are often shorter than the defaults (50) used by GLAM2 and MEME for the ‘maximum number of aligned columns’ and ‘maximum width’, respectively. It is often advisable for you to reduce those parameters to much smaller values (e.g. in the range 10–20) by entering a new value in the appropriate input box on the web form.
Finally, GLAM2 lets you specify the minimum number of input sequences that must contribute a motif occurrence. This is a generalization of MEME's OOPS (one occurrence per sequence) and ZOOPS (zero or one occurrence per sequence) options. GLAM2 cannot consider more than one occurrence per sequence.
When interpreting GLAM2 output, note that it will always report the best motif it can find, even if you give it random sequences. Thus, it may be wise to rerun GLAM2 on negative control (e.g. shuffled) sequences and compare the resulting scores with the original scores. The GLAM2 input form contains a checkbox (on the lower right-hand side) that will cause the characters in the input sequences to be shuffled before being input to GLAM2.
USING AND ANALYZING MOTIFS
Once you have discovered a collection of motifs, you may wish to perform additional analyses to better characterize those motifs. The MEME Suite provides three types of tools for carrying out such analyses. First, the MEME Suite can compare your DNA motifs to known compendia of motifs (such as JASPAR, Flyreg and DPINTERACT) to see if your motif is similar to a known regulatory motif. This type of analysis is done using Tomtom. Second, the MEME Suite can attempt to determine what types of regulatory functions your motif might be involved in. This assignment is done using the GOMO tool to determine if your motif matches upstream regions of many sequences with the similar Gene Ontology (GO) annotations. Third, the MEME Suite can search a sequence database for additional occurrences of your motif.
Comparing DNA motifs with known regulatory motifs
Often, your first question after finding a DNA motif will be, ‘Is this a novel motif?’ Thus, it may be useful to learn whether a motif found by MEME is similar to other motifs, particularly motifs with known biological functions. Tomtom (4) quantifies the similarity between two motifs, and can be used to search a database of known motifs for matches to motifs found by MEME. Tomtom not only provides a numeric score for the match between two motifs, but also provides an estimate of the statistical significance of the score. Currently, Tomtom only supports DNA motifs.
The MEME output for each reported motif contains a button for submitting that motif directly to Tomtom. The Tomtom web application also allows the user to submit a motif by pasting in columns of base counts for each position of the motif. The user then selects the motif similarity measure to use and chooses which online motif database to search.
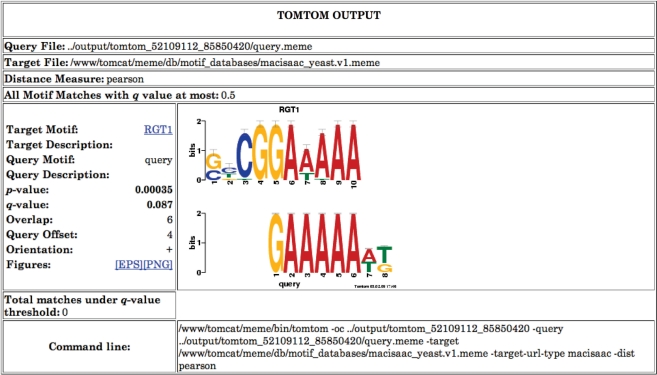
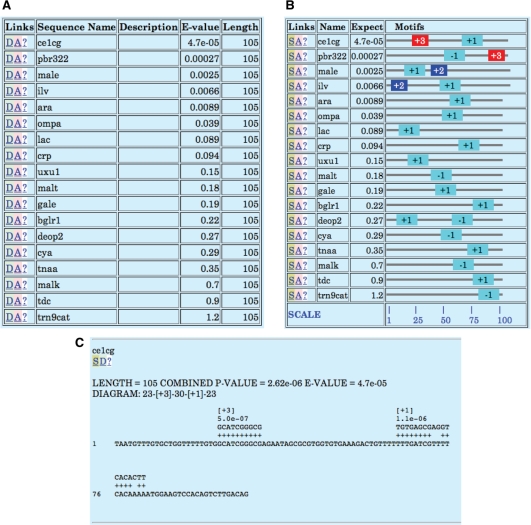
The output of Tomtom includes LOGOS representing the alignment of two motifs, the p-value and q-value [a measure of false discovery rate (10)] of the match, and links back to the parent motif database for more detailed information about the target motif. Sample Tomtom output is shown in Figure 3.
Figure 3.
Tomtom output. The figure shows the Tomtom output from searching a single DNA motif against a collection of yeast transcription factor binding site motifs identified via ChIP-seq (9). Tomtom shows that the query motif closely resembles the binding motif for transcription factor RGT1.
GO term analysis for DNA motifs
A second question you may ask is, ‘What is the functional role of this motif?’ The tool GOMO (6) is used to search a species-specific GO annotation database for GO terms that are associated with genes that a given DNA motif regulates. GOMO uses the motif models in the format generated by MEME. GOMO ranks genes by the average binding affinity of the transcription factor to the gene's upstream region and assesses GO terms associated with these genes. Gene sequences and GO annotations are linked via the sequence identifier. The latter requires a curated dataset, a selection of which are currently available covering the best annotated species with respect to GO—Escherichia coli, Drosophila, chicken, mouse, Saccharomyces cerevisiae and Schizosaccharomyces pombe.
GOMO reports for each motif the list of GO terms considered significant in descending order down to a threshold specified before. When interpreting GOMO output, note that the GO terms reported always relate to the gene the transcription factor regulates.
Sequence database search
With a set of interesting motifs in hand, an obvious next step is to look for other occurrences of these motifs. The tools FIMO and MAST are used to search sequence databases for matches to motifs discovered using MEME. The GLAM2SCAN tool is specifically designed for searching with gapped motifs of the type discovered by GLAM2. The MEME server provides web forms for performing analyses with each of these tools. As a convenience, the HTML output of MEME contains buttons for starting FIMO and MAST searches. The MEME web site provides online versions of a number of sequence databases, or users may upload their own sequence data in FASTA format.
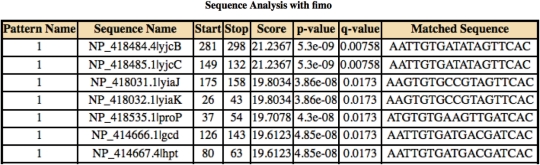
‘FIMO’ stands for ‘find individual motif occurrences’. FIMO uses the output of MEME, which may contain multiple, ungapped motifs. FIMO scores the match to each motif at each position in the sequence database. As the name of the tool suggests, each match is treated independently. The p-value for the match is computed using a dynamic programming procedure (11), and motif-specific q-values with respect to the complete set of matches are computed using a bootstrap procedure (12). The output from FIMO is a list of the matches for which the q-value is less than a user-specified threshold. Sample output from FIMO is shown in Figure 4.
Figure 4.
FIMO output.
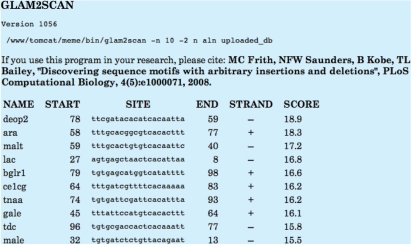
GLAM2SCAN uses the output of GLAM2, which always consists of a single motif, possibly containing gaps. GLAM2SCAN scores the match to this motif at each position in the sequence database. Like FIMO, each match is treated independently, and the output is a list of the best scoring matches. The user can adjust the number of matches reported, up to a limit of 200. Sample output from GLAM2SCAN is shown in Figure 5.
Figure 5.
GLAM2SCAN output. The figure shows the result of searching with a GLAM motif against 18 E. coli DNA sequences containing the Cyclic AMP receptor protein (CRP) binding site. Only the top 10 matches are shown.
MAST (13) also uses the output of MEME. For each sequence, MAST determines the best match in the sequence to each motif. The scores for these best sequence motif matches are combined into a score for the overall match between the complete motif set and the sequence, resulting in an E-value for each sequence. The output from MAST is a list of the sequences for which the E-value is less than a user-specified threshold. In addition to the list of sequences, the output contains a block diagram showing the relative positions of the best motif matches in the high scoring sequences, and annotated alignments of the best motif matches. The three sections of MAST output are shown in Figure 6.
Figure 6.
MAST output. The figure shows the result of searching with three MEME motifs against 18 E. coli DNA sequences containing the CRP binding site. The MAST output contains three representations of the results, excerpts of which are shown in the three figure panels. The E-value score of the overall match of the motif(s) in the input is shown in the first results section (Panel A). The second section (Panel B) displays the relative locations of significant matches of the motifs in the sequences. The third results sections gives a detailed picture of the motif matches, showing the exact location and p-value score of each motif match aligned above the target sequence.
The choice of motif search tool will depend on the goal of the analysis. MAST is ‘sequence oriented’, computing a single score for each sequence in the database. This makes MAST more suited for analyzing proteins or fixed-length sequences like upstream regions of genes. FIMO and GLAM2SCAN only provide individual motif matches, and can be used to scan genomic databases. Both FIMO and MAST require ungapped motifs, whereas searching with gapped motifs requires the use of GLAM2SCAN.
WEB SERVER AND USER SUPPORT
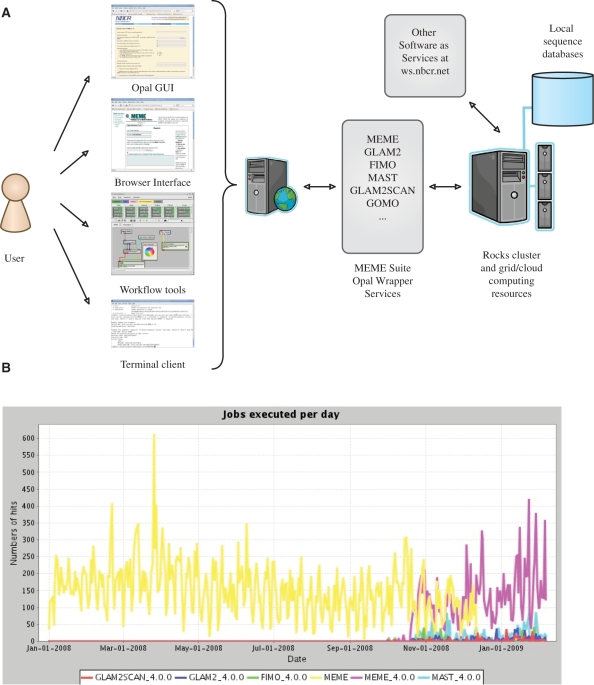
The MEME Suite web services are hosted by the National Biomedical Computation Resources (NBCR, http://nbcr.net). Since late 2007, we have adopted the Opal web service toolkit (7) to handle the computational and data management aspect of the MEME web server (Figure 7A). The Opal toolkit provides a SOAP-based interface for managing user job requests, and offers users additional means to access the MEME web services. For example, a generic Opal client may be used to send command line requests for programmatic access to any services of the MEME suite. A generic user interface may be generated automatically based upon an XML description of the command line syntax through the Opal GUI, either in a standard web browser or from a workflow program such as Vision (14), Kepler (15) or the Pipeline Pilot (16).
Figure 7.
MEME Suite deployed with Opal (A) Opal offers versatile user access options. (B) Opal dashboard provides job history data.
Customized user interfaces have been developed for the MEME Suite for enhanced user experience. All clients access the Opal services through the Opal web service application programming interface. When Opal receives a request from a client, it creates a unique working directory, transfers all the input files and dispatches the job to a local batch job scheduler, which schedules the job on an available compute node in a cluster. The adoption of Opal hides the complexity of resource management from scientific programmers, and allows the MEME Suite to take advantage of the distributed grid and the emerging cloud computing environment.
The scalable resources made available by Opal allow applications such as MEME to meet growing demand from users. The sequence databases (more than 120 GB to date) are updated on a weekly basis automatically. The server handles more than 200 user requests per day, and the Opal dashboard provides a real-time usage status update on individual applications (Figure 7B). Users are notified of their job requests with a URL for accessing the results on the web, with up to a week of data life time (a configurable option in Opal). By providing a web interface for user results, users do not have to worry about the output data size or email spam filters. In case any debugging is needed, the user may simply email meme@nbcr.net with the output URL. Some user jobs exceed the size limitations or a fair use time limit policy. These users may use the MEME roll for Rocks clusters (http://www.rocksclusters.org) or the RPM packages for i386 and x86_64 architectures. The Rocks roll automatically configures and installs MEME along with its web services with a few simple commands on a Rocks cluster. Users may also configure their own web server, and direct all MEME jobs to the NBCR server for processing. The download information is available at the MEME wiki (https://www.nbcr.net/pub/wiki/index.php?title=MEME). Additional community support for MEME is available at the MEME forum (https://www.nbcr.net/forum/viewforum.php?f=5).
FUNDING
The authors acknowledge NBCR award from NCRR, NIH P41 RR08605, for support of the MEME and MAST web site. T.L.B., C.E.G. and W.S.N. acknowledge NIH/NCRR award R01 RR021692 for support of continuing development of the MEME and related sequence analysis tools. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bailey TL, Williams N, Misleh C, Li WW. Meme: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey TL, Elkan CP. Fitting a mixture model by expectation-maximization to discover motifs in biopolymers. In: Altman R, Brutlag D, Karp P, Lathrop R, Searls D, editors. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 3.Frith MC, Saunders NFW, Kobe B, Bailey TL. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000071. e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 6.Bodén M, Bailey TL. Associating transcription factor-binding site motifs with target go terms and target genes. Nucleic Acids Res. 2008;36:4108–4117. doi: 10.1093/nar/gkn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnan S, Stearn B, Bhatia K, Baldridge KK, Li WW, Arzberger PA. Opal: simple web services wrappers for scientific applications. IEEE International Conference on Web Services. 2006 Chicago, Ill. [Google Scholar]
- 8.Bailey TL. Discovering sequence motifs. Methods Mol. Biol. 2007;395:271–292. doi: 10.1007/978-1-59745-514-5_17. [DOI] [PubMed] [Google Scholar]
- 9.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc. Natl Acad. Sci. USA. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staden R. Searching for motifs in nucleic acid sequences. Methods Mol. Biol. 1994;25:93–102. doi: 10.1385/0-89603-276-0:93. [DOI] [PubMed] [Google Scholar]
- 12.Storey JD. A direct approach to false discovery rates. J. R. Stat. Soc. 2002;64:479–498. [Google Scholar]
- 13.Bailey TL, Gribskov M. Score distributions for simultaneous matching to multiple motifs. J. Comput. Biol. 1997;4:45–59. doi: 10.1089/cmb.1997.4.45. [DOI] [PubMed] [Google Scholar]
- 14.Sanner MF. A component-based software environment for visualizing large macromolecular assemblies. Structure. 2005;13:447–462. doi: 10.1016/j.str.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Ludaescher B, Altintas I, Berkley C, Higgins D, Jaeger E, Jones M, Lee EA, Tao J, Zhao Y. Scientific workow management and the Kepler system. Concurrency Comput. Pract. Exp. 2005;18:1039–1065. [Google Scholar]
- 16.Hassan M, Brown RD, Varma-O'Brien S, Rogers D. Cheminformatics analysis and learning in a data pipelining environment. Mol. Div. 2006;10:283–299. doi: 10.1007/s11030-006-9041-5. [DOI] [PubMed] [Google Scholar]