Abstract
Exons are typically only 140 nt in length and are surrounded by intronic oceans that are thousands of nucleotides long. Four core splicing signals, aided by splicing-regulatory sequences (SRSs), direct the splicing machinery to the exon/intron junctions. Many different algorithms have been developed to identify and score the four splicing signals and thousands of putative SRSs have been identified, both computationally and experimentally. Here we describe SROOGLE, a webserver that makes splicing signal sequence and scoring data available to the biologist in an integrated, visual, easily interpretable, and user-friendly format. SROOGLE's input consists of the sequence of an exon and flanking introns. The graphic browser output displays the four core splicing signals with scores based on nine different algorithms and highlights sequences belonging to 13 different groups of SRSs. The interface also offers the ability to examine the effect of point mutations at any given position, as well a range of additional metrics and statistical measures regarding each potential signal. SROOGLE is available at http://sroogle.tau.ac.il, and may also be downloaded as a desktop version.
INTRODUCTION
The splicing machinery accurately identifies short exonic regions, typically 140 nt, in the context of intronic regions that are thousands of nucleotides long. Four core splicing signals direct the splicing machinery to the exon/intron junctions. These signals are the 5′ and 3′ splice sites (5′ss and 3′ss), which are located at the 5′ and 3′ ends of the intron; the polypyrimidine tract, located upstream of the 3′ss; and the branch site (BS), which is upstream of the PPT. These four signals are too short and too degenerate to account for precise recognition of exons. Over the last decade many studies have identified exonic and intronic sequence motifs that either boost or repress recognition of exons; these sequences presumably serve as binding sites for different splicing factors. These sequences are known as exonic or intronic splicing enhancers or silencers (ESEs/ISEs and ESSs/ISSs, respectively) and we collectively refer to them as splicing-regulating sequences (SRSs) (1,2). In order to understand the regulatory pressures to which pre-mRNA is exposed, it is necessary to obtain a comprehensive overview of the splicing signals that act on the exon and on the two introns flanking it.
Numerous methods have been devised for identifying and scoring the four splicing signals. In parallel, different groups of SRSs have been identified based on both computational and experimental methodologies. Several servers exist that provide partial information pertaining to splicing signals (http://genes.mit.edu/burgelab/software.html; http://ast.bioinfo.tau.ac.il/BranchSite.htm), and regulatory sequences (http://genes.mit.edu/burgelab/rescue-ese/; http://genes.mit.edu/fas-ess/; http://ast.bioinfo.tau.ac.il/ESR.htm; http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home). As each of these servers provides only limited data, to obtain a more comprehensive view pertaining to splicing signals within a particular sequence, a user must visit a number of websites. Moreover, even when used collectively, these servers are non-comprehensive, and provide access to only a limited number of splicing signals and SRSs. Finally, the output yielded by the different servers is often not directly interpretable. For instance, splice sites or splicing regulators are often scored based on a position specific scoring matrix (PSSM) log-odd score. For most users such a score is arbitrary and of little value.
With SROOGLE, we provide a comprehensive platform to allow biologists to visualize potential splice signals in their sequence of interest in an integrated, user-friendly and easily interpretable format. We placed emphasis on: (i) availability of data: SROOGLE allows biologists access to large sets of published data that are not available on any other public servers; (ii) integration of data: we aimed to offer an integrative overview of the signals characterizing the exon of interest, combining many different datasets of SRSs and different methodologies for scoring the four core splicing signals; (iii) intuitive statistical measures: whenever possible, we have provided percentile scores, indicating the strength of a signal with respect to scores within two large pre-compiled pools of alternatively and constitutively spliced exons; (iv) user friendliness: We put much emphasis into development of an intuitive, interactive, graphic user interface, based on dynamic client-side programming, which enables users to interactively modify their input.
USING SROOGLE
Input form
Upon entering the website, users enter the sequence of their exon and the two introns flanking it. The server will accept either consecutive stretches of DNA or stretches of DNA separated by spaces and numbers, the format obtained from the UCSC Genome web browser or GenBank. A valid DNA sequence may consist of stretches of the 4 nt (A, C, G and T), or the spacer N. Users may also choose to explore the website using our sample exons and introns by clicking on the relevant link.
Output
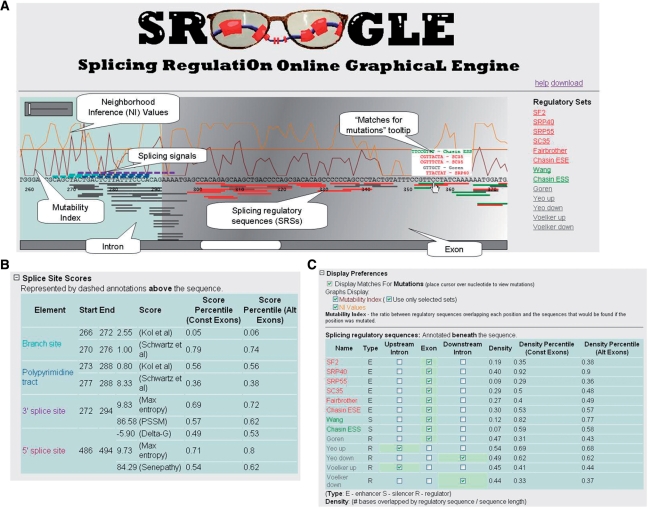
SROOGLE's output consists of a webpage with three main components: A graphic browser (Figure 1A), a table summarizing data related to the four core splicing signals (Figure 1B), and a table summarizing data related to SRSs (Figure 1C).
Figure 1.
SROOGLE output. (A) Graphic browser. The intron is presented in blue, exons in grey. Callout boxes indicate different features presented along the sequence. (B) Table summarizing data pertaining to splicing scores. The start and end position of each signal are indicated, along with its crude score, and a set of percentile scores relative to a dataset of constitutive and alternative exons. (C) Table summarizing data pertaining to splicing regulatory signals. Density scores, indicating the proportion of the exon covered by splicing signals, are presented for each signal, along with two accompanying percentile scores relative to a constitutive and alternative dataset.
Graphic browser
The graphic browser interactively displays splicing-related data along the sequences provided as input (Figure 1A). The sequences of the two introns are displayed over a light blue background and the exon over a grey one. The core splicing signals are presented as dashed lines above the sequence, whereas the SRSs are presented in solid lines below the sequence. Setting the cursor on the lines above or below the sequences displays a tooltip providing additional information regarding the nature and strength of the signals. Two plots may be displayed along the sequence: one displays neighborhood inference data and the other the mutability indexes (see below).
The various annotations and score values are computed on the web server, using an efficient matching algorithm. In order to maximize the application's responsiveness and interactivity, the visual rendering and the dynamic display behavior are implemented via client-side javascript code which controls the Microsoft Silverlight (a browser plug-in) Extensible Application Markup Language (XAML) based display structure. This design also enables offline exploring of result pages that were saved locally, and simplifies the derivation of the desktop version of the application. To view the graphic browser, Microsoft Silverlight must be installed. A link for this program is automatically provided for first-time users, if this application is not detected. This link can be used to automatically download and install browser plug-in; once installed, the browser must be refreshed.
Annotation of four core splicing signals
SROOGLE scores the four main splicing signals based on nine different algorithms. Specifically, the BS and polypyrimidine tract are detected and scored based on the algorithms described in (3) and (4). The 3′ss and 5′ss are detected and scored based on both the maximum entropy based method developed by (5) and the position-specific scoring matrix (PSSM) method as described in (6). For the 5′ss, we also implemented an additional method based on calculation of the free energy (ΔG°) of binding between U1 snRNA and a given 5′ss (7). Each of these signals is marked in the graphic browser above the sequence in dashed lines in a different color. Additional information regarding the splicing signal is presented in a table (Figure 1B), including percentile scores relative to datasets of alternatively and constitutively spliced exons (see below).
Annotation of SRSs
SROOGLE identifies and visualizes specific SRSs from 13 different datasets (Figure 1C). These include the exonic splicing enhancers identified in (8–10), exonic splicing silencers identified in (10,11), exonic regulators from (12), upstream intronic regulators from (13,14), and downstream intronic regulators from (13,14). Each SRS dataset was classified either as a splicing enhancer (marked by an E and visualized in red), silencer (S, visualized in green), or regulator (R, visualized in grey). The user can interactively select the group or groups of SRSs to display within each intron/exon/intron segment. These sequences are visualized in the browser beneath the sequence and setting the cursor on a particular sequence will provide the SRS sequence, the SRS group to which it belongs, as well as normalized rank scores, if available. The normalized rank score is a score between 0 and 1, indicating the ranking of a given sequence with respect to the different sequences identified in a given group of SRS. These scores are only available for datasets of SRSs for which either P-values or PSSM scores were provided by the studies identifying them. In each such group, we ranked sequences based on their P-values (from high to low) or on the PSSM log-odd scores (from low to high), and normalized this rank by the number of sequences in the group. Thus, a score of 1 indicates that a sequence received either the lowest P-value (i.e. most significant) or the highest PSSM log odd score with respect to all other sequences in the dataset. Finally, for each group of SRSs, two metrics are provided: exon density values and density percentile scores. Density values are calculated as the number of nucleotides within an exon covered by a group of SRSs, divided by the length of the exon. The rationale for this metric is that different splicing regulators require more than one binding site in order to mediate their function (2) and, therefore, greater binding site densities are likely to reflect increased probabilities of binding.
Percentile scores
To calculate percentile scores, we relied on two datasets. The first contains information on over 50 000 constitutively spliced exons and the second contains data on 3000 alternatively spliced exons all from the human genome and based on EST data, generated as described in (15). In each of these datasets, the splicing signals were detected and scored based on the algorithms used by SROOGLE. Based on the distribution of values for each of these signals within each of these two datasets, percentile scores were calculated. The percentile score for a user-entered sequence indicates the ranking of the user's sequence within these two pre-calculated distributions. Thus, a value of 0.95 indicates that 95% of the exons have lower scores and only 5% have higher ones.
Effect of mutations
We aimed to allow biologists to readily obtain an understanding of the potential effects of point mutations in their sequence. The user first checks the ‘Display Matches for Mutations’ box. When this is chosen, placing the cursor on a specific nucleotide along the sequence displays a window listing the various SRSs that would overlap the nucleotide if the residue were mutated to any of the three other possible nucleotides.
Neighborhood inference (NI) scores
SROOGLE reports neighborhood inference (NI) scores based on (16). Positive and negative scores indicate that a hexamer beginning at a given position resembles exonic splicing enhancers and silencers, respectively.
Mutability index
This novel index aims to provide an overview of the extent to which a given nucleotide is involved in splicing regulation. This index is calculated as (sum_nonmut − sum_mut)/(sum_nonmut + sum_mut), where sum non_mut is the numbers of SRSs overlapping a given nucleotide and sum_nonmut is the average number of SRSs overlapping that position when the nucleotide in it is mutated to each of the three other possible options. Thus, high values of this index indicate that once a given nucleotide is mutated, fewer SRSs will overlap that position, whereas low values indicate that SRSs will overlap this nucleotide no matter what its identity.
CONCLUSION
SROOGLE combines a variety of algorithms for identifying and scoring splicing signals, numerous datasets of splicing regulatory signals, and previously described and novel measures to obtain a rapid overview of splicing related data for a user-input sequence. SROOGLE also allows a user to examine the effects of mutations at any position thus providing a useful tool for experimental design. SROOGLE was built in a flexible and readily extendible platform to allow incorporation of further datasets once they are made available.
FUNDING
Israel Science Foundation (grant 1449/04); DIP; MOP Germany-Israel (to G.A.). Funding for open access charge: EURASNET.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
S.S. is a fellow of the Edmond J. Safra bioinformatic program at Tel Aviv University. This work was performed in partial fulfillment of the requirements for a PhD degree by S.S.
REFERENCES
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Ann. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 3.Kol G, Lev-Maor G, Ast G. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum. Mol. Genet. 2005;14:1559–1568. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz SH, Silva J, Burstein D, Pupko T, Eyras E, Ast G. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Gen. Res. 2008;18:88–103. doi: 10.1101/gr.6818908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmel I, Tal S, Vig I, Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004;10:828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 10.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences – the complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Voelker RB, Berglund JA. A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing. Genome Res. 2007;17:1023–1033. doi: 10.1101/gr.6017807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeo GW, Van Nostrand EL, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ram O, Schwartz S, Ast G. Multifactorial interplay controls the splicing profile of Alu-derived exons. Mol. Cell Biol. 2008;28:3513–3525. doi: 10.1128/MCB.02279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadler MB, Shomron N, Yeo GW, Schneider A, Xiao X, Burge CB. Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet. 2006;2:e191. doi: 10.1371/journal.pgen.0020191. [DOI] [PMC free article] [PubMed] [Google Scholar]