Abstract
A new web-server tool estimates Ki values from experimentally determined IC50 values for inhibitors of enzymes and of binding reactions between macromolecules (e.g. proteins, polynucleic acids) and ligands. This converter was developed to enable end users to help gauge the quality of the underlying assumptions used in these calculations which depend on the type of mechanism of inhibitor action and the concentrations of the interacting molecular species. Additional calculations are performed for nonclassical, tightly bound inhibitors of enzyme-substrate or of macromolecule-ligand systems in which free, rather than total concentrations of the reacting species are required. Required user-defined input values include the total enzyme (or another target molecule) and substrate (or ligand) concentrations, the Km of the enzyme-substrate (or the Kd of the target-ligand) reaction, and the IC50 value. Assumptions and caveats for these calculations are discussed along with examples taken from the literature. The host database for this converter contains kinetic constants and other data for inhibitors of the proteolytic clostridial neurotoxins (http://botdb.abcc.ncifcrf.gov/toxin/kiConverter.jsp).
INTRODUCTION
Some analyses of networks, pathways and metagenomics focus on identifying key proteins or polynucleic acids as targets for inhibitory compounds. Typically, high-throughput screening assays are initially used to compare and down-select potential inhibitors of enzymatic activity or macromolecule-ligand binding. Many functional assays seek a total inhibitor concentration that reduces these activities by 50% (IC50). However, the IC50 value depends on concentrations of the enzyme (or target molecule), the inhibitor, and the substrate (or ligand) along with other experimental conditions. What is required is an accurate determination of the Ki value, an intrinsic, thermo-dynamic quantity that is independent of the substrate (ligand) but depends on the enzyme (target) and inhibitor. Thus, comparisons can be more readily made among different laboratories to characterize the inhibitors. While these more time-consuming assays are usually done with the most promising candidates, accurate, initial estimates of Ki values for more of the candidates would be beneficial. A much discussed problem in the literature (1–8) is converting IC50 to Ki values because even the simplest types of inhibitory mechanisms (e.g. competitive, uncompetitive and noncompetitive) will influence the calculation.
To help address this problem, our web-server tool calculates Ki values from IC50 values using equations for enzyme-substrate and target-ligand interactions by different inhibitory mechanisms (http://botdb.abcc.ncifcrf.gov/toxin/kiConverter.jsp). Additional calculations are performed for tightly bound inhibitors of enzyme-substrate reactions in which free, rather than total, concentrations of the molecular species are calculated for nonclassic Michaelis–Menten kinetics. Similar calculations can be performed for target molecule-ligand systems. User-defined input values include total concentrations of the enzyme (or target molecule) and substrate (or ligand), the Km of the enzyme-substrate (or the Kd of the target-ligand) reaction and the IC50 value. The outputs include tabulations of the Ki values under different kinetic schemes, extensive tabulations of the results, summary histograms and the corresponding equations. Help buttons are available for Background, Assumptions, Literature, Links and Equations along with examples taken from the host database-server that contains kinetic information on neurotoxin inhibitors. An example calculation is included here for a tight-binding inhibitor of an enzyme–substrate reaction, while other enzyme inhibitor and protein–ligand–inhibitor examples are also provided. Our rationale for creating this converter is to enable end users to judge the quality of the underlying assumptions for these calculations and to help facilitate research and the development of potential therapeutic products.
METHODS
Reactions and equations
The website cited in (9) served as an initial design template for our IC50-to-Ki converter. Equations (1–4) were adapted from refs. (3), (6) and (9) whereas we derived Equation (5) for this study. The analytic expressions for Ki that are shown below were verified numerically by methods used in a previous kinetic analysis (10).
The derivations for converting IC50 to Ki values published by Brandt et al. (3) include three types of classic inhibitor mechanisms in which different relations may exist between S and Km. For tightly bound inhibitors, the equation for Ki by Copeland et al. (6) is used to take into account the larger amounts of inhibitor bound species, thus making the Michaelis–Menten assumption of the total enzyme concentration being equal invalid (5). These equations are also relevant for protein–ligand–inhibitor (P–L–I) interactions that also adhere to the above assumptions.
Enzyme–substrate–inhibitor reactions
For competitive inhibition
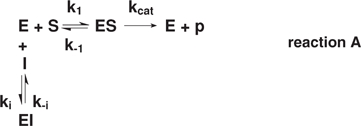 |
where Kd = k–1/k1 and Ki = k−i/ki, the classic expression is
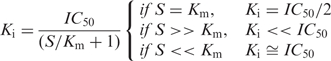 |
1a |
and for tightly bound inhibitors (5,6)
| 1b |
For uncompetitive inhibition
 |
the classic expression is:
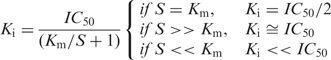 |
2a |
and for tightly bound inhibitors (5,6)
| 2b |
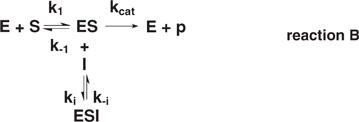
For noncompetitive inhibition (2)
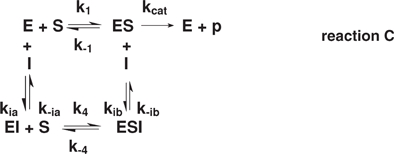 |
where Kia = k–ia/kia and Kib = k–ib/kib, the classic expression is:
| 3a |
and for tightly bound inhibitors (5,6)
| 3b |
This noncompetitive reaction also assumes that the inhibitor dissociation constants are equal: Kia = Kib = Ki. Mixed inhibition, where Kia < > Kib, is not considered here.
P–L–I reactions
For total concentrations, E is replaced by P and S is replaced by L. Additional reaction schemes are located at this tool's website. As in classic enzyme–substrate systems the relation of Ki and IC50 in competitive inhibition is:
| 4a |
For protein–ligand experiments with tight-binding inhibitors, the free rather than the total concentrations of the reactants need to be used as modified from ref. 9
| 4b |
where I50 and L50 are the free concentrations of the inhibitor and ligand, respectively, at 50% inhibition, and P0 is the free concentration of the protein in the absence of inhibitor. The concentration of the free inhibitor species is given by
| 4c |
where P0 = –((Kd + L–P) + [(Kd + L–P)2 + 4PKd]1/2)/2, PL0 = P–P0, PL50 = PL0/2, L0 = L–PL0 and L50 = L–PL50.
For this study, we derived a corresponding value of Ki for uncompetitive inhibition
| 5 |
in which the variables are the same as in Equation (4) except that L50 = – ((P–L) + [(P–L)2 + 4(PL0Kd/2)]1/2)/2. Although in this study we use the term Kd to quantify an antagonist's effect, the pharmacology-derived EC50 value is more appropriate when functional experiments are performed (11).
General assumptions and caveats
It is assumed that all of the substrate- and inhibitor-binding reactions are reversible and that they all have a one-to-one stoichiometry, i.e. no multiple binding of inhibitor molecules or any form of cooperativity, or other complex mechanisms of inhibition such as partial or mixed types (3). It is also assumed that in the enzymatic reactions enzyme autocleavage did not occur and that when substrates for fluorescence resonance energy transfer were used, appropriate corrections for inner filter effects were performed. Comparison of Km or IC50 values for a set of inhibitor candidates is only assumed to be valid when they are evaluated under identical experimental conditions. In most experimental studies of enzyme kinetics, the total concentrations of substrate and inhibitor used are in excess of the enzyme concentration to make their free and total concentrations essentially the same (1). Under the conditions of some ligand-receptor (e.g. protein)-binding studies, the free concentrations also become sufficiently important to require modifications of these equations (1, 2), and (9).
Description of the web server
The IC50-to-Ki tool is implemented as a web resource using an Oracle database (Oracle9i Enterprise Edition Release 9.2.0.4.0), Java (JDK 1.5.0) and Apache web server components including Tomcat 4.1. Information on candidate inhibitors of the botulinum neurotoxins was collected by mining the biomedical literature including searches with botXminer (12) using the National Library of Medicine's MEDLINE®/PubMed® (13). Experimental data (IC50 values) and accompanying assay information were manually extracted from primary literature results and other relevant databases: JCVI-Pathema-Clostridium (13), Brenda (14) and Protein Data Bank (15).
USAGE
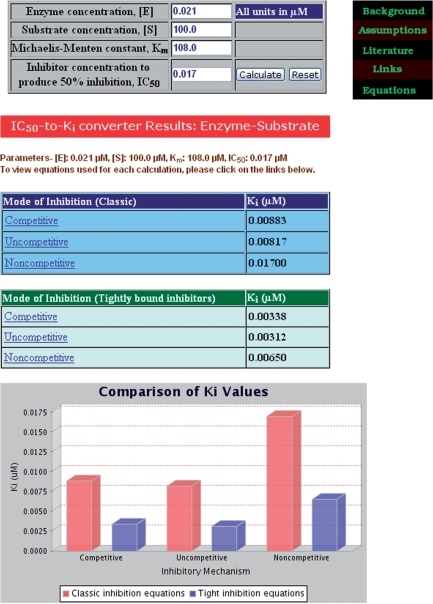
An internal link to the user-accessible converter is also located on the left side of the BotDB home page. The four required inputs for E, S, Km and IC50 are indicated with default settings for several examples. After submitting these values by using the ‘calculate’ button, these input data are returned along with the Ki results for the example cases.
An illustration is provided for a tight-binding inhibitor of an enzyme–substrate (E–S) reaction (Figure 1). The values for this example are from data using cimoxatone, a tight-binding inhibitor of monoamine oxidase (16). The four inputs for E, S, Km and IC50 are 0.021, 100, 108 and 0.017, respectively, in micromolar units. The Ki results for three modes of inhibition are returned on a new page. The top block of results corresponds to the solutions for a classic inhibitor (i.e. Michaelis–Menten kinetics). The second block represents the corrections made to the first set of equations [Equations (1b–3b)] for tightly bound inhibitors when there is substantial inhibitor depletion (5,6). Equations can be viewed by clicking on a label for a mode of inhibition. Below these two tables, histograms plotting the six results are shown for a visual comparison. In this example, the results from the classic and corrected equations are quite different. This difference in Ki values enables the user to conclude that not all of the assumptions underlying classic Michaelis–Menten equations are being obeyed and that the data are consistent with the kinetics of a tight-binding inhibitor.
Figure 1.
Results page from the IC50-to-Ki web tool for a tight-binding inhibitor of monoamine oxidase. The top table contains sample input data obtained from ref. 16. The middle table contains the results for a classic inhibitor that follows Michaelis–Menten kinetic Equations (1a–3a) for three kinetic reactions. The bottom table contains the results for nonclassic, tight-binding inhibitor uses Equations (1b–3b) for the same three reactions. The histograms summarize these results. Equations for each displayed mode of inhibition can be viewed by clicking on its label. A help list located on the upper right side is available for more detailed information about this tool.
Two other examples of enzyme inhibitors are also available for users to examine at the IC50-to-Ki tool website. For classic inhibition, data values using a candidate inhibitor of botulinum neurotoxin type A (17) are used as inputs: E, S, Km and IC50 (in micromolar units) 0.0067, 300, 1300 and 3.2, respectively. In contrast to the tight-binding inhibitor example, the returned values for Ki are similar for classic and tight-binding kinetics indicating that this data set is consistent with classical kinetics.
In another example of a potentially cooperative inhibitor of CYP3A4 (18), the input data for E, S, Km and IC50, in micromolar units, are 0.1, 50, 51 and 0.05, respectively. This example returns an error message from the converter that states that the 1:1 stoichiometry assumption may have been violated and requests the user to enter different values.
Finally, Equations (4) and (5) are used to calculate Ki values for reactions involving inhibitors of P–L-binding reactions. For this case, a user interface similar to the enzyme–substrate page is produced (see website). The values for a tight-binding inhibitor of an apoptosis-related protein from ref. 9 are used as an example calculation. The inputs are labeled P, L, Kd and IC50. In this case, only the competitive and uncompetitive modes of inhibition are considered. The tabulated output includes the free concentrations of protein and ligand species in the absence of an inhibitor (P0 and L0, respectively). The free concentrations at 50% inhibition are also returned for the protein, ligand, inhibitor, protein–ligand complex and P–L–I complex (P50, L50, etc.). As with the tight-binding enzyme inhibitor calculations, the summary histograms again indicate that these data are consistent with the kinetics of a tight-binding inhibitor.
It is our intent for this general tool to provide results for classic and tight-binding inhibitors of enzyme activity and ligand-binding reactions that are assumed to follow relatively simple kinetic schemes. These different sets of kinetic results will allow investigators to decide whether additional experiments are required to understand better the kinetic behaviors of their candidate inhibitors for further research or therapeutic product development.
FUNDING
The Defense Threat Reduction Agency Joint Science and Technology Office-Chemical Biological Defense (project 3.10043_07_RD_B to F.J.L.); and by the National Cancer Institute, National Institutes of Health (contract No. HHSN261200800001E). Funding for open access charge: Defense Threat Reduction Agency.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Drs. James J. Schmidt, John H. Cardellina and S. Ashraf Ahmed for their insightful comments on early drafts of this manuscript. Opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the US Army. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
REFERENCES
- 1.Munson PJ, Rodbard D. An exact correction to the “Cheng-Prusoff” correction. J. Recept. Res. 1988;8:533–546. doi: 10.3109/10799898809049010. [DOI] [PubMed] [Google Scholar]
- 2.Cheng Y, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 3.Brandt RB, Laux JE, Yates SW. Calculation of inhibitor Ki and inhibitor type from the concentration of inhibitor for 50% inhibition for Michaelis–Menten enzymes. Biochem. Med. Metab. Biol. 1987;37:344–349. doi: 10.1016/0885-4505(87)90046-6. [DOI] [PubMed] [Google Scholar]
- 4.Lazareno S, Birdsall NJ. Estimation of competitive antagonist affinity from functional inhibition curves using the Gaddum, Schild and Cheng-Prusoff equations. Br. J. Pharmacol. 1993;109:1110–1119. doi: 10.1111/j.1476-5381.1993.tb13737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson PJ. A linear equation that describes the steady-state kinetics of enzymes and subcellular particles interacting with tightly bound inhibitors. Biochem. J. 1972;127:321–333. doi: 10.1042/bj1270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copeland RA, Lombardo D, Giannaras J, Decicco CP. Estimating KI values for tight binding inhibitors from dose-response plots. Bioorg. Med. Chem. Lett. 1995;5:1947–1952. [Google Scholar]
- 7.Cheng HC. The power issue: determination of KB or Ki from IC50. A closer look at the Cheng-Prusoff equation, the Schild plot and related power equations. J. Pharmacol. Toxicol. Methods. 2002;46:61–71. doi: 10.1016/s1056-8719(02)00166-1. [DOI] [PubMed] [Google Scholar]
- 8.Huang X. Fluorescence polarization competition assay: the range of resolvable inhibitor potency is limited by the affinity of the fluorescent ligand. J. Biomol. Screen. 2003;8:34–38. doi: 10.1177/1087057102239666. [DOI] [PubMed] [Google Scholar]
- 9.Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal. Biochem. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 10.Lebeda FJ, Adler M, Erickson K, Chushak Y. Onset dynamics of type A botulinum neurotoxin-induced paralysis. J. Pharmacokinet. Pharmacodyn. 2008;35:251–267. doi: 10.1007/s10928-008-9087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig DA. The Cheng–Prusoff relationship: something lost in the translation. Trends Pharmacol. Sci. 1993;14:89–91. doi: 10.1016/0165-6147(93)90070-z. [DOI] [PubMed] [Google Scholar]
- 12.Mudunuri U, Stephens R, Bruining D, Liu D, Lebeda FJ. botXminer: mining biomedical literature with a new web-based application. Nucleic Acids Res. 2006;34:W748–W752. doi: 10.1093/nar/gkl194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greene JM, Collins F, Lefkowitz EJ, Roos D, Scheuermann RH, Sobral B, Stevens R, White O, Di Francesco V. National Institute of Allergy and Infectious Diseases bioinformatics resource centers: new assets for pathogen informatics. Infect. Immun. 2007;75:3212–3219. doi: 10.1128/IAI.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barthelmes J, Ebeling C, Chang A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA: the enzyme information system in 2007. Nucleic Acids Res. 2007;35:D511–D514. doi: 10.1093/nar/gkl972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berman HM, Westbrook JD, Gabanyi MJ, Tao W, Shah R, Kouranov A, Schwede T, Arnold K, Kiefer F, Bordoli L, et al. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res. 2009;37:D365–D368. doi: 10.1093/nar/gkn790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fowler CJ, Strolin Benedetti M. Cimoxatone is a reversible tight-binding inhibitor of the A form of rat brain monoamine oxidase. J. Neurochem. 1983;40:510–513. doi: 10.1111/j.1471-4159.1983.tb11312.x. [DOI] [PubMed] [Google Scholar]
- 17.Burnett JC, Opsenica D, Sriraghavan K, Panchal RG, Ruthel G, Hermone AR, Nguyen TL, Kenny TA, Lane DJ, McGrath CF, et al. A refined pharmacophore identifies potent 4-amino-7-chloroquinoline-based inhibitors of the botulinum neurotoxin serotype A metalloprotease. J. Med. Chem. 2007;50:2127–2136. doi: 10.1021/jm061446e. [DOI] [PubMed] [Google Scholar]
- 18.Maréchal JD, Yu J, Brown S, Kapelioukh I, Rankin EM, Wolf CR, Roberts GC, Paine MJ, Sutcliffe MJ. In silico and in vitro screening for inhibition of cytochrome P450 CYP3A4 by comedications commonly used by patients with cancer. Drug Metab. Dispos. 2006;34:534–538. doi: 10.1124/dmd.105.007625. [DOI] [PubMed] [Google Scholar]