Abstract
Signal transducer and activator of transcription (Stat) proteins are latent transcription factors that reside in the cytoplasm before activation. On cytokine-induced tyrosine phosphorylation, these molecules dimerize and accumulate transiently in the nucleus. No specific signals mediating these processes have been identified to date. In this report, we examine the nuclear export of Stat1. We find that treatment of cells with the export inhibitor leptomycin B does not affect steady-state localization of Stat1 but impedes nuclear export after IFNγ-induced nuclear accumulation. We identify a conserved leucine-rich helical segment in the coiled-coil domain of Stat1, which is responsible for the efficient nuclear export of this protein. Mutation of two hallmark leucines within this segment greatly attenuate the back transport of Stat1 in the cytoplasm. When fused to a carrier protein, the Stat1 export sequence can mediate nuclear export after intranuclear microinjection. We show that prolonging the nuclear presence of Stat1 by inhibiting nuclear export reduces the transcriptional response to stimulation with IFNγ. These data suggest that Stats are actively exported from the nucleus via several separate pathways and link this activity to transcriptional activation.
Signal transducers and activators of transcription (Stats) form a family of eukaryotic transcription factors that is conserved from Dictyostelium to humans (1). They transduce signals that originate from cell-surface receptors. Binding of cytokines or growth factors to their cognate receptors initiates a series of tyrosine phosphorylation events carried out by members of the Janus family of kinases (Jaks), which leads to the phosphorylation of Stats on a single tyrosine. This process, commonly termed Stat activation, triggers the dimerization of Stat proteins through reciprocal phosphotyrosine/SH2 interactions and subsequently the fast and efficient translocation of the cytoplasmic molecules into the nuclear compartment, where they activate specific genes (reviewed in refs. 2–4). Typically, cytokine-induced transcription is a transient process, lasting only minutes to hours (1). Therefore the removal of activated Stats from the nucleus is required. Cells with aberrantly high and prolonged Jak or Stat activation are subject to transformation and are associated with abnormal development (5). Two kinds of intranuclear events have been implicated in the inactivation of Stat signaling: targeting of nuclear Stats by the proteasome with their subsequent degradation (6), as well as repeated cycles of tyrosine phosphorylation and dephosphorylation with concurrent nuclear entry and exit (7–11). Although cycling asks for an active nuclear exit of Stat molecules, no pathway or signals associated with this process are known. By analyzing the time course of nucleocytoplasmic shuttling of green fluorescent protein-tagged Stat1 (Stat1-GFP), we found a markedly decreased rate of nuclear export of Stat1 in the presence of leptomycin B (LMB). This drug is known to suppress nuclear export of a variety of proteins by inhibiting the binding of the export receptor CRM1 to leucine-rich export signals (12, 13). Inspection of Stat1 for putative leucine-rich export signals led us to investigate the heptad repeats in the N-terminal region (14). Here we report the identification of a functional LMB-sensitive nuclear export signal (NES) in helix 4 of the coiled-coil domain of Stat1. This NES plays a major role in the efficient and fast removal of Stat1 from the nucleus after Stat activation with IFNγ.
Materials and Methods
Cell Culture.
Human HeLa S3, U3A, and 293T cells were grown at 37°C in a humidified 7% CO2 atmosphere in DMEM containing 10% FCS (Biochrom, Berlin). The complete medium (growth medium) also included streptomycin, penicillin, and amphotericin (all from Biochrom). 293T cells were grown on poly-l-lysine-coated glass coverslips and were transiently transfected with Lipofectamine plus (GIBCO) in 12-well plates (1 μg DNA/well) according to the manufacturer's instructions. Twenty-four hours posttransfection, cells were stimulated for 30 min with growth medium supplemented with both human IFNγ (5 ng/ml; GIBCO) and cycloheximide (CHX; 10 μg/ml), as indicated in the figures. After being washed with growth medium, the cells were further incubated in the continued presence of CHX for the indicated times. LMB (a kind gift of M. Yoshida, University of Tokyo) was used at a concentration of 10 ng/ml and added 1 h before stimulation with IFNγ. The proteasome inhibitor MG132 (Calbiochem) was added to the cells at a concentration of 50 μM starting 30 min before IFNγ stimulation.
Plasmid Construction.
Various portions of a human Stat1 cDNA (a kind gift of J. E. Darnell, Jr., The Rockefeller University, New York) were amplified by PCR by using Vent polymerase (NEB, Beverly, MA) with specific sets of primers (MWG-Biotech AG, Ebersberg, Germany) to generate artificial BamHI (5′) and EcoRI (3′) restriction sites at the ends. The following segments of Stat1 were prepared: N-domain [amino acids (aa) 1–129) and helix 1 (aa 127–188), −2 (aa 183–254), −3 (aa 254–292), and −4 (aa 289–314) of the coiled-coil domain by using the following primer sets: N-domain: 5′-atataaggatccccatgtctcagtggtacgaacttcagc-3′ and 5′-atataagaattctattccccgactgagcctgatt-3′; helix 1: 5′-atataaggatcccctcggggaatattcagagcacagtg-3′ and 5′-atataagaattcttgccacaccattggtctcgtg-3′; helix 2: 5′-atataaggatccccgagaccaatggt-gtggcaaag-3′ and 5′-atataagaattctagcattgggcggccccccaatac-3′; helix 3: 5′-atataaggatcccc-gcttgcttggatcagctgcaga-3′ and 5′-atataagaattctgtcatgttcgtaggtgtatttc-3′; helix 4: 5′-atataaggatccccacctacgaacatgaccctatcac-3′ and 5′-atataagaattctctgaatgagctgctggaaaagac-3′ (restriction sites underlined). After cleavage of the PCR products with restriction enzymes, the fragments were ligated into the BglII/EcoRI sites of pEGFPC2 (CLONTECH) for transfections of 293T cells or into the BamHI/EcoRI cloning sites of a bacterial expression vector (pGST-GFP; see below), which are situated between the cDNAs for glutathione S-transferase (GST) and GFP for microinjection of fluorescent fusion proteins with Stat1 fragments (GST-Stat-GFP) into HeLa S3 cells. Additionally, DNA fragments coding for the N-terminal (h4N; aa 289–301) and C-terminal (h4C; aa 302–314) halves of helix 4 were generated by annealing complementary oligonucleotides harboring BamHI and EcoRI overhangs and by cloning them into pGST-GFP. The oligonucleotides had the following sequence: 4N: 5′-gatccccacctacgaacatgaccctatcacaaaaaacaaacaagtgttagg-3′ and 5′-aattcctaacacttgtttgttttttgtgatagggtcatgttcgtaggtggg-3′; 4C: 5′-gatcccctgggaccgcaccttcagtcttttccagcagctcattcaggg-3′ and 5′-aattccctgaatgagctgctggaaaagactgaaggtgcggtcccaggg-3′. Mutations in 4C (codons for residues 308, 309, 312, and 313 of the full-length Stat1 sequence are underlined) were introduced by exchanging the indicated codons with the alanine codon [GCA]. pGST-GFP was constructed after an idea described by Eguchi et al. (15) with the following modifications: a portion of pEGFPN1 (CLONTECH) was PCR amplified (Vent polymerase) by using the primer pair 5′-atatatagaattcagatggtgagcaagggcgaggagctg-3′ and 5′-gattatgatctagagtcgcggccgc-3′. The resulting fragment representing the cDNA of GFP was cut with EcoRI (restriction site underlined) and NotI and ligated into these sites of pGEX5X-2 (Pharmacia) to yield pGST-GFP. A Stat1α-GFP fusion gene (pStat1-GFP) for eukaryotic expression was generated by PCR amplifying a Stat1α cDNA (aa 1–746) with the primer pair 5′-atatatgaattcatgtctcagtggtacgaacttcag-3′ and 5′-atatatggatccatcatactgtcgaattctac-3′. After cleavage with EcoRI and SmaI (restriction sites underlined) of the resulting PCR product, the DNA fragment comprising the N-terminal part of Stat1 was introduced into the corresponding sites of pEGFPNI. The vector that resulted from this cloning step was ligated (into its BamHI/SmaI sites) with a DNA fragment representing the residual C-terminal part of Stat1α, which was released from the above PCR product by digestion with BamHI and SmaI. This generated pStat1-GFP. Plasmids for the site-directed mutants are: (i) Stat1L308A-GFP, and (ii) Stat1L308,312A-GFP, in which (i) Leu308 or (ii) both Leu308 and Leu312 were replaced by alanines and were constructed in consecutive steps by using the Quick-Change mutagenesis kit (Stratagene) and the mutagenic sense primers (i) 5′-ggaccgca-ccttcagtGCAttccagcagctcattc-3′ and (ii) 5′-cagtGCAttccagcagGCCattcagagctcgtttg-3′ (mutated codons capitalized) together with the respective reverse complementary antisense primers. Stat expression constructs for luciferase assays were generated by cloning the Stat1 full-length coding region in the BamHI and NotI sites of the vector pcDNA3 (Invitrogen). The export mutant Leu308,312Ala was constructed by exchanging the HindIII fragment of pcDNA3-Stat1 with the HindIII fragment from the pEGFPNI construct containing those mutations.
Preparation of GST-Stat-GFP Fusion Proteins.
GST-Stat-GFP fusion proteins were expressed and purified from Escherichia coli strain BL21 by affinity chromatography on glutathione-Sepharose 4B according to the manufacturer's recommendations (Pharmacia). Eluted proteins were concentrated to ≈1 mg/ml (Bio-Rad protein assay) by ultrafiltration with a centriprep 50 device (Amicon) and dialyzed extensively against injection buffer (20 mM Hepes/110 mM K-acetate/2 mM Mg-acetate/0.5 mM EDTA/0.5 mM DTT, pH 7.5). The proteins were snap frozen on dry ice and stored at −80°C until microinjection.
Microinjections.
Microinjections of GST-Stat-GFP fusion proteins into HeLa S3 cells were performed by using the Transjector 5246 (Eppendorf). Twenty-four hours before injection, the cells were passaged onto poly-l-lysine-coated Cellocate coverslips (Eppendorf). GST-Stat-GFP fusion proteins were delivered into the cytosol or the nucleus of about 60 cells/fusion protein in a 20-min period. As an internal injection control, the fusion proteins were coinjected with a TRITC-coupled goat IgG (0.2 mg/ml final; Molecular Probes). Three hours after injection, the cells were fixed in 4% paraformaldehyde/PBS for 10 min at room temperature, and the nuclei were subsequently stained with Hoechst 33258 (5 μg/ml in H2O for 3 min; Sigma). After being washed with H2O, the cells were mounted in Dako fluorescence mounting medium (Dako) for fluorescence microscopy.
Luciferase Assays.
U3A cells were transiently transfected on 24-well plates by the Lipofectamine method. The following amounts of DNA were used per well: 250 ng of Stat1 expression plasmids, 50 ng of β-galactosidase reporter plasmid (Stratagene), and 200 ng of IFNγ reporter gene. The IFNγ reporter gene contained a triple Ly6E Stat-binding site in front of a luciferase reporter (16). Thirty-six hours after transfection, cells were left untreated or treated with IFNγ (5 ng/ml) and LMB (10 ng/ml) for 6 h. Whole-cell extracts were prepared and measured for luciferase (Promega) and β-galactosidase activity (Stratagene). The data are normalized for the expression of β-galactosidase and represent mean and standard deviation of four independent transfections in a single experiment done three times with identical results for each construct.
Electrophoretic Mobility-Shift Assay.
U3A or 293T cells were transiently transfected with pcDNA-Stat or pEGFPN1-Stat1 or the respective Leu308,312Ala mutant by the Lipofectamine method (GIBCO) and were left to recover for 24 h before pools of transfected cells were transferred onto several 10-cm plates (corresponding to the number of time points investigated) and stimulated 15 h later with IFNγ (5 ng/ml) for 30 min. Nuclear extracts were prepared as described (7) at the time points indicated. Four microliters of the nuclear extract collected from a 10-cm Petri dish (200 μl total) were incubated with 1 ng of [32P]-labeled M67 probe (7) for 5 min at room temperature. The assay was carried out on 4% 29:1 acrylamide/bisacrylamide gels at 4°C, as described previously (17). Binding activity was visualized with a phosphoimaging system (Storm 820; Molecular Dynamics).
Western Blotting.
The remainder of the nuclear extracts used for electrophoretic mobility-shift assay were analyzed further by Western blotting. Twenty microliters of nuclear extract was mixed with SDS sample buffer and resolved on 7% SDS polyacrylamide gels. Proteins were transferred to nitrocellulose (semidry blot; Bio-Rad), blocked with 4% BSA (Boehringer Mannheim) in Tris-buffered saline plus 0.1% Tween-20. The blot was incubated subsequently for 1 h with a polyclonal phosphospecific Stat1-Tyr701 antibody (New England Biolabs) and reprobed with a Stat1-specific polyclonal antibody (Santa Cruz Biotechnology). Before reprobing, the blots were stripped off bound antibodies (40 min at 50°C in 62.5 mM Tris⋅HCl, pH 6.8/2% SDS/0.7% β-mercaptoethanol). Blots were developed with horseradish peroxidase-conjugated secondary antibodies (Dako) and enhanced chemiluminescence (Amersham).
Detection and Recording of Fluorescence.
Fluorescence analysis was performed by using a Leica DMLB (Deerfield, IL) microscope equipped with A, N2.1, and I3 fluorescence filters. Images were recorded with a cooled Sensicam CCD camera and were processed by using the axiovision (Zeiss) and adobe photoshop (Adobe Systems, Mountain View, CA) packages.
Results
The Nuclear Export of Stat1 Is Sensitive to LMB.
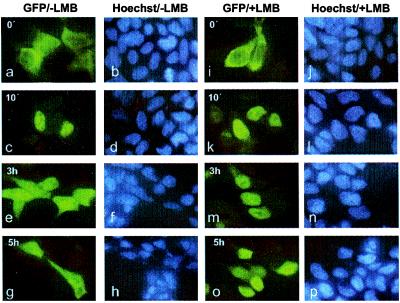
We made use of a C-terminal fusion of Stat1α with GFP (Stat1-GFP) to visualize directly the cytokine-dependent intracellular redistribution of Stat1. The validity of this approach for Stat1 has been demonstrated before (18). We initially tested the influence of the proteasome inhibitor MG132 on nuclear depletion of Stat1. MG132 was added at a concentration of 50 μM, but nuclear Stat1 disappeared at the normal rate (data not shown). To demonstrate the potential contribution of de novo Stat1 synthesis for its reemergence in the cytoplasm hours after interferon treatment, we included the translation inhibitor cycloheximide (CHX) in the medium during and after stimulation with IFNγ. Again, we could detect no changes in Stat1 dynamic redistribution in the presence of CHX. To test whether the loss of nuclear Stat1 and its reappearance in the cytoplasm after IFNγ treatment were because of active transport mediated by a NES, we followed the relocation of nuclear Stat1 in the presence of LMB. LMB has been shown to bind specifically to the export receptor CRM1 (also named exportin 1), thus preventing the formation of stable NES-CRM1 complexes (12, 13, 19, 20). This makes LMB a general inhibitor of a multitude of NES-mediated nuclear export pathways (21) in 293T cells. Fig. 1 a–h shows the typical time course of Stat1 redistribution after IFNγ stimulation. Ten minutes after a half-hour stimulation with IFNγ, almost all of Stat1 was nuclear. After 3 more hours, we detected approximately equal fluorescence in cytoplasmic and nuclear compartments, and only after 5 hours did the nucleus appear to contain fewer Stat1 than the surrounding cytoplasm. Addition of LMB for 1 hour before IFNγ treatment influenced neither Stat1 cytoplasmic localization in the resting state nor its nuclear accumulation on activation (Fig. 1 i–l). The subsequent nuclear export, however, was suppressed markedly (Fig. 1 m–p). Even 3 hours after IFN stimulation, the majority of Stat1 was still residing in the nucleus, and it took 5 hours until significant amounts of Stat1 appeared in the cytosol. Thus to a considerable extent, nuclear export of Stat1 is sensitive to the action of LMB. Taken together, these findings suggest strongly that a NES-mediated transport pathway contributes to the removal of Stat1 from the nucleus. However, even though LMB attenuates the nuclear export of Stat1, the drug does not entirely suppress the back transport of this protein. Moreover, given the size of Stat1 and the speed of this residual export activity, a further active transport mechanism rather than passive diffusion must be responsible.
Figure 1.
LMB inhibits nuclear export of Stat1. 293T cells transiently expressing Stat1α-GFP fusion proteins are shown before (a, b, i, j) and after the addition of IFNγ. Cells were stimulated with IFNγ for 30 min and fixed at the indicated time points (10 min, 3 h, and 5 h) after withdrawal of the cytokine. The subcellular localization of Stat1 in the absence (a–h) and presence (i–p) of 10 ng/ml LMB was determined by fluorescence microscopy. LMB does not influence nuclear accumulation of the fusion protein (c, k) but reduces the rate of cytoplasmic translocation (compare e and g to m and o).
Amino Acids 302–314 in Helix 4 of the Coiled-Coil Domain Function as an Export Signal.
The N-terminal third of Stat1 contains a number of helices that harbor amino acid sequences, which on visual inspection resemble putative export signals because of their high content of leucines and other hydrophobic residues. To identify an NES conveying LMB sensitivity on Stat1 nuclear export, we synthesized various portions of Stat1-cDNA by PCR and ligated them into the vector pGST-GFP. Bacterial expression generated fluorescent fusion proteins of Stat1 peptides, as described above. The affinity-purified fusion proteins were injected into the cytoplasm and nuclei of HeLa cells, and their subcellular localization was followed over several hours (Fig. 2). It was found that the transport behavior of all fusion proteins examined was not influenced by the addition of IFNγ. None of the chimeric constructs (N-domain; helices 1–4 of the coiled-coil domain) displayed a tendency to translocate from the cytoplasm into the nucleus (not shown). In contrast, when we injected the recombinant proteins into the nucleus, helix 4 of the coiled-coil domain of Stat1 exhibited translocation activity and showed strong cytoplasmic staining within a 3-h incubation period (Fig. 2 d–f). All other constructs stayed in the nucleus (shown for helix 1; Fig. 2 a–c).
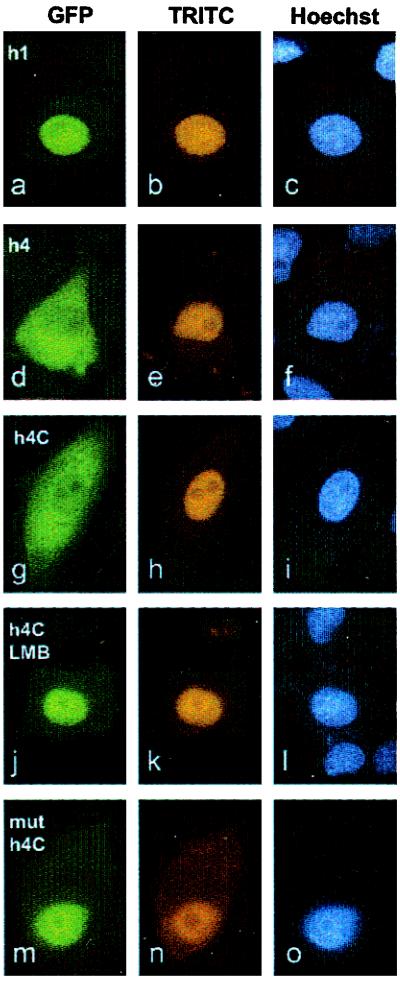
Figure 2.
Stat1 contains a NES at position 302–314. Affinity purified fusions of GFP and GST linked to Stat1 helix 1 (a–c), helix 4 (d–f), the C terminus of helix 4 comprising amino acids 302–314 (g–l), and a Leu308Ala mutant of the NES (m–o) were comicroinjected with TRITC-labeled goat immunoglobulins into the nuclei of HeLa S3 cells. Cells treated with LMB starting 1 h before microinjection are shown in j–l. After 3 h incubation at 37°C, the cells were fixed and the nuclei stained with Hoechst 33258 dye. The intracellular distribution of the GFP fusion proteins harboring Stat1 fragments, the TRITC-conjugated injection control, and positions of nuclei are indicated in the fluorescence micrographs for each Stat1 derivative. Only fusion proteins containing the intact NES from helix 4 of the coiled-coil domain of Stat1 are transported into the cytoplasm. The presence of LMB (j) and the mutation of leucine308 (m) inhibit nucleocytoplasmic translocation.
To narrow down the region required for the NES activity of helix 4, we constructed GST-GFP fusions with the N- or C-terminal half of helix 4, respectively (GST-h4N/h4C-GFP). Only the C-terminal half comprising amino acids 302 to 314 (GST-h4C-GFP) retained nuclear export ability (Fig. 2 g–i). Further truncation by five residues at the N terminus of helix 4C abolished the activity of the peptide (not shown). The nuclear export activity of helix 4C was blocked completely by LMB. When HeLa cells were incubated in the presence of LMB, the microinjected fusion protein with helix 4C stayed in the nucleus for hours (Fig. 2 j–l). Mutational analysis of the amino acid sequence of helix 4C revealed the importance of hydrophobic residues for export activity. We initially introduced substitutions to alanine in corresponding positions Leu308, Phe309, Leu312, and Ile313 of Stat1 in helix 4C. The mutated helix 4C no longer possessed export activity and the microinjected GST-GFP fusion protein remained nuclear (not shown). Moreover, a single amino acid exchange of Leu308 with alanine in helix 4C had the same effect, and the mutated GST-h4C-GFP protein no longer translocated into the cytoplasm but remained at the injection site in the nucleus (Fig. 2 m–o).
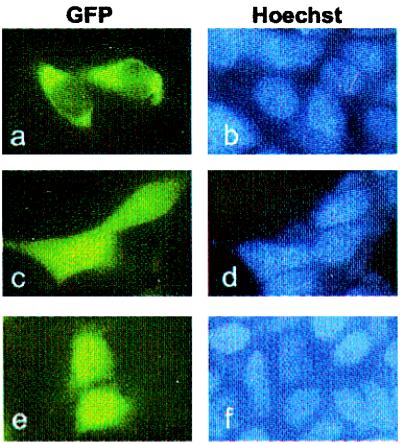
To determine the subcellular localization of a fusion protein of the Stat1-NES with a heterologous protein under steady-state conditions, we constructed a fusion gene encoding helix 4 (amino acids 292–314 of Stat1) and GFP. Because the molecular mass of the expressed fusion protein (<30 kDa) is small enough to allow passage into the nucleus by diffusion (22), the GFP fusion protein can be used as a reporter for active nuclear export. As expected, equal distribution between cytoplasm and nucleus was observed for GFP alone (Fig. 3e) or a fusion with Stat1 amino acids 137–183 (helix 1; Fig. 3 c and d). In contrast, the fusion protein of helix 4 with GFP was absent from the nucleus and clearly localized to the cytoplasm (Fig. 3 a and b). The above results indicate that a linear sequence of residues (amino acids 302–314) from helix 4 of the coiled-coil domain of Stat1 acts as a transferable nuclear export signal.
Figure 3.
The NES in helix 4 of the coiled-coil domain of Stat1 leads to nuclear exclusion of a fusion protein. The fluorescence micrographs of transiently transfected 293T cells expressing GFP linked to helix 4 (a, b), helix 1 (c, d), or GFP alone (e, f) reveal the different subcellular distribution of chimeric constructs of low molecular weight. Only the fusion protein linked to helix 4 is excluded from the nucleus (a). The localization of GFP and the GFP fusions are depicted in a, c, and e, and the corresponding positions of nuclei stained with Hoechst 33258 are documented in b, d, and f.
The Stat1 NES Is Conserved in the Stat Family but Deviates from Other NES Sequences.
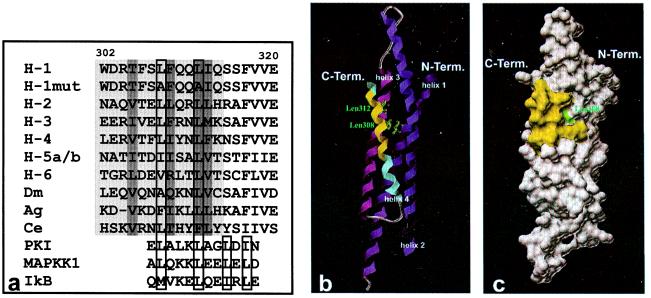
A comparison of the homologous amino acids in position 302–314 of Stat1 across the Stat family of transcription factors reveals a moderate level of sequence identity. The positions of large and hydrophobic residues are generally conserved (Fig. 4a). In positions 308 and 312, there are invariantly hydrophobic residues, predominantly leucines. Mutation to alanine in these positions in the fusion protein of Stat1 helix 4C with GST-GFP leads to a loss of export ability (Fig. 2). The spacing of these two leucines is characteristic for NES sequences found in other proteins like MAPKK1, PKIα, or IκBα (ref. 23; Fig. 4a). A distinguishing feature of known leucine-rich NESs is a conserved consensus motif of four essential hydrophobic amino acids, with typically two of them forming a core tetramer at the C-terminal end of the NES (21). This core tetramer is not required for the Stat1 export signal to function. Although there are hydrophobic residues downstream from the Stat1 NES, neither their side chains nor spacing relative to the upstream consensus motif matches the specifications delineated for leucine-rich NES motifs. As shown in Fig. 4 b and c in a model based on dimeric Stat1, only the leucine in position 308 of the critical leucines in position 308 and 312 is not part of the heptad repeats of hydrophobic residues that engage in mutual interactions to form a four-helix coiled-coil domain. While the side chain of residue 308 is exposed partially at the exterior of helix 4, the side chain of leucine 312 is buried entirely and is not accessible from the outside.
Figure 4.
(a) Aligned sequences of wild-type and NES mutant Stat1 (amino acids 302–320), of homologous regions of Stat family members, and of known leucine-rich export signals of other proteins. The NES sequence of Stat1 between amino acids 302 and 314 and the putative NESs of other Stat proteins are highlighted in light gray. Residues in the interior of the coiled-coil domain are marked with dark gray. Highly conserved hydrophobic residues in position 308 and 312 of the Stat1 NES and important hydrophobic residues in known NES sequences including hallmark residues indicative of the core tetramer are boxed. The following sequences from the National Center for Biotechnology Information Data Library are depicted: H-1, human Stat1, accession no. P42224; H-1mut, human Stat1 mutated in positions 308 and 312; H-2, human Stat2, no. P52630; H-3, human Stat3, no. NP_003141; H-4, human Stat4, no. Q14765; H-5a/b, human Stat5a/b, no. NP_003143/P51692; H-6, human Stat6, no. NP_003144; Dm, Drosophila melanogaster Stat, no. Q24151; Ag, Anopheles gambiae Stat, no. AJ010299; Ce, Caenorhabditis elegans Stat, no. AAD45535; PKI, human cAMP-dependent protein kinase inhibitor α, no. NP_006814; MAPKK1, human mitogen-activated protein kinase kinase 1, no. NP_002746; IkB, human IκBα, no. CAB65556. b and c depict the localization of the NES in the coiled-coil domain (amino acids 136–317) of Stat1. The ribbons drawing (b) shows the four-helix bundle, with amino acids 302–314 of the NES sequence in helix 4 (blue) marked in yellow. The side chains of the leucine residues in position 308 and 312 are included as atomic stick figures. A surface rendering of b is presented in c. The contributions to the surface of the coiled-coil domain by side chains of the NES sequence are highlighted in yellow, with the water accessible area of leucine 308 shown in green.
Mutations in the Export Signal Inhibit Nuclear Exit of Stat1 After Interferon Stimulation.
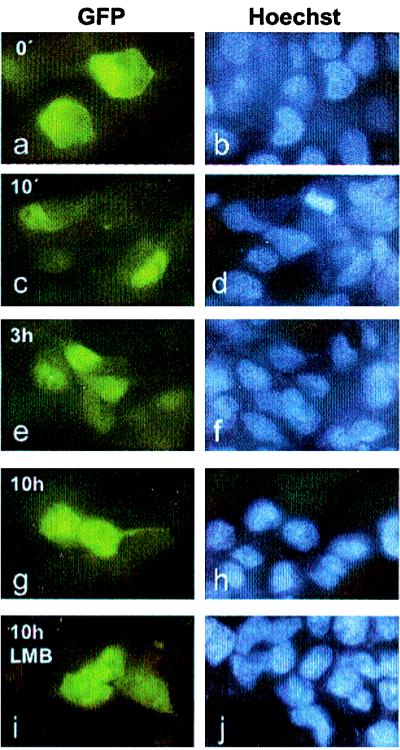
To examine the role of the export signal for nucleocytoplasmic relocation of full length Stat1, we expressed transiently mutated variants of GFP-tagged Stat1 in 293T cells. We introduced a single leucine-to-alanine mutation in position 308 as well as double leucine-to-alanine mutations in positions 308 and 312. Both proteins were expressed well and localized to the cytoplasm before the addition of IFNγ (Fig. 5 a and b). On treatment with IFNγ, the nuclear accumulation of the mutants progressed with unaltered kinetics (Fig. 5 c and d). The protein with a mutation only in position Leu308 left the nucleus with wild-type kinetics (not shown). However, the export of the double-mutant Leu308,312Ala markedly differed from wild-type Stat1 (Figs. 1 and 5). After 3 hours, most of the Stat1 NES-double mutant was still sequestered in the nucleus, with very little GFP fluorescence in the cytoplasm (Fig. 5 e and f). To better visualize the residual export activity of the mutant Stat1, we incubated the cells for 10 h after IFNγ treatment (Fig. 5 g–j). Only at this time point past interferon stimulation was a considerable fraction of Stat1 detectable outside the nucleus (Fig. 5 g and h). Furthermore, the addition of the export inhibitor LMB did not further decrease the export rate of this Stat1 mutant (Fig. 5 i and j). These results reveal that the NES in helix 4 of the coiled-coil domain of Stat1 contributes significantly to the fast removal of this transcription factor from the nucleus. Also, because export is slowed down but does not cease entirely, other export pathways must function in parallel. Because the export of the NES mutant is no longer responsive to LMB, a further export mechanism that does not depend on CRM1 as an export receptor is proposed.
Figure 5.
Mutation of two hallmark leucines in the NES impedes the nucleocytoplasmic transport of Stat1. 293T cells were transiently transfected with DNA constructs coding for a NES-minus Stat1 Leu308,312Ala double mutant linked to GFP. The cells were either left untreated (a, b) or were stimulated with IFNγ for 30 min (c–j) and fixed at the indicated time points after cytokine removal (10 min, 3 h, and 10 h). To investigate the influence of LMB on nuclear export of the Stat1 mutant, cells were incubated in the continuous presence of LMB starting 1 h before interferon treatment and were fixed 10 h after cytokine stimulation (i, j). The micrographs show the subcellular localization of the Stat1 fusion proteins (a, c, e, g, i) and the corresponding positions of nuclei stained with Hoechst 33258 (b, d, f, h, j).
The Stat1 Export Mutant Accumulates in the Unphosphorylated State.
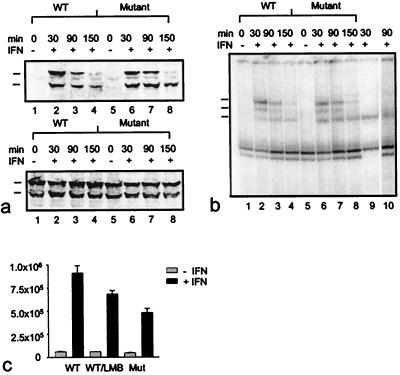
After we established the delayed nuclear exit of the export mutant, we were interested to see whether the mutant protein was activated and dephosphorylated at the same rates as the wild-type protein, to rule out the possibility that defects in the tyrosine phosphorylation cyclus were responsible for the slow nuclear exit of the mutant. We used gel retardation assays and Western blotting to investigate the time course of activation and inactivation in pools of transfected cells. As shown in Fig. 6, there were no differences in the activation/inactivation cycle of mutant and wild-type proteins. Both proteins were tyrosine phosphorylated rapidly and became dephosphorylated over a 1-h period after activation with IFNγ (Fig. 6a). As shown in Fig. 6b, this was also reflected in the DNA-binding activities of the two proteins, which reveal no appreciable differences. The mutant protein just as the wild-type Stat1 is induced rapidly to bind to DNA, followed by a fast decay of this activity in both proteins. Using supershift assays with GFP specific antibodies, we demonstrated the ability of the recombinant GFP-Stat1 proteins to form homodimers as well as heterodimers with the endogenous Stat1 protein. The DNA-binding activities of the recombinant proteins appeared to decay more rapidly than those of the endogenous Stat1. We attribute this phenomenon to the higher concentration of the endogenous Stat1 in the extracts, which leads to a saturation of the available Stat1-binding sites in the gel-shift assay. We next addressed the physiological function of cytoplasmic relocalization of Stat1 by determining whether the prolonged nuclear presence of the double mutant led to an altered transcriptional activity of this protein. It is currently unclear whether tyrosine dephosphorylation or nuclear export is required for the down-modulation of gene transcription after stimulation with interferon. If indeed nuclear exclusion of Stat1 was necessary for the termination of Stat1 signaling, blocking the export should result in a heightened level of gene transcription. For these experiments, we used constructs that were devoid of the GFP domain at the Stat1 C terminus, because in initial experiments we had observed only a low (maximum 4- to 5-fold) transcriptional activation with the GFP constructs, which made interpretations difficult. As shown in Fig. 6c, the Stat1 Leu308,312Ala mutant retains its transactivation potential, and treatment of U3A cells expressing the mutant Stat1 leads to a 10- to 11-fold increase in transactivation of the Stat1 reporter. However, in repeated sets of experiments it became apparent that the mutant protein is clearly less potent in its transactivation ability. The Stat1 Leu308,312Ala mutant consistently did not exceed 70% of the activation level that the wild-type protein achieved, which generally led to a 15- to 17-fold increase in transactivation of a reporter gene containing a triple Stat-binding site. This reduced rate of transactivation could not be attributed to defective phosphorylation of the essential residue Ser727 (16), which is phosphorylated constitutively in the export mutant as revealed by Western blot analysis with an anti-Stat1 phosphoserine antibody (not shown). To assess independently the importance of nucleocytoplasmic relocation for a full transcriptional response of Stat1, we analyzed additionally the effect of the export inhibitor LMB on IFNγ-stimulated transcriptional activity. A comparison between transcription rates of wild-type Stat1 in the absence and presence of LMB reveals clearly an inhibitory effect of the export inhibitor on IFNγ-activated transcription. The level of reduction is similar to the effect of the Leu308,312Ala mutations. We cannot rule out the possibility that the two point mutations in the four-helix bundle of Stat1 exert a direct effect on gene induction. But given the identical expression levels and phosphorylation rates of the two proteins, these data suggest that an extended nuclear stay of Stat1 results in an aborted transcriptional activity.
Figure 6.
Kinetics of tyrosine phosphorylation (a), DNA binding activity (b), and transcriptional activation of an IFNγ-responsive luciferase reporter gene (c) displayed by wild-type and export mutant Stat1. (a) 293T cells were transiently transfected with wild-type or Leu308,312Ala double mutant GFP-Stat1 DNA and pooled. Subsequently, equal numbers of cells were plated and 15 h later treated with or without IFNγ for 30 min. At this time point (30 min) as well as 1 hour (90 min) and 2 hours (150 min) later, the cells were lysed, and nuclear extracts were collected. Nuclear proteins were resolved on SDS/PAGE gels and blotted. Shown is a Western blot with a phosphospecific Stat1-Tyr701 antibody (Upper) and a reprobing with Stat1 antibody (Lower). The positions of the transfected proteins (upper mark) and the endogenous Stat1 (lower mark) are indicated. (b) Nuclear extracts prepared from the same cells described in a were subjected to DNA-binding analysis. Extracts were incubated with radiolabeled M67 probe and separated on 4% nondenaturing polyacrylamide gels. The positions of homodimers of endogenous Stat1 (bottom mark), of homodimers of transfected GFP-constructs (top mark), and of heterodimers of native and recombinant Stat1 proteins (middle mark) are indicated at the left edge of the gel. In lane 9, the DNA-binding pattern of interferon-stimulated (30 min) untransfected 293T cells is shown. Supershifting with a polyclonal GFP antibody (CLONTECH) of the same extract used in lane 7 specifies GFP-containing complexes (lane 10). (c) Effects of Leu308,312Ala mutations and LMB on transcription activation. Wild-type (WT) and mutant proteins (Mutant) were coexpressed with an IFNγ-inducible reporter plasmid in U3A cells. Luciferase activity was determined 36 h posttransfection in cells that had been stimulated with or without IFNγ for 6 h in the presence or absence of LMB. Error bars represent standard deviations for four independent experiments.
Discussion
Currently the details of the dynamic redistribution of Stat proteins triggered by cytokine activation are largely unknown. Although the striking nuclear accumulation of Stats and their subsequent clearing from the nuclear compartment were noted early on (24), no signals that regulate these processes have been identified so far. Using a Stat1-GFP fusion protein, we find no evidence for a significant contribution of proteasome-mediated degradation or de novo cytoplasmic synthesis to the nucleocytoplasmic redistribution of Stat1. Exposure of cells to the nuclear export inhibitor LMB, on the other hand, clearly attenuates the reappearance of Stat1 in the cytoplasm and prolongs nuclear accumulation. The results obtained by microinjection and transfection of Stat1-derived peptides fused to reporter proteins indicate the existence of a transferable nuclear export signal in the coiled-coil domain, amino acids 302–314, of Stat1.
The activity of the isolated Stat1 NES can be blocked completely by the export inhibitor LMB (Fig. 2). Moreover, the kinetics of export inhibition by LMB of the full-length wild-type protein resemble closely the time course of nuclear clearance of a loss-of-function NES mutant of Stat1. Additionally, the mutant no longer displays sensitivity to the effects of the export inhibitor LMB (compare Figs. 1 and 5). Interestingly, neither disruption of the Stat1 NES nor prolonged treatment with LMB for up to 15 h (Figs. 1 and 5 and data not shown) lead to nuclear accumulation of the protein in unstimulated cells. This might be taken as an indication that Stat1 is cytoplasmic in the resting state and does not shuttle permanently in and out of the nucleus, with the net export exceeding import. A clear picture will have to await the characterization of the residual export activity of the NES mutant. As to the nature of this activity, LMB insensitivity indicates the existence of additional export mechanisms that do not rely on the export receptor CRM1. The continued, albeit impeded, nuclear export of a NES-minus Stat1 molecule reveals a first glimpse of a rather intricate degree of regulatory means determining nuclear transport of Stat1. Obviously, the prompt removal from the nucleus of this transcription factor is assured by parallel and independent mechanisms to achieve maximum capacity.
An important aspect concerning export rate modulations is the role of cytokine regulation of nucleocytoplasmic travel of Stat1. Although nuclear entry is triggered by Stat activation on cytokine treatment of the cell, we do not know whether the export is at the same time also subject to cytokine control. It is conceivable that the rate of export is influenced by the accessibility of the nuclear export signal. As shown in Fig. 4c, only the leucine in position 308 of the hallmark leucine residues of the NES is accessible from the outside and therefore available for interactions with molecules of the export machinery. Experiments exploring the role of a nuclear tyrosine phosphatase in the signal inactivation of Stats as well as the time course of dephosphorylation of the mutant protein presented here suggested that monomeric Stat1 is the substrate for export (8). We cannot therefore be certain whether the position of the NES in the actual “export-conformation” is reflected accurately in the structure representation of Fig. 4, which is modeled on the available dimeric protein (14).
Our data show that the mutant Stat1 is promptly tyrosine dephosphorylated (Fig. 6a). This result was anticipated, because the current model of Stat1 inactivation localizes the inactivating phosphatase in the cell nucleus (7). Importantly, the transcription data highlight a direct coupling between the speed of nuclear export and the magnitude of transcription activation. Inhibition of Stat1 export, either by means of mutation of Stat1 or by blocking chemically the export receptor, is accompanied by a lowered transcriptional output (Fig. 6c). This suggests that the time until tyrosine dephosphorylation limits the “lifespan” of the transcriptionally active Stat1, whereas the length of time that the protein stays in the nucleus determines the overall transcriptional yield. The reduced pool of cytoplasmic Stat1 that is available for rephosphorylation might therefore be the limiting factor for the transcriptional response.
A comparison of the Stat1 NES sequence with the homologous amino acids of the other family members suggests the presence of a NES motif in all of them (Fig. 4a). The key feature of the Stat1 NES, two leucines spaced by three residues, is present almost without exception. The surrounding sequence is far less conserved, as commonly found in other NESs, but additional hydrophobic and bulky residues are generally found. These amino acid sequence characteristics as well as LMB sensitivity place the Stat1 NES among the class of leucine-rich CRM1-dependent export signals. A comparison with other members of this class indicates that the Stat1 export signal is missing one typical motif of classic NESs, which is an arrangement sometimes termed the “core tetramer” at the C terminus of the NES (23). Usually, mutation of two leucines in the core tetramer disrupts NES function. This does not hold true for the Stat1 NES, which lacks the cognate core tetramer motif. That is not without precedent, because not all leucine-rich NES sequences described so far conform to the general consensus (19, 25, 26).
Taken together, the data presented in this paper indicate that Stat1 returns to the cytoplasm from the nucleus after cytokine stimulation via a CRM1-dependent pathway. The experiments leading to the localization of a conserved NES motif in the coiled-coil domain of STAT1 reveal the existence of active nuclear export for a member of the Stat family of transcription factors. Notably, a reduced rate of back transport of Stat1 into the cytoplasmic compartment resulted in an aborted transcriptional response to stimulation with IFNγ. In addition, the present findings suggest a multipathway mechanism for the export of Stat1 from the nucleus.
Acknowledgments
We thank Dr. M. Yoshida, University of Tokyo, for the kind gift of Leptomycin B and Marc Fuccillo for expert technical assistance. U.V. expresses his gratitude to his mentor, Dr. J. E. Darnell (The Rockefeller University, New York) for his very generous supply of cDNAs and reagents.
Abbreviations
- Stat
signal transducer and activator of transcription
- Jak
Janus family of kinases
- GFP
green fluorescent protein
- LMB
leptomycin B
- NES
nuclear export signal
- GST
glutathione S-transferase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190318397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190318397
References
- 1.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 3.Leonard W J, O'Shea J J. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 4.Hoey T, Schindler U. Curr Opin Genet Dev. 1998;8:582–587. doi: 10.1016/s0959-437x(98)80015-4. [DOI] [PubMed] [Google Scholar]
- 5.Frank D A. Mol Med. 1999;5:432–456. [PMC free article] [PubMed] [Google Scholar]
- 6.Kim T K, Maniatis T. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 7.Haspel R L, Salditt-Georgieff M, Darnell J E., Jr EMBO J. 1996;15:6262–6268. [PMC free article] [PubMed] [Google Scholar]
- 8.Haspel R L, Darnell J E., Jr Proc Natl Acad Sci USA. 1999;96:10188–10192. doi: 10.1073/pnas.96.18.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C K, Bluyssen H A, Levy D E. J Biol Chem. 1997;272:21872–21877. doi: 10.1074/jbc.272.35.21872. [DOI] [PubMed] [Google Scholar]
- 10.Callus B A, Mathey-Prevot B. Blood. 1998;91:3182–3192. [PubMed] [Google Scholar]
- 11.Yu C L, Burakoff S J. J Biol Chem. 1997;272:14017–14020. doi: 10.1074/jbc.272.22.14017. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 13.Wolff B, Sanglier J J, Wang Y. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Jr, Kuriyan J. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 15.Eguchi H, Ikuta T, Tachibana T, Yoneda Y, Kawajiri K. J Biol Chem. 1997;272:17640–17647. doi: 10.1074/jbc.272.28.17640. [DOI] [PubMed] [Google Scholar]
- 16.Wen Z, Zhong Z, Darnell J E., Jr Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 17.Fried M, Crothers D M. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Köster M, Hauser H. Eur J Biochem. 1999;260:137–144. doi: 10.1046/j.1432-1327.1999.00149.x. [DOI] [PubMed] [Google Scholar]
- 19.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 20.Ossareh-Nazari B, Bachelerie F, Dargemont C. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 21.Elfgang C, Rosorius O, Hofer L, Jaksche H, Hauber J, Bevec D. Proc Natl Acad Sci USA. 1999;96:6229–6234. doi: 10.1073/pnas.96.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paine P L, Moore L C, Horowitz S B. Nature (London) 1975;254:109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- 23.Johnson C, van Antwerp D, Hope T J. EMBO J. 1999;18:6682–6693. doi: 10.1093/emboj/18.23.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindler C, Shuai K, Prezioso V R, Darnell J E., Jr Science. 1992;257:809–813. [PubMed] [Google Scholar]
- 25.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]