Abstract
With the rapid progress of biological research, great demands are proposed for integrative knowledge-sharing systems to efficiently support collaboration of biological researchers from various fields. To fulfill such requirements, we have developed a data-centric knowledge-sharing platform WebLab for biologists to fetch, analyze, manipulate and share data under an intuitive web interface. Dedicated space is provided for users to store their input data and analysis results. Users can upload local data or fetch public data from remote databases, and then perform analysis using more than 260 integrated bioinformatic tools. These tools can be further organized as customized analysis workflows to accomplish complex tasks automatically. In addition to conventional biological data, WebLab also provides rich supports for scientific literatures, such as searching against full text of uploaded literatures and exporting citations into various well-known citation managers such as EndNote and BibTex. To facilitate team work among colleagues, WebLab provides a powerful and flexible sharing mechanism, which allows users to share input data, analysis results, scientific literatures and customized workflows to specified users or groups with sophisticated privilege settings. WebLab is publicly available at http://weblab.cbi.pku.edu.cn, with all source code released as Free Software.
INTRODUCTION
To explore mechanisms underlying complex biological processes, high-throughput analysis techniques and multidisciplinary approaches are becoming main aspects of current biological research. Rapid growth of biological research places great demands on an integrative bioinformatic workbench to help biological researchers to mine knowledge from complex heterogeneous data.
Several bioinformatic analysis systems with intuitive user interface have been implemented in recent years (1–13). While some of them are designed as wrappers for a few specified software packages, a number of systems provide further support to popular bioinformatic analysis tools. Several systems including Taverna (3–5), BioManager (6), Galaxy (7,8), PISE (9), MOWServ (11) and HNB (13) support workflow-based analysis to make complex analysis much easier for non-experts. Moreover, Taverna (3–5), BioManager (6), Galaxy (7,8), PISE (9) and WildFire (10) also allow users to create workflows, increasing the flexibility and customizability.
On the other hand, while the importance of team work for research success is being widely recognized (3,14–16), few existing systems provide enough support for collaborative team work. Some systems allow users to store their input data and analysis results online (1,2,6–8,11–13), and BioManager (6), Galaxy (7,8) also support users to share their stored data and workflows. Moreover, with the help of some ‘Web 2.0’ websites, researchers can upload and share their annotation information and workflow online (14,16–18). However, to our best knowledge, no bioinformatic analysis platform with comprehensive supports for data managing, analyzing and sharing in a web-based integrative environment is publically available to the research community yet.
Here, we have developed a data-centric knowledge-sharing platform WebLab to support biological researchers to efficiently manage, analyze and share their data in an easy-to-use integrative environment. As a data-centric platform, WebLab provides dedicated user space to store and manage input data, analysis results and scientific literatures online. Supports for searching against full text, extracting citation information from PubMed, and exporting citations to EndNote and BibTeX are provided for literature, which is missing in other existing systems. By assembling customized workflows from 260+ integrated bioinformatic tools, complex analysis tasks could be performed automatically. In order to facilitate team work, WebLab provides powerful and flexible sharing mechanism and group strategy. Users can share their data, literatures and customized workflows with specific users or user-groups with sophisticated privilege settings. WebLab is publicly accessible at http://weblab.cbi.pku.edu.cn, with all source code available for downloading freely.
DESIGN AND SYSTEM ARCHITECTURE
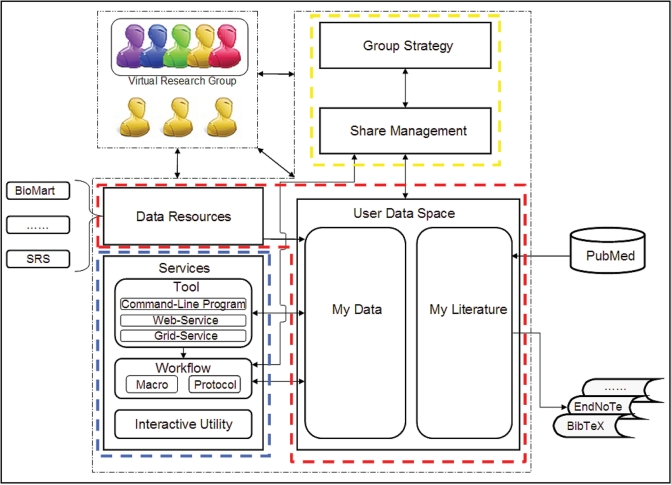
To be flexible for further extension and development, WebLab is designed with a modularization approach including three main modules: data management, analysis service and team work (Figure 1).
Figure 1.
Overview of WebLab architecture. WebLab is comprised of three main functional modules. (i) Data management module (in red frame) maintains the user data space (My Data and My Literature), and also provides supports for accessing remote databases through BioMart and SRS; (ii) Analysis service module (in blue frame) provides a uniform framework to integrate more than 260 popular bioinformatic analysis tools (including command-line program, Web-service and Grid-service), and supports workflow in both Protocol and Macro models; (iii) Team work module (in yellow frame) keeps track of user shared data, literatures and workflows. Virtual research group (VRG) is designed to help collaborators share their works easily.
Data management
As a data-centric platform, WebLab provides a powerful data management system for users to store and manage their data and scientific literatures online.
In their own data space (‘My Data’), users can create a new entry by uploading a file from local disk or retrieving from remote databases through BioMart (19) and SRS (20). After data type for the newly created file is specified, WebLab can recognize the format and automatically detect available analysis tools in a context-sensitive approach.
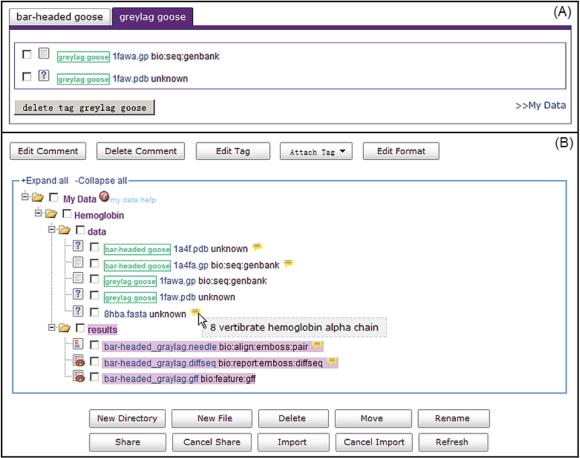
The entries in My Data are presented in hierarchical tree structure as in daily-used local file system. Users can create, rename and delete files and directories (folders) in My Data by simple mouse clicks. Moreover, users could also associate user-defined labels (Tags) or comments to entries in My Data, to classify and organize them in flexible and intuitive ways (Figure 2).
Figure 2.
My Data—user-dedicated space to store and manage input data and analysis results. The entries in My Data are presented in the hierarchical tree structure as in local file system. (A) WebLab provides an option to add user-defined labels (Tags) to each item in My Data as an annotation. Entries can later be classified and organized according to associated Tags in ‘tag view’. (B) For each entry, users can create comment, and the detailed content for a comment will be displayed when users put mouse over the yellow icon. The comment will also be shared out along with the data entries.
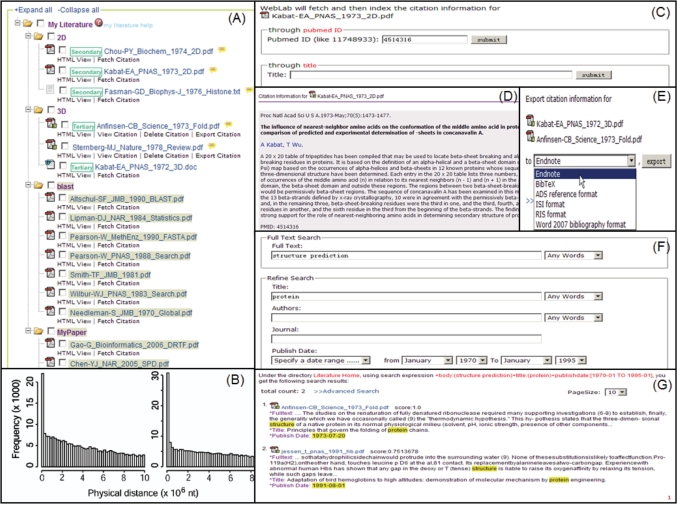
In addition to conventional operations supported in My Data, rich literature-specified functions are provided in ‘My Literature’. After uploading literature, WebLab automatically generates HTML preview for a quick check of the paper's content in browser without downloading the whole article. Then, WebLab extracts and indexes full text contents for uploaded articles. When the indexing is done, users can do simple keyword search or complex query search against full text of literatures existing in My Literature. Moreover, citation information could be fetched from NCBI according to PubMed ID or title and to be further associated to respective article in My Literature. All citation information could be easily exported into various well-known citation managers such as EndNote and BibTex (Figure 3).
Figure 3.
WebLab provides rich supports for scientific literatures. (A) Literatures in PDF, Microsoft Word and plain text formats could be stored and managed in My Literature. In addition to conventional operations in My Data, rich literature-specified functions are provided in My Literature. (B) WebLab provides ‘HTML View’ for a quick check of the contents in browser without downloading. (C) (D) Users can fetch citation information from NCBI PubMed repository through PubMed ID or title. (E) Citation information can be batch exported in well-known bibliography formats such as Endnote, BibTeX, ADS reference, ISI, RIS and Word 2007 bibliography format. (F) (G) After the index is built, users can perform search against all literatures in My Literature. The matched literatures will be sorted by their relevant scores and the searched key words are highlighted.
Analysis service
As an integrative bioinformatic analysis platform, WebLab integrates numerous analysis tools within a uniform framework. In addition to command-line programs, Web-services and Grid-services are also integrated in WebLab with full interoperation (Table S1).
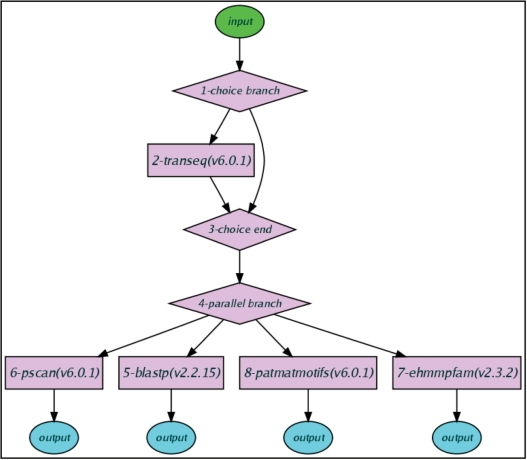
By organizing different tools into a workflow (21), complex analysis tasks are performed in one run. In a workflow, several analysis tools are launched according to previously user-defined rules. Currently, two workflow models with different user interaction abilities are available. In the Protocol model, a workflow is executed stepwise, and the user can tune parameters or options in each step, thus providing maximum flexibility. On the other hand, in the Macro model, after mandatory parameters are first inputted by the user, each tool in a workflow will be sequentially executed. Thus, Macro is more suitable for routine analysis. Moreover, an existing Macro could be re-used and treated as a standard analysis tool to define new workflow (recursive definition), which further simplifies users’ daily work and increases flexibility. Besides defining their own workflows, users can also use several pre-defined workflows for common analysis tasks such as phylogenetic tree construction and protein function analysis (Figure 4).
Figure 4.
A workflow for protein function analysis. The workflow is represented as Directed Acyclic Graph (DAG) in WebLab. The rectangular node of the DAG means a single tool or a defined Macro; the diamond node stands for operators which control the execution of workflows. WebLab supports two types of operators: condition operator (for example, nodes 1 and 3 in figure) determines following analysis depending on a given condition (for example, the workflow in figure will try to translate the input sequences by transeq for DNA sequences); parallel operator (for example, node 4 in figure) is used to share incoming data among several parallel analysis tools or combine several incoming data into one sink.
A few popular client-side utilities including Sequence Manipulation Suite (SMS) (22), WebMol (23), Dotlet (24) and JalView (25) are also integrated into WebLab for users to perform interactive work such as editing multiple sequence alignment or visualizing structure. While those utilities could not be incorporated into the workflow like other standard analysis tools due to their interactive nature, they are proved to be useful in daily work. Moreover, users also can keep their favorite analysis tools in their ‘My Toolbox’ for quick access.
Team work
Collaborations among several researchers in various fields and different locations are recently becoming more and more common, and also crucial for research success. To facilitate collaborative team work, WebLab provides flexible sharing mechanism and group strategy for users to share their data and knowledge.
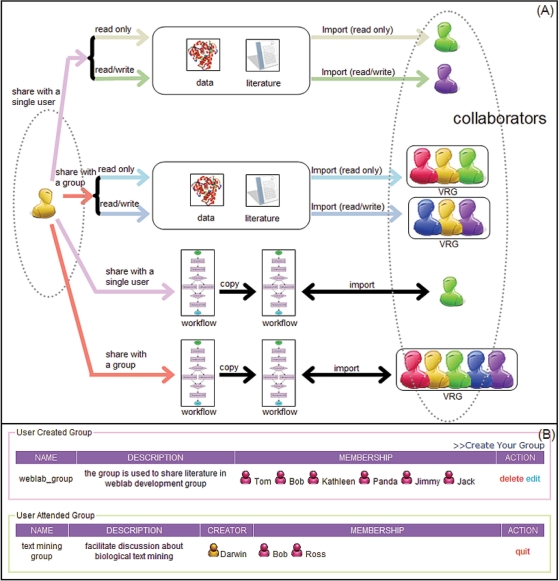
In WebLab, a user can share almost everything he owns with other users. For entries in My Data and My Literature, both ‘read only’ and ‘read and write’ sharing privileges are provided. By employing the reference-count based sharing model, changes in these shared contents will be seen by all collaborators simultaneously to assure efficient cooperation among all partners. On the other hand, once a user-defined workflow is shared out, a copy will be made which can be modified without altering the original one, to prevent possible flaw caused by recursive definition of workflow (Figure 5).
Figure 5.
Team work support in WebLab. (A) In WebLab, a user can share with his colleagues almost everything he owns. Analysis data and scientific literatures can be shared out with ‘read only’ or ‘read and write’ privileges. On the other hand, a different sharing strategy is applied for workflow to prevent possible flaw caused by recursive definition of workflow, i.e. once a user-defined workflow is shared out, the collaborators will get a copy of this workflow, which can be edited without altering the content of the original one. (B) Through group strategy provided by WebLab, users can create their own virtual research groups (VRG) in web environment. The group creator is privileged to edit or to delete the whole group. And a group member can also choose to quit from the group freely.
Groups are designed for colleagues who work closely together. Any user can set up a new virtual research group (VRG) and invite other users to join the new VRG. A member of a VRG can also share with other members in the VRG, by employing similar operations used for sharing with the normal user (Figure 5).
IMPLEMENTATION AND AVAILABILITY
Given the heavy computational load, WebLab is implemented as a loosely coupled distributed system. The portal server holds the web interface and acts as a proxy to users’ requests. With dispatch daemon running, several backend computing servers execute the required operations following the request from WebLab portal server. The results will be sent back to the portal server after the analysis is finished and saved into database maintained by the portal server. Call-back mechanism is widely used in WebLab system to increase the flexibility. Adding a new tool does not require writing additional codes besides changing an XML format configuration file.
WebLab was developed using Java 1.5, providing it with the platform-independent advantage. WebLab uses Apache Tomcat as container for Java Servlet and JSP, MySQL as backend database system to store user data and other necessary information. WebLab also uses Graphviz (http://www.graphviz.org) to produce figures, and Lucene (http://lucene.apache.org) as information retrieval library to build index and search information.
WebLab is publicly accessible at http://weblab.cbi.pku.edu.cn and is compatible with the most common web browsers such as Mozilla Firefox (version 2 and 3) and Internet Explorer (version 6, 7 and 8). Online HTML and video tutorials are being actively maintained and updated. The source code of WebLab is released as ‘Free Software’ under the GNU General Public License version 3 (GPLv3), and freely available for downloading.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
China National High-tech 863 Programs [2006AA02Z334, 2006AA02Z314, 2006AA02A312]; China high-tech platform program. Funding for open access charge: China National High-tech (863) Program (2006AA02Z334).
Conflict of interest statement. None declared.
Supplementary Material
Footnotes
Present address: Jianmin Wu, Genome-Scale Biology Program, Institute of Biomedicine, University of Helsinki, Haartmaninkatu 8, Helsinki 00014, Finland
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Carver T, Bleasby A. The design of Jemboss: a graphical user interface to EMBOSS. Bioinformatics. 2003;19:1837–1843. doi: 10.1093/bioinformatics/btg251. [DOI] [PubMed] [Google Scholar]
- 2.Sarachu M, Colet M. wEMBOSS: a web interface for EMBOSS. Bioinformatics. 2005;21:540–541. doi: 10.1093/bioinformatics/bti031. [DOI] [PubMed] [Google Scholar]
- 3.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanzen A, Oinn T. The Taverna Interaction Service: enabling manual interaction in workflows. Bioinformatics. 2008;24:1118–1120. doi: 10.1093/bioinformatics/btn082. [DOI] [PubMed] [Google Scholar]
- 5.Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock MR, Wipat A, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20:3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- 6.Cattley S, Arthur JW. BioManager: the use of a bioinformatics web application as a teaching tool in undergraduate bioinformatics training. Brief Bioinform. 2007;8:457–465. doi: 10.1093/bib/bbm039. [DOI] [PubMed] [Google Scholar]
- 7.Blankenberg D, Taylor J, Schenck I, He J, Zhang Y, Ghent M, Veeraraghavan N, Albert I, Miller W, Makova KD, et al. A framework for collaborative analysis of ENCODE data: making large-scale analyses biologist-friendly. Genome Res. 2007;17:960–964. doi: 10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia Castro A, Thoraval S, Garcia LJ, Ragan MA. Workflows in bioinformatics: meta-analysis and prototype implementation of a workflow generator. BMC Bioinformatics. 2005;6:87. doi: 10.1186/1471-2105-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang F, Chua CL, Ho LY, Lim YP, Issac P, Krishnan A. Wildfire: distributed, Grid-enabled workflow construction and execution. BMC Bioinformatics. 2005;6:69. doi: 10.1186/1471-2105-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navas-Delgado I, Rojano-Munoz Mdel M, Ramirez S, Perez AJ, Andres Leon E, Aldana-Montes JF, Trelles O. Intelligent client for integrating bioinformatics services. Bioinformatics. 2006;22:106–111. doi: 10.1093/bioinformatics/bti740. [DOI] [PubMed] [Google Scholar]
- 12.Shah SP, He DY, Sawkins JN, Druce JC, Quon G, Lett D, Zheng GX, Xu T, Ouellette BF. Pegasys: software for executing and integrating analyses of biological sequences. BMC Bioinformatics. 2004;5:40. doi: 10.1186/1471-2105-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crass T, Antes I, Basekow R, Bork P, Buning C, Christensen M, Claussen H, Ebeling C, Ernst P, Gailus-Durner V, et al. The Helmholtz Network for Bioinformatics: an integrative web portal for bioinformatics resources. Bioinformatics. 2004;20:268–270. doi: 10.1093/bioinformatics/btg398. [DOI] [PubMed] [Google Scholar]
- 14.Waldrop M. Big data: Wikiomics. Nature. 2008;455:22–25. doi: 10.1038/455022a. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert W. Towards a paradigm shift in biology. Nature. 1991;349:99. doi: 10.1038/349099a0. [DOI] [PubMed] [Google Scholar]
- 16.Roure DD, Goble C, Stevens R. Third IEEE International Conference on e-Science and Grid Computing (e-Science 2007). Bangalore, India: 2007. Designing the my Experiment virtual research environment for the social sharing of workflows; pp. 603–610. [Google Scholar]
- 17.Huss JW, 3rd, Orozco C, Goodale J, Wu C, Batalov S, Vickers TJ, Valafar F, Su AI. A gene wiki for community annotation of gene function. PLoS Biol. 2008;6:e175. doi: 10.1371/journal.pbio.0060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durinck SMY, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;15:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 20.Zdobnov EM, Lopez R, Apweiler R, Etzold T. The EBI SRS server—recent developments. Bioinformatics. 2002;18:368–373. doi: 10.1093/bioinformatics/18.2.368. [DOI] [PubMed] [Google Scholar]
- 21.Tiwari A, Sekhar AK. Workflow based framework for life science informatics. Comput. Biol. Chem. 2007;31:305–319. doi: 10.1016/j.compbiolchem.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 23.Walther D. WebMol—a Java-based PDB viewer. Trends Biochem. Sci. 1997;22:274–275. doi: 10.1016/s0968-0004(97)89047-0. [DOI] [PubMed] [Google Scholar]
- 24.Junier T, Pagni M. Dotlet: diagonal plots in a web browser. Bioinformatics. 2000;16:178–179. doi: 10.1093/bioinformatics/16.2.178. [DOI] [PubMed] [Google Scholar]
- 25.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.