Abstract
LitInspector is a literature search tool providing gene and signal transduction pathway mining within NCBI's PubMed database. The automatic gene recognition and color coding increases the readability of abstracts and significantly speeds up literature research. A main challenge in gene recognition is the resolution of homonyms and rejection of identical abbreviations used in a ‘non-gene’ context. LitInspector uses automatically generated and manually refined filtering lists for this purpose. The quality of the LitInspector results was assessed with a published dataset of 181 PubMed sentences. LitInspector achieved a precision of 96.8%, a recall of 86.6% and an F-measure of 91.4%. To further demonstrate the homonym resolution qualities, LitInspector was compared to three other literature search tools using some challenging examples. The homonym MIZ-1 (gene IDs 7709 and 9063) was correctly resolved in 87% of the abstracts by LitInspector, whereas the other tools achieved recognition rates between 35% and 67%. The LitInspector signal transduction pathway mining is based on a manually curated database of pathway names (e.g. wingless type), pathway components (e.g. WNT1, FZD1), and general pathway keywords (e.g. signaling cascade). The performance was checked for 10 randomly selected genes. Eighty-two per cent of the 38 predicted pathway associations were correct. LitInspector is freely available at http://www.litinspector.org/.
INTRODUCTION
Literature research is an indispensable but a time-consuming task in scientific projects. Since the availability of online databases like PubMed (1), this work has become much easier already. Nevertheless, the impressive and continuously growing number of PubMed entries, currently more than 18 million, demands elaborate search strategies. The PubMed Web site itself offers a straightforward and fast search interface. The search results are plain publication abstracts, which still require a lot of text scanning by the reader. The question is how to support the scientist and how to speed up literature research.
For this purpose, several tools are available that perform automatic text mining within the PubMed database, e.g. LitMiner (2), PubGene (3), iHOP (4), EBIMed (5) and PolySearch (6). These tools offer diverse search strategies, for instance, automatic gene recognition and color coding of keywords.
LitInspector was developed with the aim of a high gene-recognition performance, a straightforward HTML-based Web interface and a quick response rate. The outputs are sentences from PubMed abstracts where genes, transcription factors and keywords such as tissues and diseases are highlighted. Additionally, links to further gene information are provided. Color coding enhances the readability and speeds up literature inquiries. The evaluation of the LitInspector results showed a high performance especially in regard to the resolution of homonyms. As the original sentences remain available the scientist can directly verify the automatically generated results.
A special focus of LitInspector is signal transduction pathway mining. Gene-pathway associations are based on a manually generated database of 180 signaling pathways and general pathway keywords that cannot be found elsewhere on the Web in this form.
PROGRAM DESCRIPTION
LitInspector allows input of gene synonyms (e.g. WT1) or Entrez Gene (1) gene IDs (e.g. 7490) and free text (e.g. binding site) (Figure 1). At least one gene synonym or free text word or phrase has to be submitted. In addition, the query can be restricted to those abstracts for which defined keyword categories (tissue, disease or pathway) were identified. This keyword tagging is based on Genomatix synonym catalogs. The tissue catalog contains over 3000 entries, the disease catalog over 11 000 entries. Both catalogs were generated mainly based on MeSH terms and other freely available tissue and disease lists that were complemented by the respective plural terms. The general pathway keyword list is a manually generated list of 100 phrases indicating signal transduction. Filtering for the MeSH categories tissue and disease is also supported.
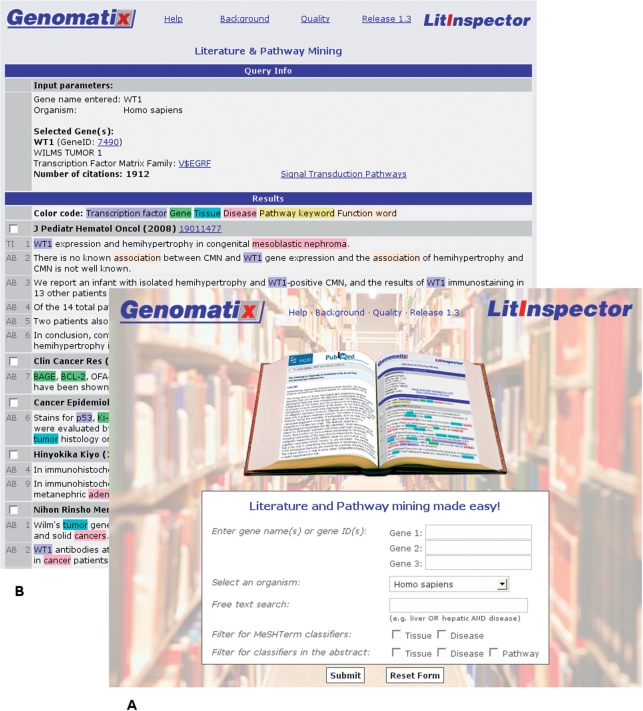
Figure 1.
Screenshot of the LitInspector user interface (A) and a results page (B). (A) The user interface allows input of up to three gene synonyms (e.g. WT1, PAX2) or gene identifiers (e.g. 7490) and free text (e.g. binding site). Default organism is H. sapiens, other organisms can be chosen from a pop-up menu. The search can be filtered for the occurrence of tissue, disease and pathway keywords in the abstract or tissue and disease annotations provided with the MeSH terms. (B) Each result page starts with query info where the user input, the number of citations found and, in case of a gene search, a link to the signal transduction pathway associations is displayed. The results are color-coded sentences wherein genes (green), transcription factors (purple), or keywords (tissue, cyan; disease, pink; pathway, yellow) are highlighted. Genes and transcription factors are hyperlinked to NCBI's Entrez Gene for further information. The output is ordered by the PubMed identifiers (latest publications coming first) which directly link to NCBI's PubMed. The user can select individual abstracts and retrieve them in batch.
Gene search
Usually, a single gene synonym is entered in the LitInspector form. One advantage of LitInspector over a PubMed search is that the submission of a single synonym will automatically consider all synonyms of this gene. Each of the more than 28 000 human genes is represented by six or seven synonyms on average. Entering a gene synonym like CARD4 (caspase recruitment domain 4) will automatically consider all synonyms of this gene, for example, NOD1 (nucleotide-binding oligomerization domain containing 1). In the same way, entering the gene ID 10392 will consider the synonyms CARD4 and NOD1 as well as all other synonyms of this gene. The user is given the possibility to search only for a special synonym by using the free text search (e.g. input of CARD4 as gene name and as free text).
If the entered gene synonym is unique (e.g. WT1), LitInspector directly displays the search results. If the entered synonym is a homonym, i.e. the same synonym is used for two or more distinct genes (e.g. MBP), an intermediate page allows selection of the required gene(s).
LitInspector allows searching for up to three different genes. Boolean OR or AND combination of the different query genes is possible. Free text searches can always be combined with gene searches. LitInspector supplies additional filters that facilitate the identification of abstracts containing certain keyword categories like tissue, disease or pathway.
The gene recognition in LitInspector is based on the comprehensive gene synonym lists provided by NCBI's Entrez Gene (1). These are complemented by our own synonym databases, which were assembled over the last years, containing additional synonyms as well as deprecated synonyms, which were realized to result in predominantly wrong taggings. For example, the synonym CO2 for complement component 2 (gene ID 717) was deprecated because it is mainly used with the meaning carbon dioxide in the literature.
Homonyms and ambiguous synonyms
Many gene synonyms are ambiguous, i.e. the same synonym is used for multiple genes or even in a completely unrelated, ‘non-gene’ context. For instance, the synonym MBP is mentioned in several thousands of PubMed abstracts. MBP is a homonym, it is used for three different genes (Table 1). Moreover, MBP is used in the scientific literature as abbreviation in a ‘non-gene’ context with various meanings (Table 1).
Table 1.
The gene synonym ‘MBP’—a homonym
| Gene synonym/ abbreviation | Gene name (Gene ID)/meaning | Number of citations |
|---|---|---|
| MBP | Myelin basic protein (4155) | 3000 |
| MBP | Major basic protein (5553) | 300 |
| MBP | Mannose binding protein (4153) | 100 |
| MBP | Mean blood pressure | 500 |
| MBP | Monobutyl phthalate | 40 |
| MBP | Megabase pairs | 10 |
MBP is used for three different genes and, in addition, in a ‘non-gene’ context as abbreviation with several different meanings.
Even human experts may have difficulties in resolving some homonyms and ambiguities. Therefore, a main challenge of automatic literature mining is the disambiguation especially of short gene synonyms. LitInspector is based on a combination of automatic disambiguation modules, manually curated context databases, and semi-automatically generated and manually refined filtering lists. Disambiguation of gene homonyms makes use of the occurrence of further gene descriptions in the same abstract as well as automatically and manually generated gene context lists.
Assignment of organism information
LitInspector makes use of the organism information annotated by the MeSH (http://www.nlm.nih.gov/mesh/) consortium provided within the MeSH terms. However, for the most recent abstracts, the MeSH annotation is lagging behind, and in other publications, organism information is not provided at all. To make sure that no publications are skipped because of missing organism annotations, LitInspector applies only soft criteria for the organism assignment. In case of mammalian gene taggings, LitInspector takes all abstracts and excludes only those for which a ‘non-mammalian’ organism (e.g. Caenorhabditis, Xenopus or plants) is annotated in the MeSH terms. An identified gene synonym will be annotated for all mammalian organisms for which this synonym is known, e.g. WT1 for Homo sapiens (gene ID 7490), Mus musculus (gene ID 22431) and Rattus norvegicus (gene ID 24883).
Signal transduction pathway mining
The LitInspector signal transduction pathway mining is based on a proprietary and manually curated database of pathway names (e.g. wingless type), pathway components (e.g. WNT1, FZD1), and general pathway keywords (e.g. signaling cascade). Currently, the database comprises 100 general pathway keywords and 180 pathway names to which 850 pathway components are assigned. For pathway mining, the PubMed database is scanned for co-occurrence of the user input gene with the LitInspector pathway components and general pathway keywords in the same sentence. Most of the 180 signaling pathways are hyperlinked to canonical pathways from BioCarta (http://cgap.nci.nih.gov/Pathways/BioCarta_Pathways), STKE (http://stke.sciencemag.org/), or KEGG (7). These hyperlinked graphics only provide an overview; they may not necessarily contain the query genes because LitInspector extracts additional pathway associations.
An identified association of the query gene to a pathway can have several possible meanings: the query gene may be part of the signaling pathway, it may regulate the pathway, it may be regulated by the pathway, it may regulate a different pathway which in turn cross talks to the mentioned pathway. It is also possible that the query gene was experimentally found to be NOT associated with that pathway. The pathway mining does not yet indicate a direction of the gene–pathway associations.
The advantage of automatic pathway mining compared to manually curated databases and static pathway associations is that the results are always up to date and may contain additional data that are hard to find in the established models. The LitInspector pathway mining provides an actual overview of possible pathway associations and potential interactions of the query gene. It also provides the literature references which allow direct verification by the scientist (Figure 2).
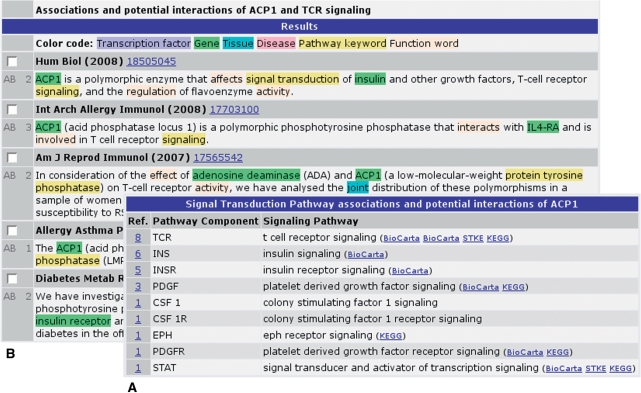
Figure 2.
Screenshot of the LitInspector signal transduction pathway mining output for the gene ACP1 (acid phosphatase 1). (A) List of the found signaling pathways sorted by the number of references. The user has full access to the color-coded PubMed sentences from which the pathway associations have been derived from. (B) In case of ACP1 most references were found for TCR (T-cell receptor) signaling (eight references).
EVALUATION
Gene recognition performance
The quality of the LitInspector results was assessed with the published dataset of 181 PubMed sentences already used as gold standard for the evaluation of iHOP and PolySearch. LitInspector achieved a precision of 96.8%, a recall of 86.6% and a F-measure of 91.4% thus outperforming both other programs (Table 2). The LitInspector evaluation results are provided as Supplementary Data.
Table 2.
Evaluation of LitInspector using a dataset of 181 PubMed sentences compared to two other programs
| Program name | Precision (%) | Recall (%) | F-measure (%) |
|---|---|---|---|
| LitInspector | 96.8 | 86.6 | 91.4 |
| PolySearch | 90.1 | 85.3 | 87.6 |
| iHOP | 87.1 | 81.8 | 84.4 |
The main reasons for the high gene recognition quality of LitInspector are the strategies used for homonym resolution and rejection of ‘non-gene’ abbreviations. In order to demonstrate this, LitInspector was compared to three other literature search tools (iHOP, EBIMed, PubGene) using the homonym ‘MIZ-1’ (gene IDs 7709 and 9063). LitInspector correctly resolved 87% of the synonyms, whereas the other tools achieved recognition rates between 39% and 67% (Table 3). In another example, the recognition of the synonym CPAP (gene ID 55835), frequently used as abbreviation for ‘continuous positive airway pressure’, was correct in all 19 abstracts found by LitInspector. EBIMed, in contrast, had 1870 matches and only 0.5% were true positives. iHOP had 80 matches, 17.5% were correct and PubGene had 71 matches, 30% were correct. Thus, LitInspector provides the most reliable results.
Table 3.
Evaluation of the LitInspector homonym resolution compared to three other data mining tools by means of the homonym MIZ-1
| Text mining software | MIZ-1 Myc-interacting zinc finger-1 (Gene ID: 7709) |
MIZ-1 Msx-interacting-zinc finger-1 (Gene ID: 9063) |
Unresolved homonyms | Correctly resolved homonyms (percent) | ||||
|---|---|---|---|---|---|---|---|---|
| Number matches | Correctly resolved | False positives | Number matches | Correctly resolved | False positives | |||
| LitInspector | 53 | 44 | 2 | 36 | 27 | 2 | 7 | 87% |
| EBIMed | 11 | 10 | 1 | 17 | 9 | 8 | 0 | 67% |
| PubGene | 45 | (9) | (1) | 34 | (2) | (8) | (0) | (55%a) |
| iHOP | 36 | 14 | 0 | 31 | 12 | 6 | 35 | 39% |
The programs were used with default parameters. All numbers refer to abstracts, i.e. sentences with the same identified gene from one paper were counted only once for this gene. The evaluation was performed in July 2008.
aPubGene provides only 10 example papers for each search, therefore, an evaluation of the complete results is not possible. This evaluation was performed using the 20 (2 × 10) abstracts available.
LitInspector scans the complete PubMed database and provides all abstracts found. In contrast, iHOP restricts the number of abstracts in the output selected by certain quality criteria. For example, an iHOP search for WT1 (gene ID 7490) displays only 319 abstracts, whereas LitInspector provides 1912 abstracts, which is the complete search result.
Pathway mining performance
The performance of the LitInspector signal transduction pathway mining was manually checked for 10 randomly selected human genes (gene IDs: 52, 55, 92, 463, 527, 1750, 1844, 3767, 6494, 9751). These randomly selected genes did not overlap with any of the pathway components. Eighty-two per cent of the 38 predicted pathway associations were found to be correct in a manual evaluation of the predicted references, i.e. the query gene was described to be part of the signaling pathway, to regulate the pathway, or to be regulated by the pathway. An example is shown in Table 4.
Table 4.
Signal transduction pathways of ACP1 (gene ID 52) found by LitInspector pathway mining
 |
For each pathway, a reference sentence from PubMed is shown (pathway keywords are highlighted in yellow, genes in green).
DISCUSSION
The usefulness of text mining tools critically depends on the reliability and verifiability of the results. Automatic extraction of gene relations for instance is very helpful, but only if the basic gene synonym identification is correct. Although best effort is made to resolve ambiguities, it is unavoidable that data mining programs will have a certain error rate. LitInspector showed a comparable or better gene recognition performance than other text mining tools. An advantage of LitInspector over solely graphical or tabular representations of other tools (e.g. PubGene) is that the scientist retains full control over the reliability of the results as the reference sentences are directly verifiable. In many cases a human expert will then recognize wrongly assigned synonyms by scanning the context.
We tried to compare the results of the LitInspector signal transduction pathway mining with another pathway mining tool. We entered the ten randomly selected genes used for the LitInspector evaluation into PolySearch (6) using default parameters. For most of the genes pathways called ‘gene expression’, ‘differentiation’, or ‘survival’ were identified. As these general terms do not correspond to common signal transduction pathways we claim that LitInspector is the first approach that is able to extract valuable signal transduction pathway information from the literature.
The LitInspector gene tagging method is the basis for the literature co-citation networks that can be analysed with the program BiblioSphere (8,9). It is also used for the manual evaluation of gene relations that are displayed as expert annotations in BiblioSphere.
LitInspector will be constantly refined, based on user requests regarding the recognition of genes or the user interface. Any feedback is appreciated at litinspector@genomatix.de.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Genomatix Software GmbH.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37(Database issue):D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maier H, Döhr S, Grote K, O'Keeffe S, Werner T, Hrabé de Angelis M, Schneider R. LitMiner and WikiGene: identifying problem-related key players of gene regulation using publication abstracts. Nucleic Acids Res. 2005;33(Web Server issue):W779–W782. doi: 10.1093/nar/gki417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat. Genet. 2001;28:21–28. doi: 10.1038/ng0501-21. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann R, Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics. 2005;21 (Suppl. 2):ii252–ii258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- 5.Rebholz-Schuhmann D, Kirsch H, Arregui M, Gaudan S, Riethoven M, Stoehr P. EBIMed–text crunching to gather facts for proteins from Medline. Bioinformatics. 2007;23:e237–e244. doi: 10.1093/bioinformatics/btl302. [DOI] [PubMed] [Google Scholar]
- 6.Cheng D, Knox C, Young N, Stothard P, Damaraju S, Wishart DS. PolySearch: a web-based text mining system for extracting relationships between human diseases, genes, mutations, drugs and metabolites. Nucleic Acids Res. 2008;36(Web Server issue):W399–W405. doi: 10.1093/nar/gkn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanehisa M, Araki Mm, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res., 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scherf M, Epple A, Werner T. The next generation of literature analysis: integration of genomic analysis into text mining. Brief Bioinform. 2005;6:287–297. doi: 10.1093/bib/6.3.287. [DOI] [PubMed] [Google Scholar]
- 9.Epple A, Scherf M. BiblioSphere – hypothesis generation in regulatory network analysis. In: Krawetz S, editor. Bioinformatics for Systems Biology. Heidelberg, Germany: Humana Press; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.