Abstract
HotSpot Wizard is a web server for automatic identification of ‘hot spots’ for engineering of substrate specificity, activity or enantioselectivity of enzymes and for annotation of protein structures. The web server implements the protein engineering protocol, which targets evolutionarily variable amino acid positions located in the active site or lining the access tunnels. The ‘hot spots’ for mutagenesis are selected through the integration of structural, functional and evolutionary information obtained from: (i) the databases RCSB PDB, UniProt, PDBSWS, Catalytic Site Atlas and nr NCBI and (ii) the tools CASTp, CAVER, BLAST, CD-HIT, MUSCLE and Rate4Site. The protein structure and e-mail address are the only obligatory inputs for the calculation. In the output, HotSpot Wizard lists annotated residues ordered by estimated mutability. The results of the analysis are mapped on the enzyme structure and visualized in the web browser using Jmol. The HotSpot Wizard server should be useful for protein engineers interested in exploring the structure of their favourite protein and for the design of mutations in site-directed mutagenesis and focused directed evolution experiments. HotSpot Wizard is available at http://loschmidt.chemi.muni.cz/hotspotwizard/.
INTRODUCTION
Protein engineering represents a powerful approach for production of useful enzymes (1,2). Directed evolution and rational protein design are two distinct technologies currently used in protein engineering. While directed evolution randomly modifies the residues throughout the entire protein structure, the rational protein design and focused directed evolution targets only selected residues, called ‘hot spots’. Modification of ‘hot spots’ often leads to desired changes in protein properties and significantly reduces the number of mutants that must be biochemically characterized to discover an interesting variant (3,4).
The functional amino acid residues that mediate the substrate binding, transition-state stabilization or product release, i.e. the residues located in the active sites and lining the access tunnels, are frequently selected as ‘hot spots’ for modification of enzyme catalytic properties (4–12). However, some functional residues may be indispensable for catalysis and their replacement leads to complete loss of enzyme function. Information about the evolutionary conservation of individual residues can serve as a useful indicator for their mutability. Mutagenesis targeting the functional residues located in highly variable positions is an effective strategy for altering catalytic properties with reduced risk of losing a catalytic activity (9).
This strategy is implemented in HotSpot Wizard, which automatically identifies the functional residues for engineering of catalytic properties and estimates their mutability. For this purpose, HotSpot Wizard integrates several bioinformatics databases and computational tools. Structural analyses are conducted to identify the residues that potentially come into contact with the substrates or products. The mutability of individual amino acid residues is derived from their conservation level. Higher mutability implies a better chance that the amino acid replacement will not compromise the enzyme function. HotSpot Wizard assigns all functional residues with high mutability as ‘hot spots’. HotSpot Wizard results, including the annotated structure, sequence and summary tables of ‘hot spots’, functional residues, pockets and tunnels, can be interactively visualized in the web browser or downloaded to a local computer. For individual residues, information about their mutability, structural location, functional role and annotations are provided.
A variety of computational tools has been developed to assist rational design of proteins and recently reviewed (13). Novelty of HotSpot Wizard lies in a tight integration of functional, evolutionary and structural information. This integration enables automation of the protein engineering protocol and makes our tool accessible to wider scientific community.
METHODS
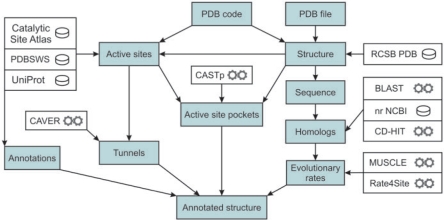
Flow chart of the HotSpot Wizard is depicted in the Figure 1. HotSpot Wizard works with a PDB file, which is either downloaded from the RCSB PDB (14) or provided by the user. The PDBSWS database (15) is used to identify UniProt entries (16) corresponding to individual PDB chains. If the structure is provided by the user, corresponding entries in UniProt and RCSB PDB are identified by the BLAST sequence search (17). The UniProt entries are retrieved by the WSDbfetch web service (18). Residue annotations are extracted from the UniProt fields ‘Active site’, ‘Binding site’, ‘Mutagenesis’ and ‘Natural variant’ to determine amino acid residues indispensable for enzymatic function and to collect available information about mutagenesis experiments and naturally occurring variants. PDBSWS is then used for the second time to map the indexes of extracted UniProt annotations to the corresponding PDB residues. The JAligner pairwise alignment tool (http://jaligner.sourceforge.net/) is used to map the annotations to the structures provided by the user. Information about catalytic residues is also downloaded from the Catalytic Site Atlas (19). The catalytic residues are assigned to the individual catalytic sites according to the Catalytic Site Atlas and their spatial proximity. In the next step, HotSpot Wizard searches for residues located in the active site pockets and access tunnels. CASTp (20) is used to find all pockets present in the query structure and was selected because it offers online, fast and reliable computation of pockets and precise measurements of their volumes and areas of their respective surfaces. Among the calculated pockets, the active site pockets are selected using the information about the catalytic residues and the volume of individual pockets. For each identified active site, a center of mass for the catalytic residues is computed, where unitary weight is assigned to each residue. In the next step, another point is calculated where (i) a sphere circumscribed around this point intersects no atom in a protein structure and (ii) the distance between this point and the center is minimal, which ensures its positioning within the space occupied by the substrate molecule. The CAVER (21,22) calculation is then performed to automatically identify tunnels connecting this point with the outside solvent.
Figure 1.
Flow chart of the HotSpot Wizard calculation. The input is either a PDB code or a protein structure in PDB format. The output is an annotated structure. The ‘hot spots’ for mutagenesis are selected through the integration of structural, functional and evolutionary information obtained from the bioinformatics databases (database symbol) and the tools (computational tool symbol).
In the final step of the HotSpot Wizard protocol, the BLAST search against the non-redundant database at NCBI (23) is performed to gather protein sequences similar to the query. Sequences are clustered by CD-HIT (24) and representatives of the clusters aligned using MUSCLE (25). Both tools were chosen for their ability to process large datasets within a short time period with required accuracy. The multiple sequence alignment is used for estimating the site-specific conservation scores. For this purpose, the approach exploiting the information about evolution of the analyzed sequences was selected. Evolutionary rates of individual amino acid positions are estimated by the empirical Bayesian method (26) as implemented in the Rate4Site program (27). The evolutionary rates are converted to the ConSurf conservation scale (28), which is then used to assign mutability to individual residues of the query enzyme. Users are notified about the finished calculation by an e-mail containing the results of the calculation in a text file and a link to relevant web pages.
DESCRIPTION OF THE SERVER
Input
HotSpot Wizard requires a structure of the query protein, provided as a PDB code or a PDB file, and an e-mail address as the only obligatory inputs. Additionally, a number of optional settings are available to control the calculation. The user may specify protein chains of interest. By default, the application attempts to extract information about the biologically relevant molecule from the PDB file. If this information is not available, the first chain of the PDB file is used for the calculation. Another option enables the selection of the resource for annotation of the catalytic residues or the manual input of the catalytic residues. Two parameters can be set for the calculation of tunnels: (i) the minimal tunnel radius specifying the minimal tunnel width (default 1.4 Å) and (ii) the minimal starting radius specifying the required free space around the tunnel starting point (default 1.6 Å). The calculation of evolutionary conservation is, by default, conducted on the dataset obtained by the BLAST search. The appropriate setting of the E-value (default 1E–12) enables the user to perform the analysis on enzymes with desired similarities or function. The user can also change the maximum number of sequences to be analyzed (default 50) and thus potentially increase the accuracy of the evolutionary rate estimation. However, demand on calculation time grows with increasing number of sequences, and it is currently not recommended to analyze more than 100 sequences at once. Results of the conservation analysis are stored on the server for 3 months. If a new job with identical parameters is submitted during this period, the precalculated results are used instead of conducting the conservation analysis. The usage of precalculated results may be disabled. Users can also upload their own multiple sequence alignment and the phylogenetic tree. Users interested only in the structural annotation or some specific analysis may skip the conservation analysis as it is the most time-consuming part of the HotSpot Wizard calculation.
Output
The ‘Job’ panel appears when the calculation is finished and enables navigation through the obtained results. The results may be accessed either by an interactive web application called the ‘Results Browser’ (Figure 2) or downloaded to a local computer. Moreover, the ‘Job’ panel provides links to the external databases and servers with further information about the query protein.
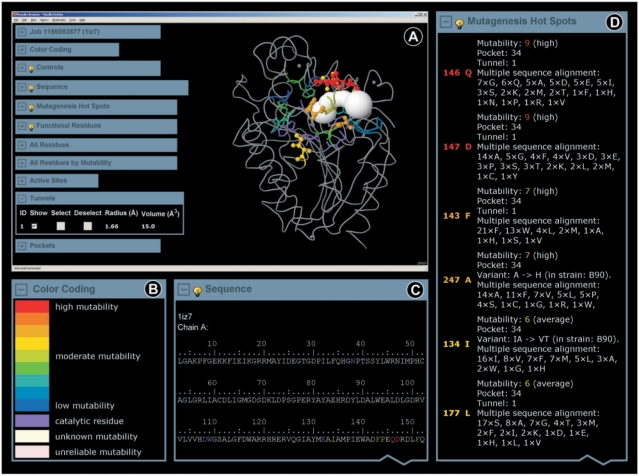
Figure 2.
Graphic interface of the HotSpot Wizard results. (A) The ‘Results Browser’ includes the embedded Jmol applet enabling visualization of the annotated structure, ligands and identified tunnels. ‘Hot spots’ and functional residues are highlighted and colored according to their estimated mutability. The ‘Results Browser’ further offers the ‘Job’ panel enabling navigation through the results, ‘Control’ panel providing basic operations for manipulating the structure, annotated sequence and summary tables of ‘hot spots’, functional residues, all residues, active sites, pockets and tunnels. (B) The mutability color scale is defined in the ‘Color coding’ panel. (C) The ‘Sequence’ panel is interactively interconnected with all other sections of the ‘Results Browser’. (D) The output is summarized in the ‘Mutagenesis Hot Spots’ table listing all identified ‘hot spots’ ordered by their mutability. For individual residues, information about their mutability, structural location, functional role and annotations are provided.
Residue annotations
For each residue, information about its mutability, potential function (catalytic, located in the active site or access tunnel), available mutagenesis data, existing sequence variants and amino acids occurring at the corresponding position of the sequence alignment is provided. The mutability of individual sites, which estimates the tolerance of a given position to amino acid replacement, is derived from evolutionary conservation. The mutability scale ranges from 1 (low mutability) to 9 (high mutability). For visualization purposes, mutability is converted to a color scale ranging from blue (low mutability) to red (high mutability). The color scale is additionally extended by the violet, pink and ivory colors to indicate catalytic residues, residues with unreliable assignment and residues with missing information about evolutionary conservation, respectively. It is important to note that low mutability of a particular position does ‘not’ necessarily mean that the corresponding residue is immutable. However, the residues at such positions are often essential for maintaining structural or functional properties of an enzyme and should not be mutated without careful consideration of the potential effects of these mutations. To prevent mutagenesis at the sites indispensable for a function, catalytic residues are assigned as immutable.
Summary tables
The main output of the HotSpot Wizard application is summarized in the ‘Mutagenesis Hot Spots’ table, listing the residues assigned as ‘hot spots’, based on their high mutability (6–9) and structural location. ‘Hot spots’ are ordered by their mutability, starting with the most mutable ones, and accompanied by the residue annotations. For some proteins, only very few ‘hot spots’ are identified by HotSpot Wizard, because functional residues of the protein are highly conserved. The users are encouraged to use the ‘Functional residues’ table to see annotations for all active site and tunnel residues identified in the query structure. However, selecting the targets for mutagenesis from this table increases a risk of producing inactive mutants, especially when the positions with low mutability are targeted. The ‘Functional residues’ table is also useful for users who want to select their own ‘hot spots’ based on other criteria than mutability. The ‘All Residues’ and ‘All Residues by Mutability’ tables provide information about each individual residue of the query enzyme.
Annotated structure
HotSpot Wizard results are mapped on the enzyme structure and are visualized directly in the web browser using the Jmol applet (http://jmol.sourceforge.net/). Besides the enzyme structure, ligands and identified tunnels may be displayed as well. The default visualization highlights the ‘hot spots’ and the functional residues. The residue annotations may be invoked by positioning the mouse cursor over a given residue in the Jmol applet. Users can select predefined groups of residues and change their coloring and visualization styles using the ‘Controls’ panel. The Jmol applet is interactively interconnected with all other sections of the ‘Results Browser’ also enabling selection from the ‘Sequence’ panel and the summary tables.
Annotated sequence
The panel ‘Sequence’ displays the annotated sequence of the query enzyme. Its functionality comprises selections of residues and displaying the residue annotations. The coloring of the sequence is synchronized with the structure and the summary tables.
Results for download
The results may be downloaded to a local computer all in one package or as individual files. The package includes the summary tables in a text format for direct reading or software parsing, the original outputs obtained from individual servers and databases, and the Python script enabling visualization and analysis of the HotSpot Wizard results in PyMOL (http://pymol.sourceforge.net/).
Limitations
HotSpot Wizard processes information from databases and computational tools in a highly automated manner. A specification of the query protein structure is the only required input information. Tight integration of individual components in the HotSpot Wizard workflow may result in the propagation of incorrect results from one tool to the remaining components of the cascade. (i) Currently, the most critical step of the protocol is the assignment of the catalytic residues. Information about the catalytic residues is used to avoid mutagenesis at the residues critical for the enzyme function, but also for the assignment of the active site pocket and the starting point for the calculation of tunnels. If catalytic residues are not found in databases either for the query sequence or for homologous sequences, users must specify this information manually. Otherwise, the pocket with the largest volume will be assumed to contain the active site and no tunnels will be computed. (ii) The identification of the active site pocket is another critical step of the computation. Surface residues can be incorrectly assigned as part of the active site pocket. This sometimes happens at the interface of two subunits forming a very large pocket. Exclusion of one of the chains is a simple solution to this problem. Miss-assignment of the residues of the active site pocket happens also for proteins carrying large open depressions at the protein surface, which has to be recognized by the user, and these residues should not be considered for mutagenesis experiments. (iii) Finally, the sequence databases might not contain a sufficient number of homologous sequences for some of the proteins and their mutability thus cannot be calculated. In such cases, HotSpot Wizard lists the functional residues without mutability scores.
EXAMPLES
The ‘hot spot’ residues identified computationally by HotSpot Wizard were compared with the mutations extracted from the Protein Mutant Database (29) and the primary literature for four different proteins: (i) haloalkane dehalogenase, (ii) phosphotriesterase, (iii) 1,3-1,4-β-d-glucan 4-glucanohydrolase and (iv) β-lactamase.
Haloalkane dehalogenase
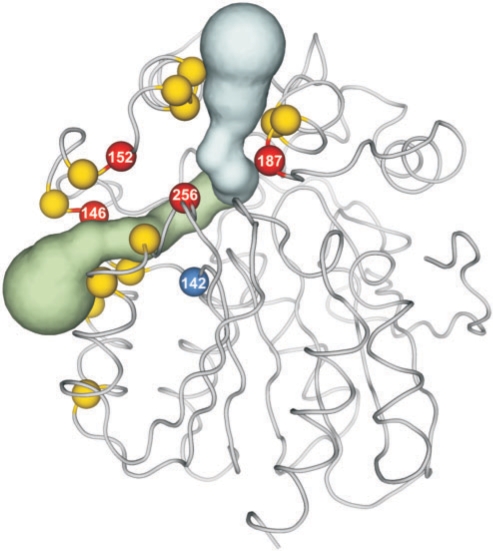
Haloalkane dehalogenase DhaA from Rhodococcus sp. catalyzes hydrolytic dehalogenation of a broad range of halogenated aliphatic hydrocarbons. The structure of the DhaA enzyme was specified by the PDB code 1BN6 (30). In further text, the numbering from the PDB file is used, while 11 residues need to be subtracted to obtain the numbering used in the literature. HotSpot Wizard identified 17 reliable ‘hot spots’ (mutability range 6–9) lining 1 active site pocket and 2 tunnels: F142, R144, I146, P147, E151, W152, F155, A156, T159, V183, K186, C187, E234, H241, V256, P259, E262 (Figure 3). Kretz et al. performed saturation mutagenesis of the entire dhaA gene and constructed its mutability map (31). While one third of the residues of DhaA could not be replaced without compromising enzyme activity according to the experiments of Kretz et al., all but one ‘hot spots’ identified by HotSpot Wizard, were able to accommodate a wide range of mutations. The only exception is the residue F142 located just next to the catalytic residue E141. It is well known that the mutations introduced to the regions closest to the catalytic residues are tricky as they can disrupt the geometry of the transition state and lead to loss of enzyme activity. In another project, error-prone PCR was used for the introduction of random mutations to the DhaA structure by Gray et al. (32) and Bosma et al. (33). The biochemical screening of 10 000 variants against toxic environmental pollutant 1,2,3-trichloropropane (TCP) provided two double-point mutants C187Y+Y284F (33) and G14D+C187F (32), respectively, showing 3.5 times and 4 times higher activities with TCP than the wild-type enzyme. These two variants obtained independently in the directed evolution experiments carry the mutation in the position C187. HotSpot Wizard identified the position C187 as the ‘hot spot’ due to its location at the tunnel opening and high mutability. Pavlova et al. (manuscript submitted) used rational design and selected five positions lining the access tunnels: I146, W152, C187, V256 and L257. The mutant of Bosma et al. (C187Y and Y284F) and the triple mutant (W152F, C187Y and Y284F) were used as templates for focused directed evolution, subjecting the positions I146, V256 and L257 to a simultaneous saturation mutagenesis. The screening of 5000 variants against TCP provided 25 unique mutants with higher activities towards TCP; the best mutant showing 32 times higher activity than the wild type. Out of the five positions rationally selected in this study, four positions were also identified as ‘hot spots’ by HotSpot Wizard.
Figure 3.
Comparison of the results from HotSpot Wizard with the experimental data. HotSpot Wizard identified seventeen ‘hot spots’ (balls) lining the active site pocket of the enzyme DhaA (PDB code 1BN6) and its two tunnels (tubes). All identified ‘hot spots’ could accommodate a wide range of mutations in the gene saturation experiment, except F142 (blue ball), which is adjacent to the catalytic residue. Four predicted ‘hot spots’ (red balls) were verified experimentally in three independent directed evolution experiments, showing a key role of these residues for the catalytic activity of DhaA. The picture was prepared in PyMOL using the Python script generated by the HotSpot Wizard server.
Phosphotriesterase
Phosphotriesterase from Pseudomonas diminuta catalyzes the hydrolysis of a variety of organophosphates, including the widely used insecticide paraoxon and the chemical warfare agent sarin. The crystal structure of the phosphotriesterase homodimer, PDB code 1I0D (34), was analyzed and nine reliably assigned ‘hot spots’ were identified: G60, L136, R139, S205, D235, A270, L271, L272, F306. The data from mutagenesis experiments were found for 4 of these residues: G60, L136, L271 and F306. Gopal et al. (35) rationally designed eight mutants, including L136Y, L271Y, F306A and F306Y, intending to enhance activity towards a chemical warfare agent VX [O-ethyl S-(2-diiso-propyl aminoethyl) methylphosphonothioate] and five other organophosphorus compounds. L136Y was the only mutation under study that leads to an increased relative activity towards VX. Five mutants, including L136Y and F306Y, displayed increased activity towards demeton-S-methyl. In another study, improvements of the catalysis of insecticide DFP up to one order of magnitude were obtained with the mutants F306Y and F306H (36). Systematic alanine-scanning of phosphotriesterase binding pocket, including residues G60, L271 and F306, was carried out to elucidate structural determinants of stereoselectivity and overall reactivity of this enzyme with different substrates (37). Substitution G60A lead to a dramatic enhancement of the chiral preference for the SP-enantiomers, increasing the enantioselectivity for several chiral substrates, e.g. from 21 to >>100 for p-nitrophenyl ethyl phenyl phosphate (6). Substitutions in L271 and F306 did not improve stereoselectivity, while the catalytic rates were decreased relative to those of the wild-type enzyme for most of the tested substrates (37).
1,3-1,4-β-d-Glucan 4-glucanohydrolase
1,3-1,4-β-d-Glucan 4-glucanohydrolase (1,3-1,4-β-glucanase) from Bacillus licheniformis is an endoglycosidase that cleaves β-glucans containing mixed β-1,3- and β-1,4- linkages. The structure of 1,3-1,4-β-glucanase was specified by the PDB code 1GBG (38). In further text, the numbering from the PDB file is used, while 29 residues needs to be added to obtain the numbering used in the literature. HotSpot Wizard identified 5 reliable ‘hot spots’: Y24, N28, M29, T69, V189. Three of them, Y24, N28 and M29, are described in the experimental study of Pons et al. (39). Pons et al. conducted alanine-scanning mutagenesis of the major loop (residues 22–38) of 1,3-1,4-β-glucanase to investigate its role in substrate binding. The substitution M29A resulted in a 3.4-fold increase in kcat/Km towards the substrate 4-methylumbelliferyl 3-O-β-cellobiosyl-β-d-glucopyranoside, while the catalytic efficiency of other mutants decreased.
β-Lactamase
The class C β-lactamase from Enterobacter cloacae P99 confers resistance to broad-spectrum β-lactam antibiotics by cleaving the amide bond in the β-lactam ring. The HotSpot Wizard calculation using the PDB code 1BLS (chain A) of the β-lactamase structure (40) suggested 7 reliable ‘hot spots’: M215, S264, T282, E285, S289, L293, L296. The 21 amino acid residues of the active site pocket of the β-lactamase, including ‘hot spots’ S289 and L293, were systematically randomized to identify the determinants of catalysis for the important antibiotic ceftazidime (41). A diverse set of substitutions was found at positions corresponding to S289 and L293 among the mutants selected for ceftazidime resistance, indicating that these residues are not essential for the binding or hydrolysis of this antibiotic. Although not critical for the function, the position L293 was shown to be important for the substrate specificity of this enzyme. Vakulenko et al. (42) conducted PCR mutagenesis, followed by the selection for cefepime resistance and obtained a mutant carrying the single-point substitution L293P. Compared to the wild-type β-lactamase, the catalytic efficiencies of the L293P mutant were increased 27- and 11-fold for cefepime and ceftazidime, respectively. In the next step, 14 additional substitutions were introduced to the position L293 by site-directed mutagenesis. More than half of the mutants conferred increased resistances towards cefepime and ceftazidime.
VALIDATION
The effectivity of mutagenesis targeting highly variable positions was assessed by comparing HotSpot Wizard results with the data obtained from the systematic mutagenesis studies: (i) 3315 mutants of lactose repressor (43), (ii) 1930 mutants of T4 lysozyme (44), (iii) 676 mutants of barnase (45) and (iv) 364 mutants of HIV reverse transcriptase (29,46). The individual mutagenesis studies differed in the activity assays and in the classification of the effects. Therefore, these data sets are not directly comparable and had to be evaluated independently. For each data set, those mutations which lead to the protein variants with very low or no activity, were assigned as deleterious. The proportion of deleterious mutations in the positions assigned as highly mutable by HotSpot Wizard, i.e. with the mutability grades from 6 to 9, were compared with proportion of deleterious mutations in the entire protein structure. Calculated ratio of deleterious mutations in the mutable positions versus in the entire protein structure were: (i) 2.7% versus 25.3% for lactose repressor, (ii) 1% versus 9.1% for T4 lysozyme, (iii) 0.4% versus 4.8% for barnase and (iv) 3.9% versus 30.7% for HIV reverse transcriptase. These results demonstrate that mutagenesis targeting the ‘hot spot’ positions identified by HotSpot Wizard provides significantly higher proportion of viable variants than blind mutagenesis. Detailed information on all presented examples can be downloaded from http://loschmidt.chemi.muni.cz/hotspotwizard/data.
CONCLUSIONS AND OUTLOOK
The development of the HotSpot Wizard server was motivated by a growing demand for comprehensive tools assisting protein engineers with the rational design of enzymes. The primary use of HotSpot Wizard is the identification of ‘hot spots’ for the site-directed mutagenesis or focused directed evolution experiments. Mutagenesis targeting the ‘hot spots’ found by HotSpot Wizard should increase the yield of active mutants with altered catalytic properties. Alternatively, HotSpot Wizard can be used for the annotation of protein structures. HotSpot Wizard automates a protein engineering protocol by the integration of several bioinformatics databases and computational tools and saves the user's time. Minimal demands on input information make this web server potentially useful for the users with no prior knowledge of structural or bioinformatics analyses. We are currently integrating additional features to HotSpot Wizard, e.g. a visualization of the residue solvent accessibility and volumes of pockets. We also aim to improve the estimation of mutability by including additional factors into the calculation of the prioritization function.
FUNDING
Czech Ministry of Education, Youth and Sport [LC06010 to E.C.H., MSM0021622412 to J.D.]; the Grant Agency of the Czech Republic [201/07/0927 to J.D.]; and the Grant Agency of the Czech Academy of Sciences [IAA401630901 to J.D.]. Funding for open access charge: LC06010 and MSM0021622412.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to express our thanks to the authors of the tools used by HotSpot Wizard and to Peter Lisak (Brno University of Technology) for the LLWS utility enabling a simple HTTP access to software installed on Linux. Access to the METACentrum supercomputing facilities provided under the research intent MSM6383917201 is highly appreciated.
REFERENCES
- 1.Bornscheuer UT, Pohl M. Improved biocatalysts by directed evolution and rational protein design. Curr. Opin. Chem. Biol. 2001;5:137–143. doi: 10.1016/s1367-5931(00)00182-4. [DOI] [PubMed] [Google Scholar]
- 2.Brannigan JA, Wilkinson AJ. Protein engineering 20 years on. Nat. Rev. Mol. Cell Biol. 2002;3:964–970. doi: 10.1038/nrm975. [DOI] [PubMed] [Google Scholar]
- 3.Chen R. Enzyme engineering: rational redesign versus directed evolution. Trends Biotechnol. 2001;19:13–14. doi: 10.1016/s0167-7799(00)01522-5. [DOI] [PubMed] [Google Scholar]
- 4.Chica RA, Doucet N, Pelletier JN. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr. Opin. Biotechnol. 2005;16:378–384. doi: 10.1016/j.copbio.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Park S, Morley KL, Horsman GP, Holmquist M, Hult K, Kazlauskas RJ. Focusing mutations into the P. fluorescens esterase binding site increases enantioselectivity more effectively than distant mutations. Chem. Biol. 2005;12:45–54. doi: 10.1016/j.chembiol.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Morley KL, Kazlauskas RJ. Improving enzyme properties: When are closer mutations better? Trends Biotechnol. 2005;23:231–237. doi: 10.1016/j.tibtech.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Zamocky M, Herzog C, Nykyri LM, Koller F. Site-directed mutagenesis of the lower parts of the major substrate channel of yeast catalase A leads to highly increased peroxidatic activity. FEBS Lett. 1995;367:241–245. doi: 10.1016/0014-5793(95)00568-t. [DOI] [PubMed] [Google Scholar]
- 8.Schmitt J, Brocca S, Schmid RD, Pleiss J. Blocking the tunnel: Engineering of Candida rugosa lipase mutants with short chain length specificity. Protein Eng. 2002;15:595–601. doi: 10.1093/protein/15.7.595. [DOI] [PubMed] [Google Scholar]
- 9.Chaloupkova R, Sykorova J, Prokop Z, Jesenska A, Monincova M, Pavlova M, Tsuda M, Nagata Y, Damborsky J. Modification of activity and specificity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26 by engineering of its entrance tunnel. J. Biol. Chem. 2003;278:52622–52628. doi: 10.1074/jbc.M306762200. [DOI] [PubMed] [Google Scholar]
- 10.Fedorov R, Vasan R, Ghosh DK, Schlichting I. Structures of nitric oxide synthase isoforms complexed with the inhibitor AR-R17477 suggest a rational basis for specificity and inhibitor design. Proc. Natl Acad. Sci. USA. 2004;101:5892–5897. doi: 10.1073/pnas.0306588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotik M, Stepanek V, Kyslik P, Maresova H. Cloning of an epoxide hydrolase-encoding gene from Aspergillus niger M200, overexpression in E. coli, and modification of activity and enantioselectivity of the enzyme by protein engineering. J. Biotechnol. 2007;132:8–15. doi: 10.1016/j.jbiotec.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Feingersch R, Shainsky J, Wood TK, Fishman A. Protein engineering of toluene monooxygenases for synthesis of chiral sulfoxides. Appl. Environ. Microbiol. 2008;74:1555–1566. doi: 10.1128/AEM.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damborsky J, Brezovsky J. Computational tools for designing and engineering biocatalysts. Curr. Opin. Chem. Biol. 2009;13:26–34. doi: 10.1016/j.cbpa.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin ACR. Mapping PDB chains to UniProtKB entries. Bioinformatics. 2005;21:4297–4301. doi: 10.1093/bioinformatics/bti694. [DOI] [PubMed] [Google Scholar]
- 16.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labarga A, Valentin F, Anderson M, Lopez R. Web services at the European bioinformatics institute. Nucleic Acids Res. 2007;35:W6–W11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: A resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J. CAVER: A new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medek P, Benes P, Sochor J. Computer Graphics and Imaging 2008. Austria: Innsbruck; 2008. Multicriteria tunnel computation; p. 5. [Google Scholar]
- 23.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayrose I, Graur D, Ben-Tal N, Pupko T. Comparison of site-specific rate-inference methods for protein sequences: Empirical Bayesian methods are superior. Mol. Biol. Evol. 2004;21:1781–1791. doi: 10.1093/molbev/msh194. [DOI] [PubMed] [Google Scholar]
- 27.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: An algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18:S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 28.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawabata T, Ota M, Nishikawa K. The Protein Mutant Database. Nucleic Acids Res. 1999;27:355–357. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman J, Peat TS, Richard R, Kan L, Swanson PE, Affholter JA, Holmes IH, Schindler JF, Unkefer CJ, Terwilliger TC. Haloalkane dehalogenases: Structure of a Rhodococcus enzyme. Biochemistry. 1999;38:16105–16114. doi: 10.1021/bi9913855. [DOI] [PubMed] [Google Scholar]
- 31.Kretz KA, Richardson TH, Gray KA, Robertson DE, Tan X, Short JM. Gene site saturation mutagenesis: A comprehensive mutagenesis approach. Methods. Enzymol. 2004;388:3–11. doi: 10.1016/S0076-6879(04)88001-7. [DOI] [PubMed] [Google Scholar]
- 32.Gray KA, Richardson TH, Kretz K, Short JM, Bartnek F, Knowles R, Kan L, Swanson PE, Robertson DE. Rapid evolution of reversible denaturation and elevated melting temperature in a microbial haloalkane dehalogenase. Adv. Synth. Catal. 2001;343:607–617. [Google Scholar]
- 33.Bosma T, Damborsky J, Stucki G, Janssen DB. Biodegradation of 1,2,3-trichloropropane through directed evolution and heterologous expression of a haloalkane dehalogenase gene. Appl. Environ. Microbiol. 2002;68:3582–3587. doi: 10.1128/AEM.68.7.3582-3587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benning MM, Shim H, Raushel FM, Holden HM. High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta. Biochemistry. 2001;40:2712–2722. doi: 10.1021/bi002661e. [DOI] [PubMed] [Google Scholar]
- 35.Gopal S, Rastogi V, Ashman W, Mulbry W. Mutagenesis of organophosphorus hydrolase to enhance hydrolysis of the nerve agent VX. Biochem. Biophys. Res. Commun. 2000;279:516–519. doi: 10.1006/bbrc.2000.4004. [DOI] [PubMed] [Google Scholar]
- 36.Watkins LM, Mahoney HJ, McCulloch JK, Raushel FM. Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase. J. Biol. Chem. 1997;272:25596–25601. doi: 10.1074/jbc.272.41.25596. [DOI] [PubMed] [Google Scholar]
- 37.Chen-Goodspeed M, Sogorb MA, Wu F, Hong SB, Raushel FM. Structural determinants of the substrate and stereochemical specificity of phosphotriesterase. Biochemistry. 2001;40:1325–1331. doi: 10.1021/bi001548l. [DOI] [PubMed] [Google Scholar]
- 38.Hahn M, Pons J, Planas A, Querol E, Heinemann U. Crystal structure of Bacillus licheniformis 1,3-1,4-beta-D-glucan 4-glucanohydrolase at 1.8 A resolution. FEBS Lett. 1995;374:221–224. doi: 10.1016/0014-5793(95)01111-q. [DOI] [PubMed] [Google Scholar]
- 39.Pons J, Querol E, Planas A. Mutational analysis of the major loop of Bacillus 1,3-1,4-beta-D-glucan 4-glucanohydrolases. Effects on protein stability and substrate binding. J. Biol. Chem. 1997;272:13006–13012. doi: 10.1074/jbc.272.20.13006. [DOI] [PubMed] [Google Scholar]
- 40.Lobkovsky E, Billings EM, Moews PC, Rahil J, Pratt RF, Knox JR. Crystallographic structure of a phosphonate derivative of the Enterobacter cloacae P99 cephalosporinase: mechanistic interpretation of a beta-lactamase transition-state analog. Biochemistry. 1994;33:6762–6772. doi: 10.1021/bi00188a004. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z, Yu Y, Musser JM, Palzkill T. Amino acid sequence determinants of extended spectrum cephalosporin hydrolysis by the class C P99 beta-lactamase. J. Biol. Chem. 2001;276:46568–46574. doi: 10.1074/jbc.M102757200. [DOI] [PubMed] [Google Scholar]
- 42.Vakulenko SB, Golemi D, Geryk B, Suvorov M, Knox JR, Mobashery S, Lerner SA. Mutational replacement of Leu-293 in the class C Enterobacter cloacae P99 beta-lactamase confers increased MIC of cefepime. Antimicrob. Agents. Chemother. 2002;46:1966–1970. doi: 10.1128/AAC.46.6.1966-1970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markiewicz P, Kleina LG, Cruz C, Ehret S, Miller JH. Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as ‘spacers’ which do not require a specific sequence. J. Mol. Biol. 1994;240:421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 44.Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J. Mol. Biol. 1991;222:67–88. doi: 10.1016/0022-2836(91)90738-r. [DOI] [PubMed] [Google Scholar]
- 45.Axe DD, Foster NW, Fersht AR. A search for single substitutions that eliminate enzymatic function in a bacterial ribonuclease. Biochemistry. 1998;37:7157–7166. doi: 10.1021/bi9804028. [DOI] [PubMed] [Google Scholar]
- 46.Wrobel JA, Chao SF, Conrad MJ, Merker JD, Swanstrom R, Pielak GJ, Hutchison CA. A genetic approach for identifying critical residues in the fingers and palm subdomains of HIV-1 reverse transcriptase. Proc. Natl Acad. Sci. USA. 1998;95:638–645. doi: 10.1073/pnas.95.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]