Abstract
MicroRNAs (miRNAs) are small (19–24 nt), nonprotein-coding nucleic acids that regulate specific ‘target’ gene products via hybridization to mRNA transcripts, resulting in translational blockade or transcript degradation. Although miRNAs have been implicated in numerous developmental and adult diseases, their specific impact on biological pathways and cellular phenotypes, in addition to miRNA gene promoter regulation, remain largely unknown. To improve and facilitate research of miRNA functions and regulation, we have developed MMIA (microRNA and mRNA integrated analysis), a versatile and user-friendly web server. By incorporating three commonly used and accurate miRNA prediction algorithms, TargetScan, PITA and PicTar, MMIA integrates miRNA and mRNA expression data with predicted miRNA target information for analyzing miRNA-associated phenotypes and biological functions by gene set analysis, in addition to analysis of miRNA primary transcript gene promoters. To assign biological relevance to the integrated miRNA/mRNA profiles, MMIA uses exhaustive human genome coverage, including classification into various disease-associated genes as well as conventional canonical pathways and Gene Ontology. In summary, this novel web server (cancer.informatics.indiana.edu/mmia) will provide life science researchers with a valuable tool for the study of the biological (and pathological) causes and effects of the expression of this class of interesting protein regulators.
INTRODUCTION
MicroRNAs (miRNAs) are a class of endogenous (nonprotein-coding), 19–24 nt single-stranded RNAs that derive from a stem–loop precursor to regulate gene expression by binding primarily to the 3′-UTR of specific ‘target’ mRNAs, resulting in the disruption of mRNA stability and/or translation (1). Due to their posttranscriptional regulatory effects, miRNAs act to ‘fine tune’ the levels of proteins involved in numerous biological processes, including embryogenesis, organogenesis, tissue homeostasis, immune system function and cell cycle control (2–4). The biosynthesis and maturation of miRNAs is quite distinct from that of small inhibitory RNAs (siRNAs), involving processing of a several kb primary RNA transcript to the 70–80 base stem loop pre-miRNA. The stem–loop precursor is then exported from the nucleus and further processed by the endoribonuclease Dicer to produce the mature, single-stranded molecule that is incorporated into the RNA-induced silencing complex (RISC) and possesses a 6 nt ‘seed’ sequence (nucleotide positions 2–7) that primarily targets complementary regions within the 3′-untranslated regions of mRNA transcripts (5). As a general rule, perfect miRNA seed-to-target complementarity results in degradation of the mRNA target transcript, but can also block protein synthesis at the level of translation (5).
Due to their integral roles in tissue generation and homeostasis, disruptions in miRNA expression have now been linked to numerous pathologies, including those related to neuronal development (6), cardiac function (7), immune system (4,8), neurodegeneration (e.g. Alzheimer's disease) (9–11), skin (e.g. psoriasis) (12) and numerous malignancies (2,4,8,13). The extensive association of miRNAs with human cancer has led to the acronym ‘oncomirs’, with the majority of these being tumor suppressors, but a significant number of oncomirs being oncogenic (13). In support of tumor suppressor function, one study found that over 52% of annotated miRNA genes reside in so-called ‘fragile sites’ of cancer-associated chromosomal instability (14). Another microarray study of 217 miRNAs in 334 primary human tumors revealed miRNA expression clusters distinct to 20 different malignancies (40). Consequently, due to the widespread association of specific miRNAs with malignant disease, these gene regulatory molecules hold promise as diagnostic, prognostic or treatment-response biomarkers, in addition to representing possible therapeutic targets.
In comparison to annotated protein-coding genes, very little is currently known regarding regulation of primary miRNA (pri-miRNA) transcripts. A handful of studies have examined epigenetic regulation of pri-miRNA genes (15–17), while Drosphila embryonic miRNAs were found to be regulated in a spatial manner (18). A more recent study utilized a transcriptionally permissive histone ‘mark’ to locate possible pri-miRNA promoters (19). However, although pri-miRNAs are transcribed by RNA polymerase II, the detailed transcriptional regulation (e.g. binding of transcription factors, coactivators, corepressors) remains unknown for the vast majority of miRNA genes. Additionally, despite the extensive role of miRNAs in development, homeostasis and numerous disease states, relatively little is known regarding mature miRNA regulation of specific biological pathways and cellular phenotypes (2). As the biological effects of miRNAs are due to their modulation of target protein expression, accurate miRNA target prediction is essential to any study of miRNA function. While databases of experimentally validated miRNA targets, including MiRecords (20) and TarBase (21), are becoming more extensive, bioinformatic algorithms remain the principal means of predicting targets of specific miRNAs. These algorithms take into account numerous parameters that influence miRNA/target interactions, including seed match complementarity, 3′-UTR seed match context, seed match conservation, favorability of free energy binding, AU content and binding site accessibility (1). Currently, three of the most widely used, and predictively accurate algorithms are TargetScan, PITA and PicTar, although more recent prediction methods, such as Miranda and mirWIP, are also increasingly being used (1).
As previously noted, perfect seed-to-target complementarity is most prominently associated with transcript degradation (as opposed to translational blockage). Consequently, target prediction algorithms requiring stringent seed matching can be used to determine likely miRNA target regulation by identifying (e.g. by microarray or high-throughput sequencing) inversely correlated expression of miRNAs and their predicted target mRNA transcripts. Moreover, by subjecting those specific inversely regulated miRNA-target transcripts to standard gene ontology and pathway analyses, determinations can be made regarding the likely biological and phenotypic effects of the misexpressed miRNAs.
While such an integrated approach (miRNA target ontology/pathway analysis) is fairly straightforward, it requires the sequential use of multiple demanding and time-consuming computational analyses. One study subjected publically available miRNA datasets, from five specific cancer types, to miRgate Gene Ontology (GO) analysis to categorize GO groups likely impacted by those cancer-dysregulated miRNAs, while also identifying miRNA target proteins affected by anticancer drugs (22). Other miRNA prediction algorithms/databases have now incorporated experimentally validated miRNA:target interactions. A previously developed database, miRGator, can classify miRNA targets (as determined by the miRanda, PicTar and TargetScanS algorithms) according to GO terms and disease associations (23). Similarly, miRNAMap 2.0 incorporates both publically available miRNA and mRNA expression databases, and provides a list of tissue-specific miRNAs, but does not include pathway or GO analyses (24), while conversely, a Microsoft Excel-based program compares experimental gene expression data to publically available miR target predictions (25). While some analysis tools, such as miRNApath (26) and DIANA-miRPath (diana.cslab.ece.ntua.gr/pathways/) support pathway analyses, for most currently available miRNA databases/target predictors, the gene expression data or predicted miRNA targets must be separately subjected to biological classification tools, such as Gene Set Analysis (GSA) (27). Likewise, unlike extensive databases of gene expression-based disease ‘signatures’ (28–30), we are aware of only one such algorithm for associating miRNA expression profiles with specific diseases (31).
Despite their noteworthy objective of miRNA target classification, several limitations remain for these previously developed tools. First, most of these utilize only mRNA microarray (or publicly available) gene expression profiles, while others are limited to providing the predicted targets of only one miRNA per analysis session. Currently available tools for analyzing multiple miRNAs cannot analyze mRNA expression data using GSA. Second, current miRNA analysis tools are restricted to supporting pathways, GO, or diseases only individually, while most mRNA analysis tools can support all three of these in one combined algorithm. Also, to our knowledge, there is currently no comprehensive tool that integrates both miRNA and mRNA expression data, in combination with pri-miRNA promoter transcription factor-binding site analysis, within a single web interface. Finally, miRNA gene set analysis based on direct relationship between a biological term and its miRNA gene list is not currently available (for example, a miRNA gene set analysis including a transcription factor (TF) term enriched in pri-miRNA regions and a disease term highly related to miRNAs based on publications). Since the roles of transcription factors in a miRNA promoter region have been elucidated in embryonic stem cells (19), miRNA/TF gene sets have now gained importance. These miRNA gene sets could be constructed similarly to other mRNA gene sets, based on either literature or experimental data.
To address these limitations, we have now designed an integrated miRNA/mRNA-analyzing web server, MMIA (microRNA and mRNA integrated analysis), that combinatorially analyzes both miRNA expression data (or miRNA gene list) and mRNA expression data, based on a recent method that can also analyze multiple miRNA genes (32). MMIA thus provides a ‘one-stop’ combined analysis of the miRNA/mRNA input data for various gene sets including pathways, GO, cancer, genetic/chemical perturbation and diseases. We have now compiled miRNA/TF gene sets and miRNA/disease gene sets for our additional analysis. In summary, we have implemented a practical, user-friendly web server for extensive biological functional analysis of multiple miRNAs. We believe MMIA will be a valuable tool, in combination with traditional mRNA GSA tools, for the biomedical research community.
MATERIALS AND METHODS
Combined analysis of miRNA and mRNA
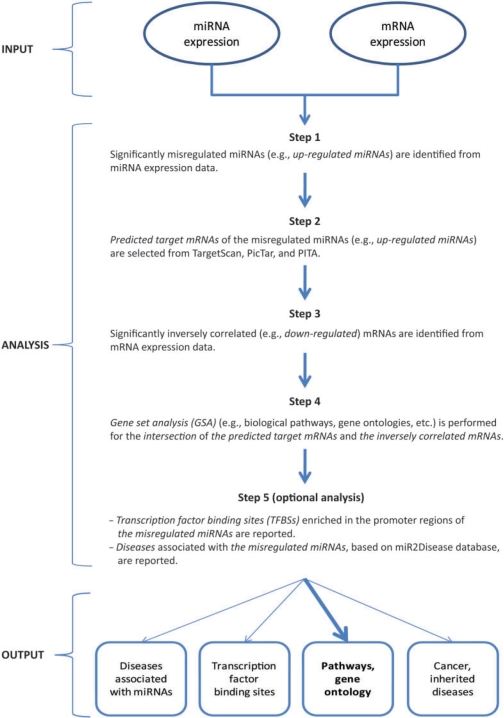
The MMIA web-server is a novel approach for integrating miRNA and mRNA expression data, based on our recent study (32). Although the precise mechanism(s) by which miRNAs induce mRNA degradation and regulate protein expression by posttranscriptional silencing have not been fully elucidated (33,34), perfect miRNA seed:target complementarity is predominantly associated with target gene destabilization (33). Thus, MMIA compares inverse expression of mRNAs and miRNAs (e.g. upregulated miRNAs and downregulation of their predicted target mRNAs). To use MMIA, the user first selects significantly up- or down-regulated miRNAs from ‘group 2’ on the input data webpage (Supplementary Figure 1). The first dataset (e.g. the control), in our customized ‘SIP’ file format (see below and documentation web page), and any subsequent replicates of that dataset, are automatically assigned to ‘group 1’, while the second (e.g. experimental) dataset (with its replicates) is designated ‘group 2’. For example, in Step 1 (MMIA web page, Supplementary Figure 1), if up-regulated miRNAs are selected, MMIA identifies the significantly (using user-defined statistical cutoff values) up-regulated group 2 miRNAs (as compared to group 1). If, however, a user selects down-regulated miRNAs, MMIA identifies significantly less expressed miRNAs in group 2. As described in Figure 1, when upregulated miRNAs are selected, MMIA identifies all significantly down-regulated mRNAs in the mRNA expression data analysis. The mRNAs common to the predicted target mRNAs and the down-regulated mRNAs are then selected, with MMIA determining the gene intersection by GSA analysis using various predefined gene set databases, including KEGG (35), MIT MSigDB database v2.5 (36) and G2D (37). GSA differs from individual gene analysis (IGA) by directly scoring predefined gene sets for misexpression (27); GSEA, one type of GSA analysis developed by Subramanian et al. (36), is a widely used algorithm that utilizes a competitive statistic (‘score function’) to represent differentially expressed genes within a specific gene set. MMIA also performs additional analyses, including transcription factor binding site identification within the pri-miRNA gene promoters of upregulated miRNAs, in addition to miRNA associations with specific diseases (31).
Figure 1.
Overview of the combined analysis of microRNA and mRNA expression data in MMIA, based on our previous published methodology (32).
Data sources
MMIA utilizes two major gene set databases, one for mRNA gene sets and the other for miRNA gene sets. The mRNA gene sets consist of canonical pathways, positional information, chemical/genetic perturbation, GO, cancer genes and inherited diseases (35,36). These mRNA sets come from KEGG (35), MIT MSigDB (36) and G2D (37). MSigDB has positional gene sets, chemical/genetic perturbation, canonical pathway gene sets, cancer gene sets and GO gene sets, while G2D contains inherited disease gene sets. Our additional miRNA gene sets are composed of transcription factor binding sites (genome.ucsc.edu) (38) in annotated pri-miRNA regions and a disease-related miRNA database, miR2Disease (31). Each miRNA gene set consists of one biological term and its corresponding miRNA gene list. The pri-miRNA regions are defined using a recent annotation method (19), and the transcription factor-binding site information was obtained from the UCSC Genome Browser TFBS Conserved track (38). These binding sites are conserved using human/mouse/rat multiple species alignments.
MMIA uses three miRNA target prediction algorithms, TargetScan (39), PicTar (PicTar 4 species conservation, PicTar 5 species conservation) (40) and PITA (probability of interaction by target accessibility) (41) that require stringent miRNA seed:target complementarity (thus favoring mRNA transcript degradation). We obtained TargetScan release 4.2 (Apr. 2008) (www.targetscan.org) and collected conserved sites, while PicTar was downloaded from the UCSC Genome Browser human NCBI Build 35. We obtained PITA (version 6, 31-Aug-08) Sites catalog (3/15 flank) from genie.weizmann.ac.il/pubs/mir07/mir07_data.html. PITA identifies initial seeds for each miRNA 3′-UTR binding site and then combines sites for the same miRNA to obtain a total miRNA:target interaction score, while also accounting for site accessibility (41). PicTar searches miRNAs for multiple alignments to potential orthologous 3′-UTR sequences and subsequently ranks genes by optimal free energies of binding, while also accounting for synergistic effects of multiple binding sites of one or more miRNAs and the number of evolutionarily conserved putative 3′-UTR binding sites (40). TargetScan, currently the most widely used miR target prediction tool, relies on strict miRNA seed region (miRNA bases 2–7) complemantarity, but also considers bases 1 and 8, in addition to the context of the miRNA-binding site, the proximal AU composition, and proximity to sites for co-clustered miRNAs (thus enhancing cooperative action) (42).
For gene expression analysis, Affymetrix chip annotation information (for 10 separate microarray platforms) was downloaded from the Affymetrix web page (www.affymetrix.com). A non-Affymetrix platform can also be selected by ‘custom platforms’ option on the MMIA web page (Supplementary Figure 1), a feature supporting different probe names, including Ensemble Transcript (www.ensembl.org), NCBI RefSeq, mRNA/protein and Entrez Gene (www.ncbi.nlm.nih.gov), Swiss-Prot (www.ebi.ac.uk/swissprot) and Gene Symbol (www.genenames.org).
The chip annotation information and miRNA target information in MMIA are stored in a MySQL database, the gene sets are stored in MIT GSEA GMT and CHIP file formats (www.broad.mit.edu/cancer/software/gsea/wiki/index.php/Data_formats), and the executables were written in Perl and R. Since the data sources utilize different gene annotations, these are converted to the nomenclature used in the UCSC Genes (43) annotation system.
WEB SERVER DESCRIPTION
Procedure
Using the MMIA tool is quite straightforward, starting with the input web page, which is composed of four mandatory steps and a fifth optional step (Supplementary Figure 1), while the procedures in MMIA follow the same order shown in Figure 1. For steps 1 and 3 of the web page, users upload miRNA and mRNA expression data, respectively. In Step 1, the user submits a miRNA gene list and is also prompted to select either up- or down-regulated miRNAs for data analysis. Step 2 provides not only selection of the desired miRNA target prediction algorithm (TargetScan, PicTar or PITA) (39–41) but also allows a combinatorial analysis, based on the intersection of two of the algorithms. Step 4 of the web page allows users to select gene sets for GSA analysis, based on our recently described method (32). Step 5 provides optional analyses of examining enriched transcription factor binding sites within miRNA primary transcript promoter regions, disease associations, and an alternative miRNA–mRNA combined analysis, using MIT GSEA software (36).
Input
The miRNA and mRNA expression data input for MMIA uses a simple tab-delimited data format, followed by two header lines for the group and dataset descriptions. We defined the tab-delimited file format called SIP. Users divide test datasets into two groups and then specify them in the first header line. The second header line consists of a descriptive name for each sample column. To provide further versatility, users can also enter a regulated miRNA list in the miRNA input field (box under ‘Regulated miRNA list’) on the web page.
Gene/probe identifiers in expression data or SIP format can consist of Ensemble Transcript (www.ensembl.org), NCBI RefSeq mRNA/protein and Entrez Gene (www.ncbi.nlm.nih.gov), Swiss-Prot (www.ebi.ac.uk/swissprot), Gene Symbol and Affymetrix probe sets (www.affymetrix.com). For non-Affymetrix mRNA expression probe sets, users should select the ‘Custom’ chip platform option in Step 3 (Supplementary Figure 1). The input expression data allows duplicated probe names in both miRNA and mRNA microarrays. Although the expression data is linear (not log-2 scale), data values can easily be converted to antilog 2. When only log fold-change values and P-values are available (in an EXCEL sheet format, for example), users can also easily convert the data into the SIP file format, as shown in the documentation web page.
Currently, MMIA supports only two-group comparison analysis. MMIA, with the first dataset in a SIP file assigned to group 1 and the other dataset to group 2. The first data samples in both miRNA and mRNA arrays belong to the same sample group. In most cases, the first sample (group 1) would correspond to a control group and the group 2 to an experimental group. The fold-change notation in MMIA is a median of group 2 samples over a median of group 1 samples. When the user pastes a miRNA gene list and uploads mRNA expression data into MMIA, by default, MMIA assumes that the miRNAs are down-regulated in group 2.
For expression data, MMIA provides a number of preprocessing and test options. The preprocessing options can be used to remove genes with extremely low or high values, data transformation into log-2 scale, and standardization of gene expression values from multiple microarrays, as based on a Bioconductor tutorial (www.bioconductor.org/workshops/2002/Summer02Course/). The test options are used to find miRNA/mRNA transcripts that are significantly differentially expressed between the two groups. When the biclustering option in the miRNA analysis is used, MMIA generates a cluster figure of significantly differentially expressed miRNAs, with three test options: user-defined fold-change, user-defined t-test statistics (P-value), or combination of the two. Users can also use a false discovery rate correction (44) in t-test statistics.
Output
The output shows not only significantly differentially expressed miRNAs and their predicted mRNA target information, but also miRNA-mRNA expression combined analysis based on one-sided Fisher's exact test for the selected gene sets. The combined miRNA and mRNA analysis is performed for the gene sets in terms of the intersection between significantly differentially expressed mRNA and predicted target mRNAs by multiple up-/down-regulated miRNA genes.
An example MMIA output, which consisting of five sections, is shown in Supplementary Figure 2. Group descriptions and sample names, based on miRNA/mRNA SIP files provided by the user, are summarized in the first section. The second section lists either up- or down-regulated miRNAs from miRNA expression data analysis, selected by the user, and also provides a hierarchical clustering figure for the miRNAs. The third section is the miRNA gene set analysis. This section describes TFBSs enriched in primary transcript regions of the selected miRNAs and diseases associated with the miRNAs. The fourth section provides a predicted target mRNA list of the selected miRNAs and also shows target mRNA expression levels. The main result of the combined analysis (based on Xin, et al.) (32), is shown in the fifth section.
CONCLUSION
MMIA is a novel web tool that analyzes both miRNA and mRNA expression data simultaneously, providing 5782 gene sets encompassing numerous pathways, functions, cancer genes, chromosomal position and inherited diseases. In addition, MMIA signal transduction, cell cycle, chromatin remodeling, TF and stemness gene sets also provide biological implications of miRNA expression. MMIA also provides miRNA gene sets based on TF binding sites in pri-miRNA regions and a manually curated disease/miRNA database, and we will compile more gene sets in the future, in addition to considering miRNA translational repression (at the protein level). In summary, we strongly believe that MMIA will be a highly informative data analysis tool for the life sciences research community, and will provide valuable information regarding the role of microRNAs in numerous normal and disease states.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (CA085289, CA113001 to K.P.N.); Walther Cancer Foundation, Indianapolis, IN (to S.N., K.P.N.); Ovar’coming Together (Indianapolis, IN; to C.B.). Funding for open access charge: National Cancer Institute grants U54 CA113001 and R01CA 085289.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Fuxiao Xin for constructive discussion in the web server development. They also thank Dr Matthew Burow for providing us with a test expression dataset.
REFERENCES
- 1.Bartel DP. MiRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asli NS, Pitulescu ME, Kessel M. MiRNAs in organogenesis and disease. Curr. Mol. Med. 2008;8:698–710. doi: 10.2174/156652408786733739. [DOI] [PubMed] [Google Scholar]
- 3.Gangaraju VK, Lin H. MiRNAs: key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao C, Rajewsky K. MiRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 5.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Divakaran V, Mann DL. The emerging role of miRNAs in cardiac remodeling and heart failure. Circ. Res. 2008;103:1072–1083. doi: 10.1161/CIRCRESAHA.108.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schickel R, Boyerinas B, Park SM, Peter ME. MiRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 9.Boissonneault V, Plante I, Rivest S, Provost P. MiRNA-298 and miRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J. Biol. Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J. Biol. Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson PT, Keller JN. RNA in brain disease: no longer just “the messenger in the middle”. J. Neuropathol. Exp. Neurol. 2007;66:461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- 12.Sand M, Gambichler T, Sand D, Skrygan M, Altmeyer P, Bechara FG. MiRNAs and the skin: Tiny players in the body's largest organ. J. Dermatol. Sci. 2009;53:169–175. doi: 10.1016/j.jdermsci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs – miRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human miRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roman-Gomez J, Agirre X, Jimenez-Velasco A, Arqueros V, Vilas-Zornoza A, Rodriguez-Otero P, Martin-Subero I, Garate L, Cordeu L, Jose-Eneriz ES, et al. Epigenetic regulation of miRNAs in acute lymphoblastic leukemia. J. Clin. Oncol. 2009;27:1316–1322. doi: 10.1200/JCO.2008.19.3441. [DOI] [PubMed] [Google Scholar]
- 16.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. A miRNA DNA methylation signature for human cancer metastasis. Proc. Natl Acad. Sci. USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber B, Stresemann C, Brueckner B, Lyko F. Methylation of human miRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6:1001–1005. doi: 10.4161/cc.6.9.4209. [DOI] [PubMed] [Google Scholar]
- 18.Biemar F, Zinzen R, Ronshaugen M, Sementchenko V, Manak JR, Levine MS. Spatial regulation of miRNA gene expression in the Drosophila embryo. Proc. Natl Acad. Sci. USA. 2005;102:15907–15911. doi: 10.1073/pnas.0507817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting miRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for miRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal miRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gusev Y. Computational methods for analysis of cellular functions and pathways collectively targeted by differentially expressed miRNA. Methods. 2008;44:61–72. doi: 10.1016/j.ymeth.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of miRNAs. Nucleic Acids Res. 2008;36:D159–D164. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of miRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–D169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creighton CJ, Nagaraja AK, Hanash SM, Matzuk MM, Gunaratne PH. A bioinformatics tool for linking gene expression profiling results with public databases of miRNA target predictions. RNA. 2008;14:2290–2296. doi: 10.1261/rna.1188208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiromatzo AO, Oliveira TYK, Pereira G, Costa AY, Montesco CAE, Gras DE, Yosetake F, Vilar JB, Cervato M, Prado PRR, et al. miRNApath: a database of miRNAs, target genes and metabolic pathways. Genet. Mol. Res. 2007;6:859–865. [PubMed] [Google Scholar]
- 27.Nam D, Kim SY. Gene-set approach for expression pattern analysis. Brief Bioinform. 2008;9:189–197. doi: 10.1093/bib/bbn001. [DOI] [PubMed] [Google Scholar]
- 28.Xia Y, Campen A, Rigsby D, Guo Y, Feng X, Su EW, Palakal M, Li S. DGEM—a microarray gene expression database for primary human disease tissues. Mol. Diagn. Ther. 2007;11:145–149. doi: 10.1007/BF03256235. [DOI] [PubMed] [Google Scholar]
- 29.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgs BW, Elashoff M, Richman S, Barci B. An online database for brain disease research. BMC Genomics. 2006;7:70. doi: 10.1186/1471-2164-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for miRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xin F, Li M, Balch C, Thomson M, Fan M, Liu Y, Hammond SM, Kim S, Nephew KP. Computational analysis of miRNA profiles and their target genes suggests significant involvement in breast cancer antiestrogen resistance. Bioinformatics. 2009;25:430–434. doi: 10.1093/bioinformatics/btn646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by miRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 34.Pillai RS. MiRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Iratxeta C, Bork P, Andrade-Navarro MA. Update of the G2D tool for prioritization of gene candidates to inherited diseases. Nucleic Acids Res. 2007;35:W212–W216. doi: 10.1093/nar/gkm223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are miRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 40.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial miRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 41.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in miRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 42.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MiRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 44.Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.