Abstract
Recent interest in non-coding RNA transcripts has resulted in a rapid increase of deposited RNA structures in the Protein Data Bank. However, a characterization and functional classification of the RNA structure and function space have only been partially addressed. Here, we introduce the SARA program for pair-wise alignment of RNA structures as a web server for structure-based RNA function assignment. The SARA server relies on the SARA program, which aligns two RNA structures based on a unit-vector root-mean-square approach. The likely accuracy of the SARA alignments is assessed by three different P-values estimating the statistical significance of the sequence, secondary structure and tertiary structure identity scores, respectively. Our benchmarks, which relied on a set of 419 RNA structures with known SCOR structural class, indicate that at a negative logarithm of mean P-value higher or equal than 2.5, SARA can assign the correct or a similar SCOR class to 81.4% and 95.3% of the benchmark set, respectively. The SARA server is freely accessible via the World Wide Web at http://sgu.bioinfo.cipf.es/services/SARA/.
INTRODUCTION
It is now known that RNA molecules are essential for a wide range of biological processes (1–6), which is changing the view of RNA as a simple vector of genetic information and reinforcing the hypothesis on the original ‘RNA world’ (7,8). Biosynthesis and transcription regulation (1–3,5), enzymatic action (5) and chromosome replication (4) are some of the functions that RNA molecules are now known to perform. RNA structure determination, which is accelerating its pace of deposition in the Nucleic Acid Database (NDB) (9) and the Protein Data Bank (PDB) (10), is thus becoming an essential and necessary tool for RNA function annotation. Although there are not standard rules to infer function, at least for proteins (11–13), structure similarity is arguably one of the most reliable methods for comparative function annotation (14,15).
Several methods have already been developed for the alignment of two or more protein 3D structures (16). However, only few are available for RNA structure comparison (17–23). The PRIMOS and AMIGOS programs identify RNA structure motifs and compare RNA structures by describing them as a set of pseudo angles from the C4’ and P atom trace (18,20). Both programs are limited to the comparison of RNA structures with the same number of nucleotides and only a newer version of AMIGOS can perform a comparison of a given structure against a set of RNA structures. The ARTS program was introduced as a general method for RNA structure alignment (17,24). ARTS describes RNA molecules with a set of ‘quadrats’ composed by four phosphate atoms of two consecutive base-pairs and uses a bipartite graph to find the maximum number of aligned ‘quadrats’ between two RNA structures. The DIAL program, developed to compare RNA structures using a dynamic programming algorithm (19), computes global, local and semi-global alignments by taking into account sequence similarity, dihedral angles and base-pair information from the two aligned structures. DIAL can also return the Boltzmann pair probabilities of the resulting alignments. However, such computation would double the runtime, hence the default in the DIAL server is not to calculate the pair probabilities. More recently, the SARSA server was developed to align two or more RNA structures using a structural alphabet of 23 nucleotide conformations (22). Both, the DIAL and SARSA servers were developed and benchmarked for their ability detecting short RNA motifs in a set of RNA structures. In contrast, the SARA program (21), which implementation for function assignment of RNA structures is here introduced, was recently developed to align two RNA structures based on a unit-vector alignment strategy (25). Given its implementation, an alignment by SARA shorter than 20 nt is likely to be indistinguishable from random structure alignments. The SARA program can be considered as an alternative to existing alignment methods such as AMIGOS, ARTS, DIAL, and SARSA.
Structure-based function assignment requires identifying structural units and classifying them into annotated functional groups. Currently, only the Structural Classification of RNA (SCOR) database (26) offers such a systematic classification for RNA structures. SCOR was designed to provide a comprehensive perspective and understanding of RNA motif structure, function, tertiary interactions and their relationships. Structure elements in the SCOR database are organized in a directed acyclic graph architecture, which allows multiple parent classes for a given structure motif. Currently, SCOR stores the structure and function classification of 3D motifs contained in 579 RNA structures. Unfortunately, the SCOR database has not been updated since May 2004 and does not include an option for automatically classifying new RNA structures. Therefore, the SCOR database does not reflect the rapid increase of deposited RNA structures in the PDB. Only recently, the DART database (27), which relies in the ARTS program for the alignment of two RNA structures, proposed an automatically generated RNA structure classification that resulted in 94 clusters containing 1333 RNA structure motifs. In contrast to SCOR, DARTS allows for automatic classification of new structures by providing a user-friendly Web interface. However, the DARTS database does not include a function-based classification similar to the SCOR database. To overcome such limitations we introduce the implementation of our SARA program (21) for automatic function assignment based on the SCOR classification.
We begin by briefly describing the benchmark sets of RNA alignments for the development and evaluation of the SARA program as well as outlining the algorithms behind the SARA program (Method outline). Next, we detail the requirements for using the SARA web server (server details). Finally, we conclude by assessing the impact of the SARA server on the automatic annotation of the RNA structure space (Conclusions).
METHOD OUTLINE
RNA structure and alignment data sets
As of March 2009, the PDB stored a total of 1534 structure files containing at least one RNA chain. The initial list of RNA structures was further filtered by: (i) removing any RNA structure with missing heavy atoms, (ii) removing any RNA structure with less than 20 nucleotides and less than 3 base-pairs and (iii) removing redundancy at 100% sequence identity. The filtered data set, called RNA09, included 451 RNA chains from 409 PDB entries. Next, we run an all-against-all comparison of the entries in the RNA09 data set using the ‘align’ program for global sequence alignment without end gap penalty and with default parameters (28). This run resulted in 50 995 pair-wise alignments with sequence identity with respect to the length of the alignment below 25%, which constituted our BgALI data set. All pairs of structures in the BgALI data set were then realigned with the SARA program to obtain a background distribution of scores for pairs of unrelated RNA structures.
To assess the accuracy of SARA in functional classification, the SCOR database (version 2.0.3, October 2004) was used as a standard of truth. Although outdated, to our knowledge SCOR is the only available function-based classification of RNA structures. Three more functional data sets were collected from the SCOR database: (i) the FSCOR data set, which includes RNA chains with more than three base-pairs that are annotated with a unique deepest SCOR functional class, (ii) the R-FSCOR data set, which includes only representative structures clustered at 90% structure identity for each class in the FSCOR data set and (iii) the T-FSCOR data set, which includes all structures in the FSCOR data set not present in the R-SCOR data set. All data sets of RNA structures and alignments are summarized in Table 1 and available for download at http://sgu.bioinfo.cipf.es/datasets/.
Table 1.
Composition of the different RNA datasets used in this work
| Datasets | Number of chains | Number of alignments | Number of different SCOR functions |
|---|---|---|---|
| RNA09 | 451 | 101 475 | |
| BgALI | 451 | 50 995 | |
| FSCOR | 419 | 168 | |
| R-FSCOR | 192 | 168 | |
| T-FSCOR | 227 | 88 |
SARA program for RNA structure alignment
The SARA program (21) is based on a unit-vector alignment strategy previously implemented for the alignment of protein structures (25,29–31). Briefly, the unit-vector representation of an RNA, originally introduced as a tool to analyze molecular dynamics trajectories (29) and fast detection of common geometric substructure in proteins (25), is calculated as follows: (i) given an RNA structure with N nucleotides, for each i = 1, …, N–1 extract the vector from the i-th to the (i + 1)-th selected atoms; (ii) normalize all obtained vectors to a unit-distance, and place the tails of all normalized vectors at the origin of a unit-sphere; the resulting collection of normalized vectors is the unit-vector representation of the input RNA; (iii) the Unit-vector RMS (URMS) distance between two input RNA structures is the root-mean-square deviation (RMSD) between their corresponding normalized vectors after determining the rotation to minimize RMSD. Specifically, SARA aligns two RNA structures by selecting its C3′ or P atoms. If secondary structure information is used, SARA selects only C3′ or P atoms involved in the base-pairing as computed by the 3DNA program (32) and omits all other atoms. In such a case, RNA structures are represented with a set of three unit-vectors for each selected atoms forming a base-pair. The SARA program calculates unit-spheres using four or eight successive atoms, depending on the existence or not of base-paring information, respectively. The SARA program cannot compute an alignment between two structures with less than nine selected atoms. The comparison of consecutive unit-spheres generates an all-against-all similarity-scoring matrix, which is used in a dynamic programming procedure for the global alignment of two RNA structures using 0 end gap penalties (33). The output alignment is then refined by maximizing the number of equivalent atoms or base-pairs within 3.5 Å RMSD using a variant of the MaxSub algorithm, which ensures that the best local alignment is contained in the resulting alignment (34).
To assess the likely accuracy of the alignments, the SARA program calculates three different raw scores: (i) percentage of structural identity (PSI), which is the percentage of superimposed C3′ nucleotide atoms within 4.0 Å with respect to the length (N) of the shorter of the two structures; (ii) percentage of secondary structure identity (PSS), which is the percentage of aligned base-pairs as defined by 3DNA within 4.0 Å with respect to the lowest number of base-pairs (NSS) in the two structures; and (iii) percentage of sequence identity (PID), which is the percentage of aligned nucleotides of the same type with respect to the length (N) of the shorter of the two structures. Additionally, the SARA program calculates P-values and their negative logarithms for the three identity scores, which estimate the probability of obtaining an equal- or better-scored alignment by chance. The logarithm of the P-value, which is independent of the raw score distributions, allows the combination of the three accuracy measures into a single score reported as the mean of the three negative logarithms for PSI, PSS and PID. The BgALI set of alignments was used to plot a background distribution of PSI and PID with respect to N and PSS with respect to NSS. Based on the analysis of the scores resulting from random structure alignments (35), such distributions were fitted into an extreme value distribution described by its mean (μ) and standard deviation (σ). Extreme value distributions has been previously used to describe the statistics of background structure alignments for both proteins (30) and RNA (21). The relationship between μ and σ and N or NSS can be extrapolated by fitting the points with high significance to the power law function Y = A × XB, where Y is μ or σ relative to PSI or PID and X is N (PSS and NSS when the alignment contains base-pairs, respectively). The values for the A and B parameters and the correlation coefficients of the fitting to an extreme value distribution are reported in Table 2.
Table 2.
Parameters for the extreme value distribution fitting
| PID |
PSS |
PSI |
||||
|---|---|---|---|---|---|---|
| μ | σ | μ | σ | μ | σ | |
| A | 75.4 | 630.4 | 444.2 | 519.7 | 644.3 | 779.4 |
| B | −0.569 | −1.132 | −0.869 | −1.148 | −0.727 | −1.059 |
| r | −0.915 | −0.947 | −0.985 | −0.946 | −0.986 | −0.934 |
A and B are the parameters that describe the power law function (Y = A × XB) and r is the associated correlation coefficient of the fitted data.
Structure-based function assignment
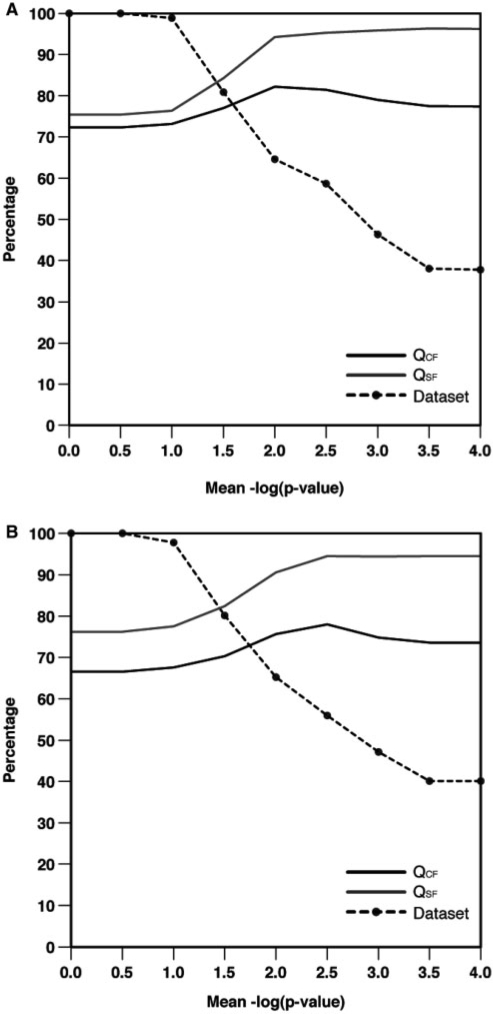
The SARA program can be used for structure-based function assignment by searching with a query structure against a representative data set of annotated RNA structures. SARA predicts the function of the query structure as the function of the top hit structure in the searched database (i.e. with the alignment that results in the largest mean of the three negative logarithms for PSI, PSS and PID). Such hit corresponds to the highest probability of correct assignment as measured by the ‘geodesic’ distance (d), which is the number of edges linking two SCOR annotations (36). Two RNA structures annotated in SCOR with the same function will have a d = 0 and two RNA structures differing at least in the deepest SCOR classification will have a d ≤ 2. To evaluate the accuracy of the SARA program for function assignment, we have performed two different tests: (i) a leave-one-out benchmark using the FSCOR data set and (ii) a benchmark annotating each of the structures in the T-FSCOR data set as query against the R-FSCOR data set. The accuracy of SARA for function assignment was evaluated by the fraction of corrected annotated SCOR functions (QCF) and the fraction of equal or similar assigned SCOR functions (QSF), which corresponds to d = 0 and d ≤ 2, respectively. The SARA program correctly assigns the exact or similar function to 81.4% and 95.3% of the FSCOR data set at the mean –log(P-value) cut-off of 2.5, respectively (Figure 1A). At this cut-off, 58.7% of the structures in the FSCOR data set were annotated by the SARA program (Figure 1A). Similar results were obtained for the benchmark using each of the structures in the T-FSCOR data set as query against the R-FSCOR data set. The accuracy of the SARA program was 78.0% and 94.5% for QCF and QSF, respectively, with 56.0% of data set coverage (Figure 1B). These results indicate that the accuracy of the SARA program for assigning the exact function decreases with the use of a representative set of the SCOR functions (i.e. the R-FSCOR data set), which has less structural diversity within each of the functional groups. However, the SARA program maintains the accuracy for assigning similar functions to those structures (i.e. d ≤ 2).
Figure 1.
Accuracy of structure-based function assignment by the SARA program. (A) QCF, QSF and dataset coverage as a function of the mean logarithm of the P-values for PSI, PSS and PID scores for the leave-one-out benchmark using the FSCOR dataset. (B) Same representation as in panel A for the T-FSCOR benchmark dataset using the R-FSCOR dataset for searching.
We have also evaluated the SARA program accuracy by calculating the area under its receiver operating characteristic (ROC) curve (AUC) (37). A ROC curve is obtained by plotting the false-positive rate against the corresponding true-positive rate for all possible cut-offs on the mean of the negative logarithm of P-values. The AUC, a threshold independent measure, is considered a robust indicator of a classifier quality given its independence from the selected threshold and its correlation with the probability of the classifier error (37). Thus, an AUC of one indicates a perfect classifier and an AUC of zero corresponds to a totally erroneous classification. The SARA program tested with the FSCOR benchmark data set resulted in an AUC of 0.61 and 0.83 for d = 0 and d ≤ 2, respectively. The prediction accuracy is maintained for the T-FSCOR benchmark set resulting in AUC values of 0.58 and 0.85 for d = 0 and d ≤ 2, respectively.
SERVER DETAILS
Pair-wise structure alignment
The SARA server for structure alignment requires the input of either two PDB/NDB codes or two coordinates files in PDB format (Figure 2A). Alternatively, the user can manually modify the default options of the SARA program by unchecking the ‘default options’ check box. Optional parameters include the open and extension gap penalties to be used during dynamic programming, the number of consecutive atoms to use in the unit-vector representation, the use of secondary structure information calculated by the 3DNA program, and the type of atom selected for calculating the unit vectors. When the secondary structure information option is selected, but the 3DNA program cannot calculate any base-pairs, SARA will use the single atom unit-vector alignment method. Moreover, the SARA server also aligns two RNA structures in the case when one of the two PDB contains only a phosphate-trace by automatically selecting the appropriate atom type. For pairs of structures under 1000 nucleotides of length, the results are normally reported within few seconds. The user is provided with a single output page that is divided in three sections containing: (i) the superposed coordinates of the two RNA input structures visualized using the Jmol applet (http://www.jmol.org) and as three downloadable files in the PDB, PIR and easy-to-read ALI formats, (ii) an easy-to-read sequence alignment that corresponds to the superposed structures, and (iii) all numerical results calculated from the superposition. Additionally, all the data shown in the output page is reported within the superposed coordinate file using the ‘REMARK’ field of the PDB format.
Figure 2.
User interface for the SARA server. (A) Pair-wise structure alignment. (B) Structure-based function assignment. Both panels include snapshots of the actual user interface as well as a flowchart of the actions taken by the back-end SARA program. User input and output are enclosed within the orange and green dashed areas, respectively.
Structure-based function assignment
The SARA server for function assignment requires the input of one PDB/NDB code or a coordinates file in PDB format (Figure 2B). Similar to the structure alignment interface of the SARA server, the user may select to change the default parameters including the possibility of selecting to do the structure search against RNA structures larger than 1000 nucleotides (i.e. including ribosomal RNA molecules). This last option requires more computational time, thus delaying the return of the results. By default, the SARA server will not include large structures and the search will be performed against a set of 162 RNA representative structures from the SCOR database (i.e. RNA structures shorter than 1000 nucleotides from the R-FSCOR data set). If the default option is changed, then the search is conducted over the whole set of 192 RNA representative structures, obtained by clustering at the 90% structure identity cut-off, all the structures within each of the SCOR functional class (i.e. the R-FSCOR data set). The search results are returned within few minutes for query structures of about 100 nucleotides. The output contains a sorted list of hits with details of each of the individual alignments such as: the alignment rank, the PDB code of the chain in the representative set, the length of the shorter of the two aligned structures, the lowest number of base-pairs between the two aligned structures, the percentage of sequence, secondary structure and tertiary structure identities (PID, PSS, PSI), the negative logarithms of the P-values relative to the three identity scores, their mean value and the probability of correct (d = 0) and similar (d ≤ 2) SCOR function assignments. The output list is sorted by the mean of the negative logarithm of the three P-values. All computed alignments and the PDB files containing the coordinates of the superimposed pairs of structures are stored for download in the SARA server for about 24 h. Finally, the SCOR functional classification of the representative RNA structures is reported in the output and reachable via a URL to the SCOR database.
Computational limitations
Due to its algorithmic implementation, the SARA program cannot align RNA structures shorter than 9 nt, which corresponds to the minimal number of atoms needed to calculate at least two sets of seven unit-vectors. Moreover, because the high computational requirements for aligning two very large RNA structures, the SARA server for both structure alignment and function assignment is currently limited to input RNA structures of less than 1000 nucleotide length, which covers 92% of all available RNA structures deposited in the PDB.
CONCLUSIONS
The SARA server, which relies in the SARA program for structure alignment, can perform two different tasks: (i) structure alignment of two input structures, which usually is returned within seconds of computational time (e.g. ∼10 s for input RNA structures of ∼100 nt) and (ii) function assignment of new RNA structures, which is returned usually within few minutes after submission (e.g. ∼5 min for input RNA structures of ∼100 nucleotides). The SARA server provides an alternative structure alignment method to ARTS (17), DIAL (19) and SARSA (22) as well as an alternative for function assignment to the DARTS database (27). The SARA program has a more general application with respect to previous structure alignment and function assignment methods, allowing the calculation of RNA structure alignment even when only the phosphate-trace is known and no secondary structure is available. In addition, it quickly reports a statistical significance of the alignment that can be used for assessing its relevance. To our knowledge, the SARA server represents the first method available on the Web for automatic classification of new RNA structures within the SCOR database. In the future, the SARA program could be used for studying the RNA structure and function space and defining a catalogue of RNA structures, which should help in characterizing how RNA molecules function.
AVAILABILITY AND REQUIREMENTS
SARA is freely available on the Internet at http://sgu.bioinfo.cipf.es/services/SARA/ and requires a Web browser that is capable of running the JMol applet. The web interface is programmed in PHP and the SARA method is programmed in Python. The SARA standalone program is available upon request.
FUNDING
Marie Curie International Reintegration Grant (FP6-039722); Spanish Ministerio de Ciencia e Innovación (BIO2007/66670). Funding for open access charge: BIO2007/66670.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Prof. Chin Lung Lu for making the SARSA benchmark data set available to us.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 3.Doudna JA. Structural genomics of RNA. Nat. Struct. Biol. 2000;7(Suppl.):954–956. doi: 10.1038/80729. [DOI] [PubMed] [Google Scholar]
- 4.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 5.Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3:e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grosshans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451:414–416. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- 7.Dennis C. The brave new world of RNA. Nature. 2002;418:122–124. doi: 10.1038/418122a. [DOI] [PubMed] [Google Scholar]
- 8.Ganem B. RNA world. Nature. 1987;328:676. doi: 10.1038/328676d0. [DOI] [PubMed] [Google Scholar]
- 9.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devos D, Valencia A. Practical limits of function prediction. Proteins. 2000;41:98–107. [PubMed] [Google Scholar]
- 12.Rost B, Liu J, Nair R, Wrzeszczynski KO, Ofran Y. Automatic prediction of protein function. Cell Mol. Life. Sci. 2003;60:2637–2650. doi: 10.1007/s00018-003-3114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedberg I. Automated protein function prediction – the genomic challenge. Brief Bioinform. 2006;7:225–242. doi: 10.1093/bib/bbl004. [DOI] [PubMed] [Google Scholar]
- 14.Whisstock JC, Lesk AM. Prediction of protein function from protein sequence and structure. Q. Rev. Biophys. 2003;36:307–340. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]
- 15.Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 2007;8:995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- 16.Carugo O. Recent progress in measuring structural similarity between proteins. Curr. Protein. Pept. Sci. 2007;8:219–241. doi: 10.2174/138920307780831839. [DOI] [PubMed] [Google Scholar]
- 17.Dror O, Nussinov R, Wolfson H. ARTS: alignment of RNA tertiary structures. Bioinformatics. 2005;21(Suppl. 2):ii47–ii53. doi: 10.1093/bioinformatics/bti1108. [DOI] [PubMed] [Google Scholar]
- 18.Duarte CM, Wadley LM, Pyle AM. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. Nucleic Acids Res. 2003;31:4755–4761. doi: 10.1093/nar/gkg682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferre F, Ponty Y, Lorenz WA, Clote P. DIAL: a web server for the pairwise alignment of two RNA three-dimensional structures using nucleotide, dihedral angle and base-pairing similarities. Nucleic Acids Res. 2007;35:W659–W668. doi: 10.1093/nar/gkm334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wadley LM, Keating KS, Duarte CM, Pyle AM. Evaluating and learning from RNA pseudotorsional space: quantitative validation of a reduced representation for RNA structure. J. Mol. Biol. 2007;372:942–957. doi: 10.1016/j.jmb.2007.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capriotti E, Marti-Renom MA. RNA structure alignment by a unit-vector approach. Bioinformatics. 2008;24:i112–i118. doi: 10.1093/bioinformatics/btn288. [DOI] [PubMed] [Google Scholar]
- 22.Chang YF, Huang YL, Lu CL. SARSA: a web tool for structural alignment of RNA using a structural alphabet. Nucleic Acids Res. 2008;36:W19–W24. doi: 10.1093/nar/gkn327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capriotti E, Marti-Renom MA. Computational RNA structure prediction. Curr. Bioinformatics. 2008;3:32–45. [Google Scholar]
- 24.Dror O, Nussinov R, Wolfson HJ. The ARTS web server for aligning RNA tertiary structures. Nucleic Acids Res. 2006;34:W412–W415. doi: 10.1093/nar/gkl312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chew LP, Huttenlocher D, Kedem K, Kleinberg J. Fast detection of common geometric substructure in proteins. J. Comput. Biol. 1999;6:313–325. doi: 10.1089/106652799318292. [DOI] [PubMed] [Google Scholar]
- 26.Tamura M, Hendrix DK, Klosterman PS, Schimmelman NR, Brenner SE, Holbrook SR. SCOR: Structural Classification of RNA, version 2.0. Nucleic Acids Res. 2004;32:D182–D184. doi: 10.1093/nar/gkh080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abraham M, Dror O, Nussinov R, Wolfson HJ. Analysis and classification of RNA tertiary structures. RNA. 2008;14:2274–2289. doi: 10.1261/rna.853208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson WR. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 29.Kedem K, Chew LP, Elber R. Unit-vector RMS (URMS) as a tool to analyze molecular dynamics trajectories. Proteins. 1999;37:554–564. [PubMed] [Google Scholar]
- 30.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yona G, Kedem K. The URMS-RMS hybrid algorithm for fast and sensitive local protein structure alignment. J. Comput. Biol. 2005;12:12–32. doi: 10.1089/cmb.2005.12.12. [DOI] [PubMed] [Google Scholar]
- 32.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 34.Siew N, Elofsson A, Rychlewski L, Fischer D. MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics. 2000;16:776–785. doi: 10.1093/bioinformatics/16.9.776. [DOI] [PubMed] [Google Scholar]
- 35.Abagyan RA, Batalov S. Do aligned sequences share the same fold? J. Mol. Biol. 1997;273:355–368. doi: 10.1006/jmbi.1997.1287. [DOI] [PubMed] [Google Scholar]
- 36.Bouttier J, Di Francesco P, Guitter E. Geodesic distance in planar graphs. Nucl. Phys. B. 2003;663:535–567. [Google Scholar]
- 37.Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]