Abstract
Homozygosity mapping is a common method for mapping recessive traits in consanguineous families. In most studies, applications for multipoint linkage analyses are applied to determine the genomic region linked to the disease. Unfortunately, these are neither suited for very large families nor for the inclusion of tens of thousands of SNPs. Even if less than 10 000 markers are employed, such an analysis may easily last hours if not days. Here we present a web-based approach to homozygosity mapping. Our application stores marker data in a database into which users can directly upload their own SNP genotype files. Within a few minutes, the database analyses the data, detects homozygous stretches and provides an intuitive graphical interface to the results. The homozygosity in affected individuals is visualized genome-wide with the ability to zoom into single chromosomes and user-defined chromosomal regions. The software also displays the underlying genotypes in all samples. It is integrated with our candidate gene search engine, GeneDistiller, so that users can interactively determine the most promising gene. They can at any point restrict access to their data or make it public, allowing HomozygosityMapper to be used as a data repository for homozygosity-mapping studies. HomozygosityMapper is available at http://www.homozygositymapper.org/.
INTRODUCTION
Homozygosity mapping, also called autozygosity mapping, is a common method for mapping recessive traits in consanguineous families. It is powerful because it does not require DNA of other family members than the affected offspring (1,2). The normal workflow consists of a genome-wide linkage analysis with microsatellites or, increasingly, SNPs (3). Especially for SNP markers, owing to their low informativity and hence the usually small number of informative meioses, this is mostly carried out with multipoint linkage analysis with software such as GENEHUNTER (4) or derivatives (5,6), Allegro (7), SIMWALK2 (8) or Merlin (9) under a recessive disease model. This is followed by the preparation of haplotypes either manually or by the software used for the analysis. Haplotypes are then manually inspected and searched for homozygous regions shared by all affected individuals who are homozygous by descent (if genotypes from ancestors are available) and are not homozygous in unaffected family members.
Currently, in some cases, when affected persons are known to be distant offspring of a consanguineous couple, the consanguineous marriage is introduced arbitrarily into a ‘virtual pedigree’. Unfortunately, wrong specification of the level of inbreeding has dramatic influence on the LOD scores obtained (10). This issue can be addressed by the inbreeding coefficient to reveal the pedigree structure and eventually permit the calculation of LOD scores (11,12). Other researchers prefer to arbitrarily introduce first or second cousin consanguinity into the pedigrees when there is evidence for inbreeding but details are not known (13). While such an approach renders the absolute value of the LOD score meaningless, it will nevertheless detect the correct genomic region when enough affected individuals (optionally supplemented by healthy siblings) are studied, albeit without any reliable measure of significance.
Computation of multipoint LOD scores and generation of haplotypes pose high demands on computational resources, are time consuming and largely depend on correct allele frequencies (14). The proper assessment of haplotypes becomes even more error-prone when no DNA from relatives is available because phase information is unknown and type I errors cannot be corrected (15). Beside heuristic approaches, most multipoint linkage applications use the Lander–Green algorithm (16) which scales linearly with the number of markers analysed and due to time constraints often only a subset (i.e. about 10 000) of the total SNPs is studied. Another drawback is that large families, especially those with a high level of inbreeding, have to be split because computational time increases drastically (17) or because the family size may simply become too large for the computational resources. Splitting pedigrees can however significantly reduce the information obtainable from them (18).
If studies comprise unrelated families with only one or few affected individuals, it may occur that they do not share a disease haplotype rare or long enough to be detectable even if they carry the same founder mutation [see the hypomagnesia example (19) on our website]. However, in consanguineous families with a rare recessive disease, it is very likely that the same disease allele has been inherited from both parents (1). Therefore, as long as the same locus is responsible for the phenotype, the proportion of homozygosity in the mutation's vicinity among individuals from different families should still be substantially higher than expected by chance. This is even the case when there is no common haplotype among different families due to ancient or even different mutations at the same locus. Especially in populations in which consanguinity is common, apparently unrelated individuals with the same phenotype are often found to be distantly related (20) and might hence share the same founder mutation albeit with only short shared disease haplotypes between families. Additionally to its presence in consanguineous families, autozygosity occurs also by chance and without known inbreeding (3,21–23) but the use of many non-related families makes it very unlikely that affected individuals from different families share the same autozygous region accidentally. It might hence even be possible to find disease genes in families with a more distant inbreeding background (13).
Several researchers have suggested methods to circumvent the problems posed by using linkage analysis software for autozygosity mappings. A simple approach is the genotyping of pooled DNA samples from affected individuals and the search for markers where only one allele is present or at least predominant (24). However, this method will fail in case of genetic heterogeneity (i.e. different homozygous genotypes), because no correlation between a single sample and a genotype is possible.
A computational solution is ExcludeAR (25), a Microsoft Excel spreadsheet, which searches for contiguous homozygous SNPs in the individuals included in the analysis. Due to its use within the spreadsheet application, the number of SNPs to be studied is limited to the number of rows supported by the version of Excel being used. Moreover, ExcludeAR can only handle data from up to four samples and copying and pasting of data is potentially error-prone and requires a manual effort for every operation making this solution suitable for small analyses but rather unfeasible for large studies and different models within one project. This approach was extended by AutoSNPa (26), a dedicated software designed to visualize homozygosity genome-wide in all patients and hence find regions of high homozygosity which may harbour the disease gene. Another method that avoids the generation of LOD scores is the detection of homozygous segments implemented in PLINK (27).
None of the alternatives to classic linkage analysis has yet become the common choice for homozygosity mappings. We believe that this is partly due to the researchers’ unwillingness to ‘risk’ the use of novel methods which might possibly be challenged by conservative reviewers. On the other hand, the software approaches mentioned above require at least some effort concerning installation, data preparation and familiarization.
To overcome the restraints posed by linkage software and the present applications for homozygosity mapping described above, we have developed a web-based approach to homozygosity mapping. In our tests with real and simulated genotypes, it always identified the same genomic regions as conventional linkage analyses. It does not require any installation or data preparation at all. All interfaces are well known HTML pages, so it is very easy and intuitive to use. Most of all, it is by orders of magnitudes faster than conventional linkage analysis. Data upload into our database and analysis of a typical project with six affected individuals with 50 000 genotypes each is completed in less than 5 min. A similar project on a 1 M array takes less than 30 min to upload and analyse the data. Benchmarks can be found on our website (http://www.homozygositymapper.org/documentation.html).
THE WEBSITE
Access
There is no login requirement to use HomozygosityMapper. However, while non-registered users can upload and analyse data, everyone else can access and also delete their data. Users wanting private access to their data or to store them, should therefore create themselves an account for HomozygosityMapper on the website. Access to a certain project can then be granted to (or revoked from, respectively) the public as well as to other registered users at will so that cooperation partners can participate in the analysis of one project without even seeing the other projects. Access permissions can be changed at any time. Also, complete projects or single analyses can be deleted by their owner at will.
HomozygosityMapper can thus be used to make the underlying data of a published study available, which is a common requirement in other fields such as gene expression studies.
Upload
Users can directly upload their genotype files to the database. In case of genotypes generated on Affymetrix platforms in which the SNP is identified by a proprietary Affymetrix ID, they must also choose the appropriate genotyping chip. The application offers the possibility to add new genotypes to existing projects. Despite the later supplement with recently genotyped samples, this also frees the users from combining different genotyping files (e.g. from the Affymetrix 100K set) into one. The users can also decide whether their data shall initially be kept confidential or made public. This setting can be changed later at any time.
During the data import, HomozygosityMapper screens all samples for blocks of homozygous genotypes in contiguous markers and stores the actual genotype together with the length of the block it resides in. To avoid false negative results, unknown genotypes are counted as homozygous here but stored as not determined. Marker positions are derived from dbSNP (28) and dubious markers are ignored.
Analysis
On the second interface, users can select one of their genotyping projects to be analysed. Here, some samples can be specified as cases (i.e. affected individuals) which will be used to calculate homozygosity scores. Other samples can be marked as controls; their genotypes are later shown in the genotype view to allow the exclusion of regions due to homozygosity in non-affected siblings and can be also used to calculate control allele frequencies. Users can decide whether they are interested in control frequencies of homozygous genotypes for the markers. These can be obtained from the HapMap project (29) for the correct population or calculated from the controls. Users can also select whether they want to restrict the maximum length of homozygous blocks considered for the determination of homozygosity scores (see below).
When the analysis starts, HomozygosityMapper reads the length of homozygous blocks in all affected samples for every marker and adds them to a homozygosity score for the respective marker. To prevent the inflation of this score by very long blocks in single individuals, any added block length is limited to an optimal value for each chip unless another length limit is specified by the users. This procedure yields homozygosity scores for each SNP which are stored in the database and can be searched for peaks. As a further measure, the observed frequency of homozygous genotypes in the affected individuals is calculated for each marker and compared with the homozygosity frequency in the controls.
Details on the analysis procedure can be found on our website.
Browsing the results
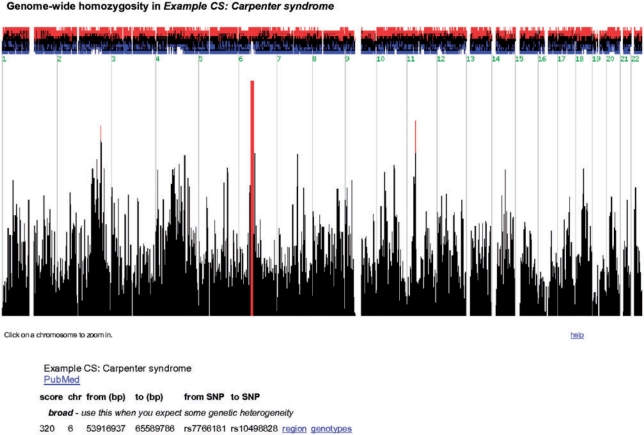
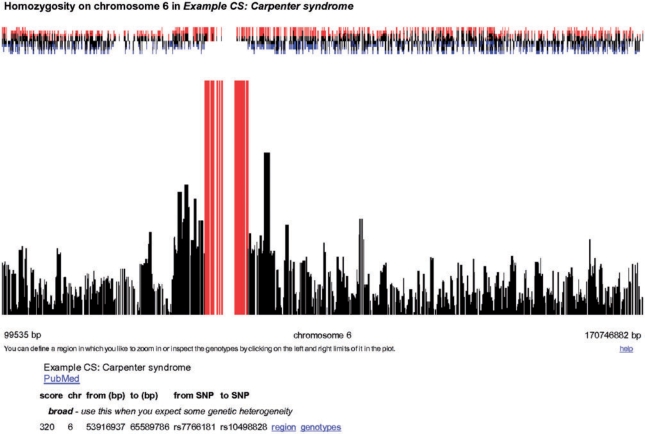
In a first step, HomozygosityMapper plots the genome-wide homozygosity (Figure 1) as bar charts together with the surplus or the shortfall of the homozygosity in affected individuals compared with the control genotypes. To emphasize interesting regions, any score higher than 80% of the maximum score reached in this project is coloured in red. Below the plot, a summary of the regions bearing the highest homozygosity scores is given together with hyperlinks to directly zoom into the region or to the underlying genotypes. Additionally, users can manually zoom into single chromosomes or regions selected with two mouse clicks. A promising region will yield a high homozygosity score over many contiguous markers (i.e. many red bars next to each other or a single thick one if markers are very close, see Figure 2) and show an excess of homozygosity in cases over controls.
Figure 1.
Genome-wide homozygosity. This screen shots show the genome-wide homozygosity scores produced by HomozygosityMapper. These are plotted as a bar chart with red bars indicating the most promising genomic regions. Clicking on a bar will zoom into the chromosome harbouring the score. Above the bar chart, the excess or shortage of homozygous genotypes in cases versus controls is depicted. Below the figure, direct links to the most interesting regions are given. All figures depict the Carpenter syndrome study (31).
Figure 2.
Homozygosity in genomic regions. Here the homozygosity within a selected genomic region is shown. The small red bars above the homozygosity bar chart indicate that there is a visible excess of homozygosity in the cases compared to the controls.
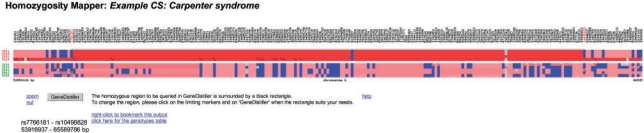
The next step is the inspection of the underlying genotypes, either by selecting a region and click on the hyperlink or by following the direct links below the plot. Here, genotypes for each sample specified as case or control are shown so that unaffected siblings can be employed to exclude regions or a possible genetic heterogeneity can be identified (Figure 3). For a quick overview, we use a colour coding scheme; unknown genotypes are displayed as grey boxes, heterozygous genotypes as blue boxes and stretches of homozygosity as red bars. The saturation of the red colour depends on length of the homozygous block. The two possible different homozygous genotypes are indicated by a small vertical bar in the less abundant genotype. This case may arise when data from different families with the same disease locus but different disease haplotypes is analysed (Figure 3).
Figure 3.
Genotypes view. HomozygosityMapper also displays the single genotypes of all samples. Here, the markers are placed on the x-axis while the samples are on the y-axis, with the patients on top and with red IDs. Genotypes are colour-coded: grey, unknown, blue, heterozygous, red, homozygous stretches (colour saturation reflects the length). This figure also reveals the presence of a single heterozygous marker within the homozygous region (possibly a genotyping error and ignored by HomozygosityMapper). The patient on the bottom is from another family than the first two and does not share the same haplotype over the whole homozygous stretch. This can be seen from the genotypes with the diagonal bar indicating the less abundant of the homozygous genotypes. Users are free to change the limits of the region and can subsequently submit this region to GeneDistiller.
Integration with GeneDistiller
Whenever a target region is displayed on the genotype level, a link to our candidate gene search engine GeneDistiller (30) is provided. Users are free to change the limiting SNPs of a region simply by clicking on the new borders in the plot. Clicking on the GeneDistiller button will then open its query interface with the limiting markers already filled in. Every input made by the user is saved in a cookie so that there is no need to fill out the interface again when another region is studied. Results from the homozygosity mapping can be used to filter and sort the genes in the candidate region. The degree of homozygosity around its position will be displayed together with a hyperlink to HomozygosityMapper that will indicate the current gene's position in the genotype view.
In reverse, users can also directly query a list of candidate genes in GeneDistiller for homozygosity (from a HomozygosityMapper analysis) and might hence detect founder mutations even when there is no clear disease haplotype.
Sustainability
Due to hardware and financial constraints, we cannot promise that every project can be kept on the website forever. Data uploaded by non-registered users will be deleted periodically. Should we have to delete data uploaded by registered users in the future, the respective data owners will be informed in advance and asked whether they still want to store their projects, or whether we can archive them and take them from the website. We guarantee that projects made public will not be deleted.
GENETIC HETEROGENEITY
In some studies, only a portion of the affected individuals share the same disease locus. As these projects often do not lead to the identification of a disease gene and are hence not published, we decided to simulate such cases. On our website, we provide an example for such a scenario: to account for naturally occurring homozygous stretches, real genotypes were used instead of purely simulated genotypes. For this, the genotypes of eight unrelated samples from the sample data for the Affymetrix GeneChip Mapping 50K Hind array were chosen and homozygous regions of 2 Mbp of length were arbitrarily introduced at various positions. Eight affected individuals were thus simulated; five of them were assigned a shared homozygous region on one chromosome, the remaining three of them on another. 0.5% of the genotypes within these regions were randomly changed to simulate genotyping errors. HomozygosityMapper correctly detected the first homozygous region but it also found the second. Details on this simulation as well as other test scenarios can be found on our website.
ROBUSTNESS AGAINST GENOTYPING ERRORS
The genotypes obtained from genotyping chips are not error-free. In fact, in our experience some chips show a rather poor performance which might be due to problems during the manufacturing but also to inappropriate genotype-calling settings or insufficient DNA quality. Genotyping errors are known to have a potentially drastic influence on the generation of multipoint LOD scores and haplotypes (15). The situation worsens when individuals without relatives (who could be used to check Mendelian inheritance or to draw phase information from them) are employed—a situation that might arise when financial constraints appear or unaffected family members decline to be involved in the study.
To circumvent this problem, HomozygosityMapper ignores single heterozygous genotypes with seven or more homozygous genotypes on each side. This setting fits chips with few markers as well as high-density arrays with usually higher error rates. Although this cannot render very bad genotyping results useful, it normally avoids type II errors introduced by genotyping chips with a more reasonable error frequency of 2% or less and hence saves the users from the effort to exclude markers by other, arguable, means such as analysis of Hardy–Weinberg equilibrium. To give the researchers full control, the original genotypes are displayed in the genotypes view (Figure 3).
Using only the genotypes provided by affected individuals without any pedigree information, HomozygosityMapper is also robust against misspecifications of the family structure.
USAGE AND VALIDITY OF OUR APPROACH
HomozygosityMapper has been used for four years now, at the beginning in the form of a stand-alone application. Over this time, more than 40 mappings were conducted with HomozygosityMapper and validated with the conventional approach or vice versa. With four or more affected individuals in the sample, HomozygosityMapper usually did not fail to identify the disease-linked region instantly. With two or three affected individuals, results sometimes required a closer inspection and, in some cases, the exclusion of regions due to presence of a homozygous stretch in unaffected siblings. However, even in such studies, HomozygosityMapper did not perform worse than linkage analysis. Linkage analysis had, on the other hand, a clear advantage when only two affected individuals were available but the information from unaffected family members could be included in the calculation.
With permission of the owners, some published studies with 10K, 50K and 250K arrays in which the disease mutation could eventually be identified, can be interactively viewed on our website: Carpenter syndrome (31), Crisponi syndrome (32), Cutis laxa (33), Dyschromatosis universalis hereditaria (34), Familial thrombocytosis (35), Hypomagnesia (19), Meckel-Gruber syndrome (36), Meckel-Gruber-like syndrome (37), Nephrotic syndrome (13), Pontocerebellar hypoplasia (38), Senior-Loken syndrome (13). These homozygosity mappings were either carried out with conventional linkage software and could be reproduced with HomozygosityMapper or vice versa. A more thorough description is given on the website.
IMPLEMENTATION
HomozygosityMapper was programmed in Perl. It makes use of a PostgreSQL 8.3 database. The web server, together with the database and the CGI scripts, runs on an Intel Xeon platform with two QuadCore processors and 48 GB of RAM under Fedora Core Linux. To grant access to non-public genotypes and results, cookies are employed. JavaScript is used to zoom into chromosomal regions and to redefine the limits of the homozygous interval. A more thorough description of the implementation can be found on the website.
The website was developed with and optimized for Mozilla Firefox. It was successfully tested with Firefox 2 and 3 (under different versions of Linux, Microsoft Windows, and MacOS) and Microsoft Internet Explorer 6, 7 and 8.
FUTURE PLANS
We are currently implementing a function to generate input files for the common multipoint linkage software GENEHUNTER and Allegro for all promising regions in one step. Users could thus run a fast genome-wide scan with HomozygosityMapper and subsequently check only the possible regions with the information provided by parents and most of all by healthy sibs and produce haplotypes using the slow but generally accepted approach and save much time compared to the conventional approach. This feature will include the option to simply download the genotypes of a whole project or a single analysis either for specific regions or genome-wide.
Another forthcoming change is the option for users to modify their user profiles and projects and analysis details with dedicated web interfaces.
Should HomozygosityMapper become a widely used repository to publish genotypes and results from homozygosity mappings, we will restructure the database and the website so that literature references and co-authors can be included more easily.
CONCLUSION
We have presented HomozygosityMapper, a web-based application aimed at autozygosity mapping. Our software is independent of parameters like family structure or allele frequencies, the ‘homozygosity score’ is calculated simply from the observed homozygosity and it is robust against genotyping errors. HomozygosityMapper is much faster than conventional linkage software. Albeit it might not fully replace these, it could be used in combination with them. In our opinion, a sensible strategy would be the rapid identification of possible disease regions using HomozygosityMapper and the subsequent generation of LOD scores and haplotypes by conventional software only for these, if necessary at all.
The integration with GeneDistiller greatly facilitates the search for promising candidate genes compared to the conventional approach. We also encourage geneticists to consider HomozygosityMapper as a public repository for genotypes and results when publishing their homozygosity mappings.
Due to its user-friendly intuitive interface and the lack of any local hardware requirements, it can be used by the geneticists themselves without the need for computer specialists.
FUNDING
Deutsche Forschungsgemeinschaft within the SFB 665 TP A6 ‘Developmental Disorders of the Nervous System’; NeuroCure Cluster of Excellence [Exc 257] at the Charité, Berlin. Funding for open access charge: Charité LOM.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Special thanks to Gudrun Nürnberg who has been a beta tester of HomozygosityMapper since its very beginnings and gave valuable advice on how to improve the usability of the application.
REFERENCES
- 1.Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 2.Smith CAB. The detection of linkage in human genetics. J. Royal Stat. Soc. Series B (Methodological) 1953;15:153–192. [Google Scholar]
- 3.Gibbs JR, Singleton A. Application of genome-wide single nucleotide polymorphism typing: simple association and beyond. PLoS Genet. 2006;2:e150. doi: 10.1371/journal.pgen.0020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am. J. Hum. Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 5.Dietter J, Mattheisen M, Furst R, Ruschendorf F, Wienker TF, Strauch K. Linkage analysis using sex-specific recombination fractions with GENEHUNTER-MODSCORE. Bioinformatics. 2007;23:64–70. doi: 10.1093/bioinformatics/btl539. [DOI] [PubMed] [Google Scholar]
- 6.Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am. J. Hum. Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudbjartsson DF, Thorvaldsson T, Kong A, Gunnarsson G, Ingolfsdottir A. Allegro version 2. Nat. Genet. 2005;37:1015–1016. doi: 10.1038/ng1005-1015. [DOI] [PubMed] [Google Scholar]
- 8.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am. J. Hum. Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 10.Miano MG, Jacobson SG, Carothers A, Hanson I, Teague P, Lovell J, Cideciyan AV, Haider N, Stone EM, Sheffield VC, et al. Pitfalls in homozygosity mapping. Am. J. Hum. Genet. 2000;67:1348–1351. doi: 10.1016/s0002-9297(07)62966-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leutenegger AL, Labalme A, Genin E, Toutain A, Steichen E, Clerget-Darpoux F, Edery P. Using genomic inbreeding coefficient estimates for homozygosity mapping of rare recessive traits: application to Taybi-Linder syndrome. Am. J. Hum. Genet. 2006;79:62–66. doi: 10.1086/504640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leutenegger AL, Prum B, Genin E, Verny C, Lemainque A, Clerget-Darpoux F, Thompson EA. Estimation of the inbreeding coefficient through use of genomic data. Am. J. Hum. Genet. 2003;73:516–523. doi: 10.1086/378207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hildebrandt F, Heeringa SF, Ruschendorf F, Attanasio M, Nurnberg G, Becker C, Seelow D, Huebner N, Chernin G, Vlangos CN, et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5:e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kruglyak L, Daly MJ, Lander ES. Rapid multipoint linkage analysis of recessive traits in nuclear families, including homozygosity mapping. Am. J. Hum. Genet. 1995;56:519–527. [PMC free article] [PubMed] [Google Scholar]
- 15.Kirk KM, Cardon LR. The impact of genotyping error on haplotype reconstruction and frequency estimation. Eur. J. Hum. Genet. 2002;10:616–622. doi: 10.1038/sj.ejhg.5200855. [DOI] [PubMed] [Google Scholar]
- 16.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc. Natl Acad. Sci. USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omran H, Fernandez C, Jung M, Haffner K, Fargier B, Villaquiran A, Waldherr R, Gretz N, Brandis M, Ruschendorf F, et al. Identification of a new gene locus for adolescent nephronophthisis, on chromosome 3q22 in a large Venezuelan pedigree. Am. J. Hum. Genet. 2000;66:118–127. doi: 10.1086/302705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goedken R, Ludington E, Crowe R, Fyer AJ, Hodge SE, Knowles JA, Vieland VJ, Weissman MM. Drawbacks of GENEHUNTER for larger pedigrees: application to panic disorder. Am. J. Med. Genet. 2000;96:781–783. [PubMed] [Google Scholar]
- 19.Konrad M, Schaller A, Seelow D, Pandey AV, Waldegger S, Lesslauer A, Vitzthum H, Suzuki Y, Luk JM, Becker C, et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am. J. Hum. Genet. 2006;79:949–957. doi: 10.1086/508617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller RF, Bishop DT. Autozygosity mapping, complex consanguinity, and autosomal recessive disorders. J. Med. Genet. 1993;30:798–799. doi: 10.1136/jmg.30.9.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broman KW, Weber JL. Long homozygous chromosomal segments in reference families from the centre d'Etude du polymorphisme humain. Am. J. Hum. Genet. 1999;65:1493–1500. doi: 10.1086/302661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McQuillan R, Leutenegger AL, bdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polasek O, Tenesa A, et al. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods CG, Cox J, Springell K, Hampshire DJ, Mohamed MD, McKibbin M, Stern R, Raymond FL, Sandford R, Malik SS, et al. Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am. J. Hum. Genet. 2006;78:889–896. doi: 10.1086/503875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nystuen A, Benke PJ, Merren J, Stone EM, Sheffield VC. A cerebellar ataxia locus identified by DNA pooling to search for linkage disequilibrium in an isolated population from the Cayman Islands. Hum. Mol. Genet. 1996;5:525–531. doi: 10.1093/hmg/5.4.525. [DOI] [PubMed] [Google Scholar]
- 25.Woods CG, Valente EM, Bond J, Roberts E. A new method for autozygosity mapping using single nucleotide polymorphisms (SNPs) and EXCLUDEAR. J. Med. Genet. 2004;41:e101. doi: 10.1136/jmg.2003.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carr IM, Flintoff KJ, Taylor GR, Markham AF, Bonthron DT. Interactive visual analysis of SNP data for rapid autozygosity mapping in consanguineous families. Hum. Mutat. 2006;27:1041–1046. doi: 10.1002/humu.20383. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The International HapMap Consortium. The International HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 30.Seelow D, Schwarz JM, Schuelke M. GeneDistiller–distilling candidate genes from linkage intervals. PLoS ONE. 2008;3:e3874. doi: 10.1371/journal.pone.0003874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins D, Seelow D, Jehee FS, Perlyn CA, Alonso LG, Bueno DF, Donnai D, Josifova D, Mathijssen IM, Morton JE, et al. RAB23 mutations in Carpenter syndrome imply an unexpected role for hedgehog signaling in cranial-suture development and obesity. Am. J. Hum. Genet. 2007;80:1162–1170. doi: 10.1086/518047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crisponi L, Crisponi G, Meloni A, Toliat MR, Nurnberg G, Usala G, Uda M, Masala M, Hohne W, Becker C, et al. Crisponi syndrome is caused by mutations in the CRLF1 gene and is allelic to cold-induced sweating syndrome type 1. Am. J. Hum. Genet. 2007;80:971–981. doi: 10.1086/516843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornak U, Reynders E, Dimopoulou A, van RJ, Fischer B, Rajab A, Budde B, Nurnberg P, Foulquier F, Lefeber D, et al. Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat. Genet. 2008;40:32–34. doi: 10.1038/ng.2007.45. [DOI] [PubMed] [Google Scholar]
- 34.Stuhrmann M, Hennies HC, Bukhari IA, Brakensiek K, Nurnberg G, Becker C, Huebener J, Miranda MC, Frye-Boukhriss H, Knothe S, et al. Dyschromatosis universalis hereditaria: evidence for autosomal recessive inheritance and identification of a new locus on chromosome 12q21-q23. Clin. Genet. 2008;73:566–572. doi: 10.1111/j.1399-0004.2008.01000.x. [DOI] [PubMed] [Google Scholar]
- 35.El-Harith E, Roesl C, Ballmaier M, Germeshausen M, Frye-Boukhriss H, von NN, Becker C, Nurnberg G, Nurnberg P, Ahmed MA, et al. Familial thrombocytosis caused by the novel germ-line mutation p.Pro106Leu in the MPL gene. Br. J. Haematol. 2009;144:185–194. doi: 10.1111/j.1365-2141.2008.07430.x. [DOI] [PubMed] [Google Scholar]
- 36.Frank V, den Hollander AI, Bruchle NO, Zonneveld MN, Nurnberg G, Becker C, Du BG, Kendziorra H, Roosing S, Senderek J, et al. Mutations of the CEP290 gene encoding a centrosomal protein cause Meckel-Gruber syndrome. Hum. Mutat. 2008;29:45–52. doi: 10.1002/humu.20614. [DOI] [PubMed] [Google Scholar]
- 37.Bergmann C, Fliegauf M, Bruchle NO, Frank V, Olbrich H, Kirschner J, Schermer B, Schmedding I, Kispert A, Kranzlin B, et al. Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am. J. Hum. Genet. 2008;82:959–970. doi: 10.1016/j.ajhg.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Budde BS, Namavar Y, Barth PG, Poll-The BT, Nurnberg G, Becker C, van RF, Weterman MA, Fluiter K, te BE, et al. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat. Genet. 2008;40:1113–1118. doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]