Abstract
We have developed a set of web-based SNP selection tools (freely available at http://www.niehs.nih.gov/snpinfo) where investigators can specify genes or linkage regions and select SNPs based on GWAS results, linkage disequilibrium (LD), and predicted functional characteristics of both coding and non-coding SNPs. The algorithm uses GWAS SNP P-value data and finds all SNPs in high LD with GWAS SNPs, so that selection is from a much larger set of SNPs than the GWAS itself. The program can also identify and choose tag SNPs for SNPs not in high LD with any GWAS SNP. We incorporate functional predictions of protein structure, gene regulation, splicing and miRNA binding, and consider whether the alternative alleles of a SNP are likely to have differential effects on function. Users can assign weights for different functional categories of SNPs to further tailor SNP selection. The program accounts for LD structure of different populations so that a GWAS study from one ethnic group can be used to choose SNPs for one or more other ethnic groups. Finally, we provide an example using prostate cancer and demonstrate that this algorithm can select a small panel of SNPs that include many of the recently validated prostate cancer SNPs.
INTRODUCTION
The completion of the International HapMap Project (1) and the development of advanced genotyping technologies have made genome-wide association studies (GWAS) possible. These studies typically genotype more than 1000 cases and 1000 controls for 300 K to 1 million SNPs. A number of GWAS have been published with many more in progress (2–4). A number of disease-associated SNPs have been identified and confirmed by these breakthrough studies with many more yet to come. Repeating GWAS in additional individuals has helped to find more disease-associated SNPs, although doing so is costly. Interestingly, the SNPs identified and subsequently confirmed in large replication samples are not always those with the smallest P-value in the GWAS, and two GWAS may have radically different P-values assigned to a confirmed SNP. For example, in prostate cancer a confirmed SNP in MSMB from the initial GWAS had a P-value of only 0.042, but the P-value was 7.31 × 10–13 in a follow up study (4,5). Thus the list of potential SNPs from any GWAS remains large. This large SNP list poses a problem for validation studies where a very large number of people are genotyped because custom arrays can cost more than standard GWAS arrays.
For many diseases there exists a rich, diverse and growing literature that can be used to identify genes and chromosomal regions of high interest. This literature includes existing genetic studies of linkage and candidate genes as well as research on disease pathogenesis. For example, information about disrupted cell signaling pathways and genomic-level expression data from comparisons of tumor and normal tissues have identified interesting candidate genes for cancer. Thus investigators may have a large but finite set of genes and genomic regions that they feel deserve particular scrutiny or they may have a special interest in certain genes or chromosomal regions.
Agnostic GWAS data provide a unique opportunity for hypothesis driven candidate gene exploration, but the sheer size and complexity of GWAS data can be difficult to manage. Although it may not be difficult to find which SNPs of a gene are directly included in a GWAS panel, it is harder to determine which additional SNPs are tagged by the panel, particularly when examining multiple ethnic groups where linkage disequilibrium (LD) structure and allele frequencies differ. There are a growing array of tools for gene annotation (e.g. identifying regulatory elements, alternative splicing, miRNA-binding sites), but many researchers may find it difficult to gather and employ these algorithms. Finally, while such tools predict putative functional regions for the Reference Sequence, they do not necessarily consider if the alternative alleles of SNPs in that sequence are likely to have different consequences.
Here we describe a comprehensive web server designed to select SNPs for genetic association studies. In designing this application we provide 3 pipelines for SNP selection with options to combine all three pipelines. The candidate gene pipeline uses both a user-provided list of candidate genes and disease-specific GWAS data [readily available from dbGaP (www.ncbi.nlm.nih.gov/sites/entrez?db=gap) and elsewhere] to select SNPs that are predicted to have functional consequences and that are in high LD with a small P-value GWAS SNP. For genes where a large proportion of the SNPs were not in LD with any GWAS SNP and thus are uninvestigated in the GWAS, the web application can pick LD tag SNPs to evaluate the untagged SNPs. The second, genomic pipeline selects SNPs with likely functional consequences from SNPs with small P-value in a GWAS and from SNPs in high LD with such SNPs. The third, linkage pipeline uses a user-provided list of linkage regions to select small P-value GWAS SNPs for each linkage region. The web application has information on all SNPs in HapMap and dbSNP and automatically constructs ethnic-specific LD relationships from both sources provided that the SNPs have population data available. In this way, SNPs that were not genotyped in a GWAS, but are in LD with a SNP that was genotyped, can be screened appropriately and GWAS data generated in one ethnic group can be used to pick SNPs in one or more other ethnic groups. We illustrate this application using prostate cancer as an example in which we start with a set of a priori candidate genes, prostate cancer GWAS data, and a set of linkage regions, and use the pipelines to select a small panel of 1361 SNPs. We evaluate the utility of the application against the results of a follow-up validation study that screened a much larger panel of ∼27 000 SNPs genotyped in ∼8000 cases and controls and find that we included five of the seven SNPs found to be associated with prostate cancer.
METHODS
Candidate gene pipeline
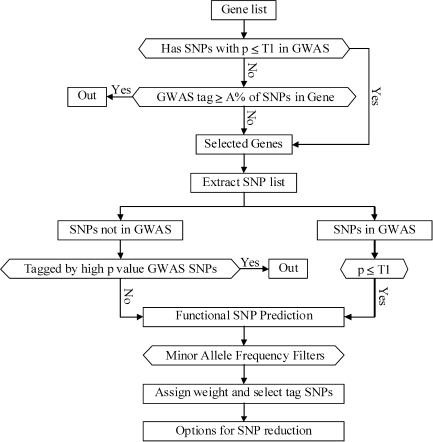
A list of candidate genes for a particular disease can be gleaned from published association studies, gene expression studies, disease pathways and the specific interests of an investigator. Such lists may be very large, so we first filter the list against GWAS results as shown in Figure 1. We use SNPs that have genotype data in dbSNP as our source of SNPs in and near a gene (for a user-specified flanking region around the gene). We keep a gene if it has at least one small P-value SNP (less than or equal to a user-specified threshold, T1) in the GWAS. We also keep genes that were not adequately represented by SNPs in the GWAS panel. The percent of common SNPs (within a gene and flanking region) in high LD (pairwise r2 ≥ a user-specified threshold) with any GWAS SNP (including GWAS SNPs outside the gene and flanking region) is calculated and genes with coverage less than a user-specified cutoff A% are retained. Genes that do not have SNPs with small P-value but do have sufficient coverage by GWAS SNPs are excluded from further analysis.
Figure 1.
GenePipe: decision tree to prioritize SNPs for candidate genes based on GWAS results, SNP functional prediction characteristics and pair-wise LD. The six-sided boxes represent decision points and rectangles represent action steps or end points.
For the candidate genes that pass the above screen we extract SNPs from dbSNP and process this list as shown in Figure 1. If a SNP was examined in the GWAS and had a P-value less than the user-specified threshold T1 it is retained. If a SNP was not in the GWAS but was in high LD with a GWAS SNP that had a P-value larger than T1 it is eliminated because we reason that it was adequately evaluated by the GWAS and found to have no association with disease. We then score all retained SNPs for functional significance and apply different minor allele frequency (MAF) filters depending on the functional category of the SNP. These user-specified MAF filters are provided because functionally important SNPs often have lower MAF due to natural selection (6) and we wish to provide extra flexibility to retain functional SNPs below the MAF filter being applied to SNPs without such function. The details of the functional predictions used in this and other pipelines are provided in a separate section below.
In the final processing step we select LD tag SNPs. Because there are certain advantages to having functional and small P-value SNPs directly assessed by the genotyping panel (instead of being indirectly assessed via LD) we provide for the assignment of user-specified weights for different categories of functional SNPs and small P-value SNPs. If weights are assigned the null value of 1, then tag SNPs are selected simply by rank order, so that SNPs that are in high LD with the largest number of SNPs are selected first and SNPs that tag only themselves (singleton tags) are selected last. If a functional SNP has a weight applied, then the weight act as multiplier of the actual number of SNPs tagged so that it is more likely to be selected early. For example, a functional SNP with a weight of two that is in LD with four SNPs (including itself) would have a weighted tag value of 2 × 4 = 8. Investigators may modify a variety of values (e.g. P-value threshold T1, LD threshold, or weights) to adjust selected SNP counts to fit their genotyping panel size and budget. We provide two options for additional SNP reduction that we think are useful: (i) Each SNP must be in LD with a user-specified minimum number of common SNPs (after multiplied by the user-assigned weights). For example, this option can be used to eliminate singleton SNPs. (ii) A user can also specify the maximum number of SNPs that are allowed for any one gene using a method which is similar to selecting the best N SNPs to optimize power (7). To insure that each gene has some coverage, we also provide a user-specified minimum number of best SNPs (in terms of number of SNPs captured at a specific LD threshold) that must be selected for each gene even if they do not meet the previous criterion for tag SNPs.
Genome pipeline
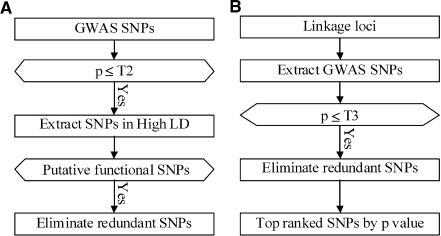
Small P-value GWAS SNPs were considered in the previous pipeline if they occur within a specified candidate gene, but for those in the remainder of the genome we provide additional means of selection based on function and evolutionary conservation (Figure 2A). In providing this screen we consider not only all the GWAS SNPs that were found to have small P-values, but the much larger set of SNPs in dbSNP that meet two criteria: (i) they are within a user-specified distance from a small P-value GWAS SNP; and (ii) they are known to be in high LD with a small P-value GWAS SNP. From this large pool we screen SNPs based on functional predictions and apply MAF filters. Finally, we eliminate redundant SNPs based on a user-defined LD threshold.
Figure 2.
(A) GenomePipe: flow chart for functional SNP selection from SNPs that are in high LD with small P-value GWAS SNPs. (B) LinkagePipe: flow chart to prioritize SNPs in linkage loci based on P-values in GWAS.
Linkage pipeline
Linkage studies of family-based samples are another valuable source for candidate regions of the genome involved in disease. GWAS panels have much higher SNP density than linkage studies, and provide finer mapping information using large population-based samples. Within each user-specified linkage region, we select small P-value GWAS SNPs at a user-specified threshold, rank them by P-value and select a user-specified number of non-redundant SNPs (based on pairwise LD) that have the smallest P-value (Figure 2B).
Functional SNP prediction
Depending on their position and flanking sequence in a gene, SNPs may have varied functional effects on protein sequence, transcriptional regulation, RNA splicing or miRNA binding. There are a variety of in silico tools available for prediction of such functional regions within genes, and we use these tools to help identify SNPs that are more likely to affect biological function. In doing so we examine not only whether a SNP occurs within a likely functional region, but also whether the alternative alleles are likely to have differential functional effects.
Coding SNPs
Within the coding region of a gene, we identify nonsense SNPs that lead to premature termination of translation and are therefore very likely to affect protein function. In addition, non-synonymous polymorphisms (nsSNPs) that lead to amino acid changes may also affect protein function depending on the location and nature of the amino acid substitution. We used two in silico classification programs, Polyphen (8) and SNPs3D (9), to predict the effect of an amino acid substitution on the structure and function of a human protein, and then classified nsSNPs as possibly or probably damaging or benign.
Transcription-factor-binding sites (TFBS)
If a SNP is located at a TFBS of a gene, it may affect the level or timing of gene expression. We identified such SNPs according to the procedure shown in Supplementary Figure 1. For each SNP within 5 kb upstream or 1 kb downstream of a transcription start site (TSS), we first extracted 29 bp DNA sequence on either side of the SNP, and then used the MATCH (10) method to predict possible TFBSs in the resulting 59 base pair sequence using each alternative allele. A SNP was classified as affecting TFBS activity if MATCH predicted a TFBS with one allele but not with the other and the difference in the matrix similarity scores (MSS) or core similarity scores (CSS) between the two alleles was ≥0.2. Possible scores for MSS and CSS range from 0 to 1 (10). We performed predictions using all the 187 position weight matrices classified as high quality non-redundant vertebrate (mouse, rat and human) matrices in TRANSFAC Release 12.1 (11). We used the default set of MATCH score thresholds provided by TRANSFAC to allow for 10% false negative results. We also filtered out SNPs in non-conserved TFBS. To find conserved TFBS, we first identified the mouse or rat homolog sequence for each predicted TFBS in the human genome based on 17-way vertebrate multiz alignment from UCSC genome bioinformatics web site. We then ran MATCH on these homolog sequences with the same position weight matrices. We categorized a TFBS as conserved if both mouse and rat homolog sequences also have the same predicted TFBS. Several studies (12–14) show that using both the predicted conserved TFBS together with the regulatory potential score (13) can improve predictions, so we also provide this option on the web server.
Splice sites
SNPs that are located within two base pairs of an intron–exon junction, or located at exonic splicing enhancer (ESE) or exonic splicing silencer (ESS)-binding sites may disrupt mRNA splicing and severely affect protein function (15). We predicted ESE and ESS sites using procedure outlined in Supplementary Figure 2. If an exon was longer than 140 base pairs, only SNPs within the first and last 70 base pairs of each exon were evaluated because the effect of alternative alleles on the activity of ESEs and ESSs decrease with distance from the splice site (16,17). We only considered a maximum of 10 base pairs on either side of a SNP because there are no significant compensatory or correlated relationships between non-overlapping ESE or ESS motifs (18). ESE sites were predicted using RESCUE ESE (19) or ESEfinder (20) methods. ESS sites were predicted using the FAS–ESS (21) method. A SNP was classified as affecting splicing activity if there was at least one predicted binding site with one allele, but none with the other allele. In order to reduce false positive results, we excluded predicted binding sites within an exon if, based on Ensembl transcripts isoform data, there were no alternatively spliced transcripts observed involving the exon. For example, suppose a gene has eight exons and five different transcript isoforms reported in Ensembl. If there was a predicted ESE or ESS-binding site in exon 3 but all five transcripts include exon 3, then we would exclude the site.
MicroRNA-binding sites
MicroRNAs (miRNA) are 21–23-base single-stranded RNA molecules that bind to the end of a messenger RNA (mRNA) and can inhibit protein translation. Human miRNA is usually complementary to a site in the 3′ UTR region of an mRNA. We extracted the 20 base pair flanking sequence on both sides of SNPs in the 3′ UTR region of genes. We search for possible miRNA-binding sites on the 41 base pair DNA sequence for each allele of a SNP using the software miRanda (22), with default parameter values. Using the procedure outlined in Supplementary Figures 3 and 4, we predicted putative miRNA-binding sites for all 677 human miRNAs in the miRBase database (23). We excluded SNPs in miRNA-binding sites that were not conserved in either the mouse or rat homolog sequences. We classified a SNP as affecting miRNA-binding site activity if the miRanda scores for the two alleles differed by ≥16, a value which is equivalent to that of a SNP in the ‘seed’ region of a miRNA-binding site.
Web server and usage
We have incorporated these methods into a user-friendly web server: SNPinfo (http://www.niehs.nih.gov/snpinfo). The web utility is supported by a set of optimized mySQL databases. Depending on the specific pipeline being used (GenePipe, GenomePipe or LinkagePipe), an investigator may input several types of data: a list of candidate genes, a GWAS SNP list of Reference Sequence (rs) numbers with associated P-value from the GWAS of interest, or a list of linkage loci.
LD relationships between SNPs may differ between ethnic groups so we have deposited, as a central resource of our web server, the information on SNP genotype data and pairwise LD for each ethnic group. This allows the user to incorporate the results of a GWAS from one ethnic group into LD tag SNP selection for one or more different ethnic groups. To evaluate LD relationships between SNPs, a user can use not only pair-wise LD data calculated from HapMap genotype data for 11 populations in HapMap Phase III, but also has the option to use pair wise LD based on all dbSNP genotype data for each of five population groups (African American, Asian, European, Hispanics and sub-Saharan African). dbSNP genotype data includes all deposited HapMap data as well as additional SNPs, individuals and ethnic groups. dbSNP includes genotype data from many different genotyping and resequencing efforts on sometimes overlapping sets of individuals. We combined genotypes for individuals of the same ethnic group. If multiple submitters genotyped the same SNP in the same person and the genotype calls are inconsistent, we assigned the person the most commonly called genotype or a missing call if they are equally split. We employed an efficient greedy algorithm that was originally implemented in TAGster (24) to select LD tag SNP for single or multiple populations.
In addition to the three pipelines the server provides three additional tools. The first of these ‘TagSNP’ allows a user to combine the SNP lists selected from different pipelines and eliminate redundant SNPs based on LD relationships and SNPs with low SNP design scores. It also allows the user to mandate inclusion of SNPs of special interest, or exclusion of undesired SNPs. This same tool may be used as a stand-alone tool to find and list SNPs, choose LD tag SNPs, and produce high quality LD or genotype figures for individual genes or chromosome regions. A second stand-alone tool, ‘FuncPred’ allows a user to query functional prediction results and ethnic group allele frequencies for all of the SNPs in a gene or chromosomal region, or for a list of input SNPs. The final tool ‘SNPseq’ allows a user to visualize SNP related information and CpG regions in DNA sequence context for an individual SNP, gene, or region of a chromosome. This is particularly useful for PCR primer design.
Example and validation
We have used the GWAS data from the Cancer Genetics Markers of Susceptibility (CGEMS) project on prostate cancer (4) to demonstrate the utility of our method. This GWAS genotyped 550 K SNPs in 1172 prostate cancer cases and 1157 controls of European origin. We used our web utility to construct a small SNP genotyping panel for a genetic association study on prostate cancer in African-American and European-American men.
Based on published candidate gene association studies, gene expression studies, and pathway analysis we constructed a list of 848 candidate genes of interest in prostate cancer. Using GenePipe, 542 genes were excluded because none of the GWAS SNPs in these genes had a P-value ≤0.05 and there were sufficient GWAS SNPs to capture (at r2 ≥ 0.8) more than 50% of common (MAF ≥ 0.05) SNPs in Europeans. For the remaining 306 genes, 822 non-redundant SNPs were selected as outlined in Figure 1 with the following GenePipe parameter values: gene upstream region = 5 kb, gene downstream region = 1 kb, MAF = 0.05 for all SNPs, weight = 3 for any predicted functional SNP and small P-value SNPs, weight = 1 for all other SNPs, r2 threshold = 0.8, minimum number of SNPs tagged by each selected tag SNP = 3, minimum number of tag SNPs/gene = 1, and maximum number of tag SNPs/gene = 5.
The CGEMS GWAS reported 6034 GWAS SNPs with P ≤ 0.01. GenomePipe identified 41755 SNPs that are in high LD with these GWAS SNPs (r2 ≥ 0.8), and from the 41755 SNPs selected 543 common SNPs (MAF ≥ 0.05) that were predicted to be functional by at least one of the prediction methods.
Published studies have identified 43 non-overlapping linkage regions for prostate cancer. As shown in Figure 2B, we used LinkagePipe to select 266 GWAS SNPs using the following parameter values (MAF = 0.05, Maximum number of SNPs/linkage locus = 7, GWAS P threshold = 0.01, LD threshold = 0.8).
The resulting SNP lists from GenePipe, GenomePipe and LinkagePipe were combined and we used TagSNP to eliminate duplicate and redundant SNPs, or SNPs with low assay design scores, yielding a set of 1361 SNPs. Of these, 709 (52%) were GWAS SNPs and the remaining 48% were new SNPs not in the GWAS which were selected to provide additional functional examination of genes.
Although the selection algorithm used the P-value data for 550 K SNPs from the CGEMS GWAS, we did not, in this example, use information from other GWAS data sets or from the validation portion of the CGEMS initial study (4). The CGEMS follow up study was particularly robust because it genotyped 26 958 SNPs, including all SNPs with P-value <0.068 from the initial CGEMS GWAS, in 3941 cases and 3964 controls (5). This provides an unbiased opportunity to evaluate whether the very small set of SNPs selected by our algorithm include the SNPs validated in a genotyping panel that was many times larger. The CGEMS validation study identified seven prostate cancer related SNPs which had P-value ranks in the initial GWAS ranging from 116 (P = 0.0004) to 24407 (P = 0.042). Our algorithms selected five (71%) of those seven SNPs. Three of the five SNPs were selected by GenePipe, one was selected by GenomePipe and three were selected by LinkagePipe. Of the two SNPs that were missed, rs10486567 in JAZF1 was not in our candidate gene list because at the time we constructed the gene list, JAZF1 had not previously been reported in the literature as having any association with prostate cancer. The other SNP, rs10896449, was not located in a known gene or linkage region. Although the very small panel of SNPs selected by the algorithm cannot substitute for massive follow-up genotyping, it performs very well with 2.5% (709 vs. 26 958) of the GWAS SNPs, and in addition dedicates almost half of the SNPs to new functional and candidate gene polymorphisms that were unexplored in the half million GWAS SNP panel.
DISCUSSION
SNP selection for an association study can be a complex problem. Decades of diverse investigation provide a tremendous amount of information on genes, pathways, and chromosomal regions that appear to be linked to disease. GWAS offers an agnostic approach to investigating SNP-disease association, and the results of such studies offers a wealth of data to inform the next generation of investigation. Here, we develop a user-friendly web server to incorporate such clinical, experimental, mechanistic, and computational information with the results of GWAS in order to organize, annotate, and select SNPs. The web server can be used for either small or large-scale SNP selection and is particularly useful for association studies. It uses both functional prediction and GWAS results to select not only SNPs included in the GWAS, but other functional SNPs in dbSNP that were not in the GWAS. Considering the varied interests and emphasis different investigators may bring to a problem, we provided many tunable parameters in each web utility, so the algorithm can be adjusted to meet different needs.
We employ several methods for functional sequence assessment and predict functional consequence of different alleles of a SNP. To reduce the number of false positive results, we perform the predictions in only the most probable genomic regions for each category of functional sequence site (such as the gene promoter region for TFBS or the 3′ UTR for microRNA-binding sites) and use phylogenetic footprint information to filter out non-conserved putative functional sequence. The SNP selection algorithm uses functional prediction results to prioritize LD tag SNP selection. These LD tag SNPs capture other unexamined SNPs in and around a gene, including SNPs with unknown or unpredicted functional consequences. The web utility options allow an investigator to choose prediction methods and assign weights to those predictions for study-specific SNP selection. Functional sequence prediction is a rapidly developing field. The web server structure allows rapid updates as better methods of functional prediction become available and it allows expansion to include predictions on other biologic functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. Funding for open access charge: National Institute of Environmental Health Sciences.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruglyak L. The road to genome-wide association studies. Nat. Rev. Genet. 2008;9:314–318. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 4.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 5.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 6.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 1999;22:231–238. doi: 10.1038/10290. [erratum appears in Nat. Genet. (1999), 23, 73] [DOI] [PubMed] [Google Scholar]
- 7.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. [see comment] Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 8.Sunyaev S, Ramensky V, Koch I, Lathe W, Kondrashov AS, 3rd, Bork P. Prediction of deleterious human alleles. Human Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 9.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformat. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elnitski L, Hardison RC, Li J, Yang S, Kolbe D, Eswara P, O'Connor MJ, Schwartz S, Miller W, Chiaromonte F. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elnitski L, Jin VX, Farnham PJ, Jones SJ. Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 2006;16:1455–1464. doi: 10.1101/gr.4140006. [DOI] [PubMed] [Google Scholar]
- 15.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, Wang HH, Yao A, Chen YT, Hsu CN. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairbrother WG, Holste D, Burge CB, Sharp PA. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLos Biol. 2004;2:E268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graveley BR, Hertel KJ, Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao X, Wang Z, Jang M, Burge CB. Coevolutionary networks of splicing cis-regulatory elements. Proc. Natl Acad. Sci.USA. 2007;104:18583–18588. doi: 10.1073/pnas.0707349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, Burge CB. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. [see comment] Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 22.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLos Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [erratum appears in PLoS Biol. (2005), 3, e264] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z, Kaplan NL, Taylor JA. TAGster: efficient selection of LD tag SNPs in single or multiple populations. Bioinformatics. 2007;23:3254–3255. doi: 10.1093/bioinformatics/btm426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.