Abstract
Despite its wide usage in biological databases and applications, the role of the gene ontology (GO) in network analysis is usually limited to functional annotation of genes or gene sets with auxiliary information on correlations ignored. Here, we report on new capabilities of VisANT—an integrative software platform for the visualization, mining, analysis and modeling of the biological networks—which extend the application of GO in network visualization, analysis and inference. The new VisANT functions can be classified into three categories. (i) Visualization: a new tree-based browser allows visualization of GO hierarchies. GO terms can be easily dropped into the network to group genes annotated under the term, thereby integrating the hierarchical ontology with the network. This facilitates multi-scale visualization and analysis. (ii) Flexible annotation schema: in addition to conventional methods for annotating network nodes with the most specific functional descriptions available, VisANT also provides functions to annotate genes at any customized level of abstraction. (iii) Finding over-represented GO terms and expression-enriched GO modules: two new algorithms have been implemented as VisANT plugins. One detects over-represented GO annotations in any given sub-network and the other finds the GO categories that are enriched in a specified phenotype or perturbed dataset. Both algorithms take account of network topology (i.e. correlations between genes based on various sources of evidence). VisANT is freely available at http://visant.bu.edu.
INTRODUCTION
One of the most widely used bioinformatics resources is the gene ontology (GO) (1), which provides hierarchically organized information about gene products, their activity, biological functions and cellular locations. Tools for network visualization and analysis often provide functions to annotate gene functions using GO. For example, Cytoscape (2) allows users to annotate genes in a network although several steps are required to load the ontology from a server and to prepare a gene-GO association file. More recently, GO has been used to find shared functions of genes in biological networks (3–6), and to direct network layout processes using overrepresented GO categories (7). Nevertheless, network topology is usually ignored in these analyses and automated integration of a user-mined network or specified gene set and the GO hierarchy as a single network—which would facilitate the intuitive interpretation, management and analysis—has not been achieved.
Among the technical barriers to a fully integrated representation framework are the different relational meanings of the GO and network edges, and the ontological structure of GO. GO terms are structured as a directed acyclic graph (DAG), where nodes represent terms and edges represent inclusive relationships between terms. A key characteristic of such representation is that a term in a DAG can have multiple parents. As a result, genes are associated with multiple biological terms and individual biological terms can also be associated with multiple genes. These ‘many-genes-to-many-terms’ (8) associations reflect the complex nature of biological processes and make visualization and modeling of the integrated network difficult (9).
A metagraph (9,10) is an advanced graph type developed in our laboratory to address these difficulties by integrating into a single network, inclusive or partially inclusive, relationships between information at different abstraction scales, the adjacent relationships between biomolecules. The inclusive relationship in a metagraph is represented by a metanode which is a special type of node that contains associated subnodes, much as a GO term contains its subterms or associated genes. A metanode has two states, expanded or collapsed; the expanded state manifests the internal subgraph (i.e. places all descendent nodes with their connections into the graph) while the collapsed state replaces this subgraph with the single node. Networks represented by a metagraph are usually termed metanetworks, and such visualization technology is often referred to multi-scale visualization because information at different abstraction scales is presented in one network. From this perspective, the implementation of metagraphs paves the way for VisANT to provide an intuitive visualization of integrated GO hierarchy and biological networks.
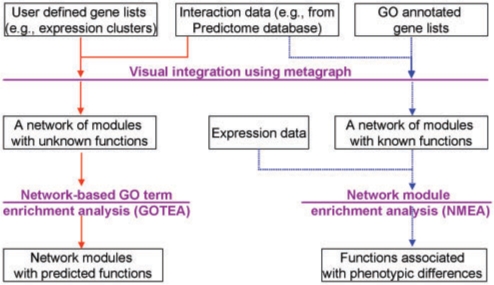
GO term enrichment analysis, or functional profiling (11), aims to determine whether particular GO terms inform the difference of molecular phenotypes in any set of user-specified genes, typically the co-expression modules (Figure 1, red lines). In a network context, the goal is to identify biological functions for a given subnetwork, or for a network module. Although many algorithms and tools (3,5,8,12–21) have been developed for GO term enrichment analysis, they generally omit correlations based on disparate and varied datasets, such as yeast two hybrid, genetic interaction, mass spectrometry (MS), and so on. Such relations may help to overcome some drawbacks in the current enrichment analysis. For example, one drawback is that all terms are weighted equally (22), while in a network module, terms annotated for highly connected genes will have more weight than those annotated for the loosely connected genes. Accuracy may also be improved if network type is considered; e.g. for a regulatory network, we probably can exclude those annotations of metabolic processes. From this perspective, flexible annotation schema will be needed to enable users to select subsets of GO annotations. Such flexibility could help determine the functions of genes in a specified network.
Figure 1.
Different roles of GO annotation in two different workflows designed for VisANT 3.5. The solid red lines represent the workflow of functional profiling where GO annotations are used to interpret the roles of a given gene set. The dashed blue lines represent the workflow of the gene set/network module enrichment analysis, where GO terms and associated genes are used to construct the functional modules.
Another typical application is the analysis of differential RNA expression patterns (e.g. tumor vs normal) determined by genome-wide association studies, to determine if one or more specified gene sets (e.g. KEGG pathways) might account for some of the differences (Figure 1, blue lines) (23–26). Gene set enrichment analysis (GSEA) (25) is probably the most used algorithm which does not take account of prior network knowledge.
Here we report VisANT 3.5, which enables visualization, analysis and inference of networks using GO. The new functions address the three key aspects discussed above: (i) integrated visualization of the GO hierarchy and user-specified networks. The inclusive relationship between the terms is represented by metanodes, where the metanode of a parent term embeds a network of connected subterms (also represented as metanodes) and/or genes annotated under the term; (ii) flexible annotation schema. We carefully evaluated all GO annotation methods and summarized them as a total of seven different annotation schemas. In addition, we developed a GO Explorer to visualize the GO hierarchies and to facilitate the easy selection of GO terms/branches for the corresponding annotation schema and (iii) GOTEA to predict the functions of network modules and network module enrichment analysis (NMEA) to test whether the modules are enriched with transcriptional changes between the control and the sample. In VisANT, a network can be constructed using the data from any combination of 60 odd methods (e.g. Y2H, Chip–Chip, MS and knock-outs) for the interested gene lists. And modules can be easily constructed as metanodes through corresponding menus, extended edge-list (http://visant.bu.edu/import.htm) and simple drag&drop operation from GO Explorer. In addition, GOTEA uses a fuzzy search (27) to detect those GO terms that are weakly enriched as individual terms, yet clearly over-represented when being evaluated together because they are very similar. Both algorithms take advantage of the extra information provided by network connectivity.
VisANT is implemented in Java and can run on any platform where Java is supported. The VisANT applet has been tested on many popular browsers including Internet Explorer, FireFox, Chrome and Netscape. VisANT relies on the Java Script to communicate between the Web pages and the applet. VisANT can also be run as a local application or through Java's Web start technology.
PROGRAM DESCRIPTION
All new functions are built based on VisANT's flexible three-tier architecture (10,28–30). Annotation hierarchy is retrieved from the Predictome database (31) and is updated monthly, so it is synchronized with the latest changes in the GO database (1). Information on genes annotated under each GO term for 13 species is obtained from the Entrez gene database (32) and is updated weekly. VisANT supports gene-based integration and has built-in functions to resolve gene names using various identification systems (30). Users are advised to resolve node names first if they use their own data to build a network. In the case of metanetworks (9,10,29), options are available under the MetaGraph menu to resolve the names of the entire network, including those hidden in collapsed metanodes.
Integrated visualization of GO annotation and the network
GO explorer
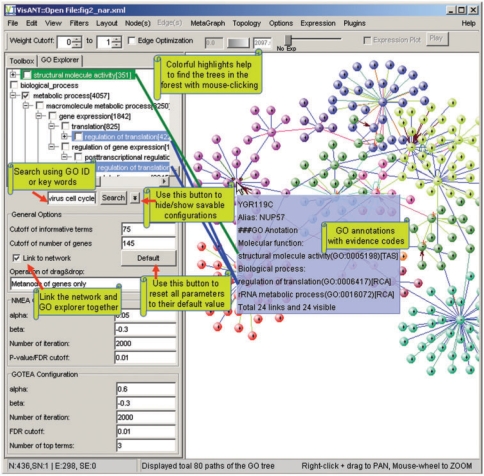
Although GO is stored as a DAG, probably the best option for navigation is to represent and visualize it using a denormalized tree structure in which a term with multiple parents is represented multiple times (22,33). The original context of the parent–child relationships of the DAG can be recovered using appropriate software functions. In the GO explorer, this is achieved by the simultaneous highlighting of all instances of the tree nodes representing the same GO term whenever one of them is highlighted. A tab control is introduced in VisANT so that the GO explorer and the toolbox remain together in the left control panel (Figure 2). The width of the panel can be changed through mouse-dragging to facilitate browsing, while the width of the toolbox remains unchanged. Clicking on the expansion symbol or double-clicking over the tree node will expand or collapse it. A database query will be sent to the VisANT server to retrieve the node's descendents. When the species changes the number of genes annotated under the branches of the corresponding descendents change accordingly. Other information, such as the number of genes directly annotated under the term, is shown in the tooltip by a mouse-over of the tree node (Figure 2). Each tree node is associated with a checkbox to allow user selection of GO branches. If the GO term appears in multiple places of the tree, selecting one of them will automatically select the rest. This also applies to node highlighting. Terms under different categories are highlighted using different color (Figure 2).
Figure 2.
Integrative visualization of GO annotation and user-mined or -specified networks. Note that YGR119C is annotated using two selected GO branches only: molecular function and metabolic process (indicated by the selected checkbox of the tree node; only the checkbox for metabolic processes is visible in the figure).
The queried GO tree is not designed to be saved locally so that whenever VisANT starts, it will always get the latest GO information from the VisANT server. The GO hierarchy stored in the Predictome database is updated monthly. The search box at the bottom of the GO explorer allows users to search the GO tree using GO IDs and key words. The options of GO-related functions can be configured by clicking the button nearby, as also shown in Figure 2.
Creating multi-scale networks
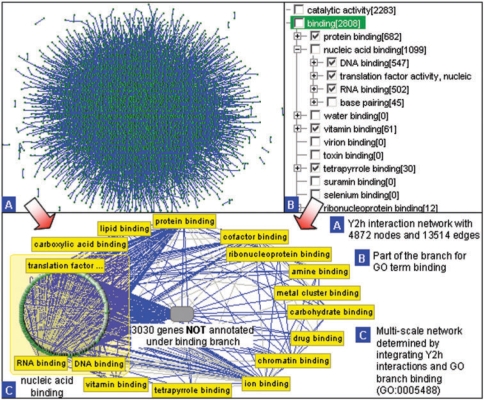
A GO term can be dragged into the network panel in VisANT to create a metanode which contains all its descendents and genes annotated directly under it, as shown in Figure 3. Other options for the drag&drop operation, such as creating a metanode containing all genes under the branch, are available in the configuration panel (Figure 2). If the genes are already in the network, the operation will group them into metanodes unless they are already in another metanode. In such cases, a duplicated node will be created and grouped in the new metanode. Once the metanode of the GO term is created in the network, users can double-click the node of its child GO term (such as RNA binding [GO: 0003723] shown in Figure 3C) to expand it if the GO term has its child term and/or genes annotated under it. Such multi-scale visualization schema classify groups of genes and/or terms as biological network modules thereby reducing the size of a large network to a manageable level, and greatly facilitating the analysis of gene-to-gene, term-to-term and gene-to-term relationships. The schema brings related genes and terms together in one place, facilitating the study of the related biological modules using the default aggregation functions of the metagraph to infer the term associations from the network of genes or their products (9).
Figure 3.
Illustration of the multi-scale network found by integrating the Y2h protein–protein interaction network of yeast and GO hierarchies. Y2h interaction network with 4872 nodes and 13 514 edges, part of the branch for GO term binding and multi-scale network resulted from the integration of y2h network and binding branch. Note that in C, the metanode contains genes annotated under nucleic acid binding (green nodes in circle), and three of its sub-terms. Only 1842 genes in the Y2H network are annotated under the GO branch binding.
Interaction between network and GO explorer
By default, a network and the GO explorer are integrated in such a way that when a GO term is selected, the corresponding metanode will be selected in the network. When a node with GO information is selected (a gene with GO annotation, a metanode of a GO term, a metanode with functions obtained from GOTEA, etc.), all possible paths from the corresponding GO terms to the root of GO tree will be shown in the GO explorer (Figure 2), with the terms being highlighted in the tree to allow users to view and explore the areas of the ontology graph surrounding the terms. Since GO terms usually have multiple parents, many paths can be associated with each term. Therefore, paths only display the necessary GO terms and will not, for brevity, show all the children of terms in the path. Users can, however, collapse and expand those terms to obtain all their descendants.
While the drag&drop function indirectly enables VisANT users to search for interactions using keywords, the interaction between a network and the GO explorer indirectly allows users to search the GO terms using the gene's name or IDs.
Flexible annotation schema
VisANT provides four basic options to annotate genes using GO annotations. Options 1–3 listed below can also be applied to the selected branches (Figure 2). These options provide users great flexibility to test various hypotheses.
Option 1: Genes are annotated with the most specific functional descriptions available.
Option 2: Genes are annotated using GO terms having more than a user-specified number of genes (34).
Option 3: Option 2 but limited to descendent terms having fewer than the specified number of genes (35).
Option 4: Genes are annotated using only selected GO terms.
Options 2 and 3 are frequently used when predicting gene functions using functional linkages. Cutoffs for both options are configurable (Figure 2). Annotations resulting from different options can coexist as node descriptions in VisANT for comparison purposes.
Enrichment analysis
GO term enrichment analysis
To take advantage of network information, a new algorithm has been developed and implemented as a VisANT plugin to find over-represented GO terms in user-specified network modules (represented as metanodes in VisANT). The function is available under the MetaGraph menu. By default, the analysis will be performed for all non-embedded metanodes; i.e. it is not performed for descendent metanodes unless they are specifically selected. Over-represented GO terms will be shown as a quick tip when the mouse is passed over a node, and clicking on a node will display the hierarchy of GO annotations. GOTEA requires genes in the modules to be annotated prior to the analysis.
For a given target GO term, the algorithm first computes the density score of each node based on the path distance (number of links) to other nodes in the same module, and the similarity between its associated GO terms and the target term. The use of a similarity score rather than an exact match enables the algorithm to give the target term a high score so long as it is functionally similar to the annotations of the genes in a module. The similarity score between two terms is calculated by aggregating the semantic contributions of their ancestor terms in the GO graph (27). The enrichment of target term is determined using statistical measurement through permutation test over the subset of the same number of genes extracted from all known genes annotated by Entrez gene database (32) with appropriate false discovery rate (FDR) (36) cutoff. Details of the algorithm can be found in the supplemental materials. Related parameters, such as the cutoff and the iteration number of the permutation test can be configured (Figure 2). By default, all terms that have the associated genes for the current species will need to be tested; users, however, may select subset of term branches in the GO explorer to speed up the analysis.
The advantage of the algorithm over similar algorithms is reflected in the computation of the density score, where the impact of one gene on another is a function of the GO term similarity, and the number of links between the genes. GO term similarity is calculated using a fuzzy search rather than a conventional exact match (27). With such a density score, a gene having many neighbors with similar GO terms will have more significant contributions to the enrichment outcome; the algorithm, therefore, leverages network topology as well as the GO hierarchy. In addition, metagraphs provide a flexible visual context to perform analysis for hierarchically organized network modules. The function is designed for workflow shown as the solid red line in Figure 1; network modules need not be limited to expression profiling.
Permutation-based algorithms tend to be computationally intensive and, therefore, time consuming. VisANT provides three options to address this shortcoming. First, VisANT implements a hypergeometric test-based algorithm to allow quick identification of the shared functions of a given gene set; second, VisANT provides an option ‘Fast GOTEA’, which only scans related GO terms for a given network module (GO terms annotated for the genes in the module and corresponding ancestor terms) and finally, macro commands have been created to allow the time-consuming GOTEA tasks to be carried out in the background with the command-line mode of VisANT.
Network module enrichment analysis
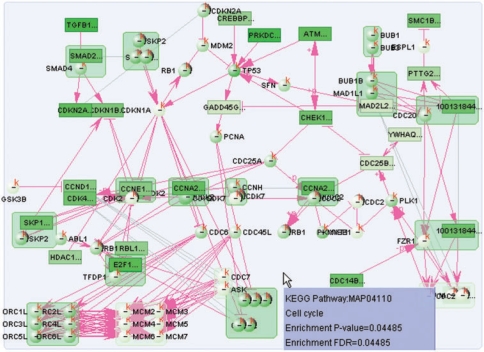
NMEA is an extension of our previous work on pathway enrichment analysis (PWEA). It is designed to find functional modules that inform phenotypic differences. For the current release, such differences are usually transcriptional activities. NMEA is implemented as a VisANT plugin and by default will be performed for all non-embedded metanodes. The function is available under the Expression menu and is similar to the GOTEA except that it requires the input of expression data (Figure 1, dashed blue lines). When execution of NMEA is complete, the nodes in the modules will be colored according to their enrichment scores (Figure 4) and an html report with detailed results for all modules will be generated.
Figure 4.
Visualization of NMEA of p53 mutation data on the cell-cycle pathway. The color of each node is in proportion to the density score according to the same color map of the expression shown in the VisANT toolbar, nodes with darker green have more contribution to the enrichment score.
The NMEA algorithm uses the same density function as GOTEA except that the P-values from t-tests on the expressions of two phenotypes replace GO term similarity. As a result, expressions of highly connected genes are weighted more in the enrichment outcome than those of loosely connected ones. Among the advantages of this approach are that it expands the flexibility and space of network modules in comparison to the available pathways; and that it improves the accuracy of enrichment analysis by introducing the impact of the network topology.
CASE STUDY
To demonstrate the functionality of the GOTEA and NMEA plugins, we first apply GOTEA to the KEGG cell-cycle pathway (hsa04110) to determine whether we can recover the pathway's functions from GO annotations of the pathway genes. We then apply NMEA to 22 microarray samples with mutations in p53 and 17 wild-type samples from GSEA Web site (http://www.broad.mit.edu/gsea/) and examine whether the NMEA algorithm can detect a phenotypic difference in the expression of genes from the cell-cycle pathway between mutated and wild-type samples. Macro files have been created and are accessible at http://visant.bu.edu to allow users to easily replay the case study.
The cell-cycle pathway for Homo sapiens can be loaded into VisANT by searching for ‘map04110’ or reading corresponding KEGG markup language (KGML) file. Each gene in the pathway is then annotated with its most specific GO annotation using the corresponding menu under MetaGraph (which will annotate all gene nodes including those hidden within collapsed metanodes). GOTEA is then performed by scanning all human-related GO terms (total 10 466), which detects 40 informative GO terms (with cutoff γ = 500). Among the 40 terms, 23 falls into the category of biological process, 12 falls into the category of molecular function and the remaining 5 belong to the category of cellular component. The informative terms are ranked according to the number of enriched GO terms (FDR < 0.01) that belong to them. As expected, the most informative term in the biological processes category is cell cycle. The most informative term under molecular function is protein kinase activity and the most informative term under cellular component is protein complex. These results agree with the definition of the pathway, which describes the reactions between protein kinases and protein complexes that take place in regulating the cell cycle. The complete list of informative nodes that were detected can be found in Table S1.
The result of using NMEA to analyze p53 mutants is also satisfactory. Since p53 mutants influence the behavior of the cell cycle, we expected that cell-cycle-related modules should be enriched. NMEA reported the cell-cycle pathway as significantly enriched with P-value 0.04 and, therefore, supports this point (Figure 4).
CONCLUSION
The new release of VisANT introduces extensive functionalities to visualize and integrate the gene ontology with biological networks using metagraph technology. The resulting multi-scale networks are more interpretive and more manageable. The new release also facilitates the biological characterization of networks (NMEA) and hypothesis testing. VisANT accepts various inputs—node or edge-lists, PSI-MI, GML and so on. The resulting network can be saved, shared and published in the different formats including VisANT XML format (VisML), edge-list and SVG. VisANT, along with the full user manual and tutorials, is available at http://visant.bu.edu.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (1R01RR022971-01A1 and 1R21CA135882-01). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues, students and other users of VisANT for their valuable comments and suggestions in improving all facets of the system.
REFERENCES
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reimand J, Tooming L, Peterson H, Adler P, Vilo J. GraphWeb: mining heterogeneous biological networks for gene modules with functional significance. Nucleic Acids Res. 2008;36:W452–W459. doi: 10.1093/nar/gkn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahlquist KD. Using GenMAPP and MAPPFinder to view microarray data on biological pathways and identify global trends in the data. Curr. Protoc. Bioinformatics. 2004 doi: 10.1002/0471250953.bi0705s05. Chap. 7, Unit 75, doi:10.1002/0471250953.bi0705s05 pmid:18428731. [DOI] [PubMed] [Google Scholar]
- 5.Khatri P, Voichita C, Kattan K, Ansari N, Khatri A, Georgescu C, Tarca AL, Draghici S. Onto-tools: new additions and improvements in 2006. Nucleic Acids Res. 2007;35:W206–W211. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 7.Garcia O, Saveanu C, Cline M, Fromont-Racine M, Jacquier A, Schwikowski B, Aittokallio T. GOlorize: a Cytoscape plug-in for network visualization with gene ontology-based layout and coloring. Bioinformatics. 2007;23:394–396. doi: 10.1093/bioinformatics/btl605. [DOI] [PubMed] [Google Scholar]
- 8.Huang da W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z, Mellor J, Wu J, Kanehisa M, Stuart JM, DeLisi C. Towards zoomable multidimensional maps of the cell. Nat. Biotechnol. 2007;25:547–554. doi: 10.1038/nbt1304. [DOI] [PubMed] [Google Scholar]
- 10.Hu Z, Mellor J, Wu J, Yamada T, Holloway D, Delisi C. VisANT: data-integrating visual framework for biological networks and modules. Nucleic Acids Res. 2005;33:W352–W357. doi: 10.1093/nar/gki431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhee SY, Wood V, Dolinski K, Draghici S. Use and misuse of the gene ontology annotations. Nat. Rev. Genet. 2008;9:509–515. doi: 10.1038/nrg2363. [DOI] [PubMed] [Google Scholar]
- 12.Zhang M, Ouyang Q, Stephenson A, Kane MD, Salt DE, Prabhakar S, Burgner J, Buck C, Zhang X. Interactive analysis of systems biology molecular expression data. BMC Syst. Biol. 2008;2:23. doi: 10.1186/1752-0509-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brohee S, Faust K, Lima-Mendez G, Sand O, Janky R, Vanderstocken G, Deville Y, van Helden J. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic Acids Res. 2008;36:W444–W451. doi: 10.1093/nar/gkn336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alibes A, Canada A, Diaz-Uriarte R. PaLS: filtering common literature, biological terms and pathway information. Nucleic Acids Res. 2008;36:W364–W367. doi: 10.1093/nar/gkn251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonov AV, Schmidt T, Wang Y, Mewes HW. ProfCom: a web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res. 2008;36:W347–W351. doi: 10.1093/nar/gkn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee T, Desai VG, Velasco C, Reis RJ, Delongchamp RR. Testing for treatment effects on gene ontology. BMC Bioinformatics. 2008;9(Suppl. 9):S20. doi: 10.1186/1471-2105-9-S9-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Wang J, Guo Z, Zhang M, Yang D, Li Y, Wang D, Xiao G. GO-2D: identifying 2-dimensional cellular-localized functional modules in gene ontology. BMC Genomics. 2007;8:30. doi: 10.1186/1471-2164-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonov AV, Tetko IV, Mewes HW. A systematic approach to infer biological relevance and biases of gene network structures. Nucleic Acids Res. 2006;34:e6. doi: 10.1093/nar/gnj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khatri P, Bhavsar P, Bawa G, Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21:1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- 24.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volinia S, Evangelisti R, Francioso F, Arcelli D, Carella M, Gasparini P. GOAL: automated gene ontology analysis of expression profiles. Nucleic Acids Res. 2004;32:W492–W499. doi: 10.1093/nar/gkh443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang JZ, Du Z, Payattakool R, Yu PS, Chen CF. A new method to measure the semantic similarity of GO terms. Bioinformatics. 2007;23:1274–1281. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
- 28.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Z, Ng DM, Yamada T, Chen C, Kawashima S, Mellor J, Linghu B, Kanehisa M, Stuart JM, DeLisi C. VisANT 3.0: new modules for pathway visualization, editing, prediction and construction. Nucleic Acids Res. 2007;35:W625–W632. doi: 10.1093/nar/gkm295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Z, Snitkin ES, DeLisi C. VisANT: an integrative framework for networks in systems biology. Brief Bioinform. 2008;9:317–325. doi: 10.1093/bib/bbn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellor JC, Yanai I, Clodfelter KH, Mintseris J, DeLisi C. Predictome: a database of putative functional links between proteins. Nucleic Acids Res. 2002;30:306–309. doi: 10.1093/nar/30.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pena-Castillo L, Tasan M, Myers CL, Lee H, Joshi T, Zhang C, Guan Y, Leone M, Pagnani A, Kim WK, et al. A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol. 2008;9(Suppl. 1):S2. doi: 10.1186/gb-2008-9-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X, Kao MC, Wong WH. Transitive functional annotation by shortest-path analysis of gene expression data. Proc. Natl Acad. Sci. USA. 2002;99:12783–12788. doi: 10.1073/pnas.192159399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.