Abstract
@TOME 2.0 is new web pipeline dedicated to protein structure modeling and small ligand docking based on comparative analyses. @TOME 2.0 allows fold recognition, template selection, structural alignment editing, structure comparisons, 3D-model building and evaluation. These tasks are routinely used in sequence analyses for structure prediction. In our pipeline the necessary software is efficiently interconnected in an original manner to accelerate all the processes. Furthermore, we have also connected comparative docking of small ligands that is performed using protein–protein superposition. The input is a simple protein sequence in one-letter code with no comment. The resulting 3D model, protein–ligand complexes and structural alignments can be visualized through dedicated Web interfaces or can be downloaded for further studies. These original features will aid in the functional annotation of proteins and the selection of templates for molecular modeling and virtual screening. Several examples are described to highlight some of the new functionalities provided by this pipeline. The server and its documentation are freely available at http://abcis.cbs.cnrs.fr/AT2/
INTRODUCTION
Sequence and structure comparisons are widely used to identify protein similarities and to derive functional or structural information (1). Indeed, the sequence–sequence software, Psi-BLAST (2), is now a standard for identifying protein homology (within five iterations by default). When various homologous sequence can be gathered, multiple alignments are also used to define conservation and deduce functional information. Distinct programs such as CLUSTALW (3), T-COFFEE (4) and MUSCLE (5) are available to perform this task rapidly and efficiently. Thanks to the gathering of protein 3D structures into available databases such as the PDB (6), sequence–structure relationships can be analysed to improve protein sequence comparisons. Remote similarities can be detected, routinely, using fold-recognition software such as FUGUE (7), SP3 (8) and many others. More recently, the efficiency of the profile-based approach was greatly improved through profile–profile comparisons (9) or similar methods based on Hidden Markov-Model such as HHSEARCH (10). These tools take into account structural information and/or protein evolution more accurately to provide better structural alignments (SAs). The latter can be evaluated and refined using various software combining alignment editing and structure visualization such as ViTO (11) or JalView (12). Further validation and analysis of SAs can be obtained through molecular modeling. Crude and partial 3D models can be extracted from 3D templates using TITO (13) in order to validate the input SA with the potential of mean force PKB (14). Alternatively, side-chain conformations can be optimized on fixed backbone using SCWRL (15). Complete modeling is routinely obtained using MODELLER (16) for each compatible template separately, or using the multi-template option. These models can be evaluated using standard sequence–structure compatibility functions, such as Verify3D (17). Clustering of modeled 3D structures using MaxCluster (18) and 3D-Jury (19) has been shown to efficiently highlight optimal 3D models or identify similar regions in related templates. All these programs (and many others) work efficiently, but their combination is often necessary in order to complete a predictive analysis. The use of multiple programs implies numerous data format changes, which tend to discourage or confuse many.
Automatic procedures are now available to provide 3D models of good quality when the sequence identity is above 30% and databases of 3D models exist (1). Important functional prediction can be also performed at much lower level of sequence similarities in the so-called twilight zone (see examples below). Indeed, it was recently shown that threading or fold recognition can be used for functional annotation and ligand-docking predictions (20). However, at lower level of similarities, consensus methods are needed and several tools have to be used for each task necessary for the completion of predictive modeling. This requires tedious changes of data formats as most software use specific input and output formats. This significantly slows down or limits an efficient use of the above-mentioned programs. Modeling protein–ligand complexes remains an even more difficult task. Template searches generally focus on sequence similarities rather than on the presence or not of a ligand bound to the template. Nevertheless, in most protein families, one template may provide the best protein scaffold, while another related template may provide a valid ligand (or a similar compound). Meanwhile, the lower the sequence conservation, the lower the model quality limiting the functional insights which can be derived. This means that at the end of an extensive sequence similarity search and molecular modeling study, virtual docking is unlikely to produce valid results. This currently limits extensive protein function annotations. Indeed, even the identification of the potential active site might be complicated.
Paradoxically, indicating a potential ligand or biological activity may help in validating the proposed sequence similarities. Improving sequence–structure alignment and simultaneously analysing interactions with potentially relevant ligands may help in refining template selection, molecular modeling and function predictions. Connecting structure analyses and functional annotations is more and more necessary for genome analysis. This has been exemplified previously even at very low level of sequence identity (10–15%; see examples below). These works relied on expertise and long-term sequence studies. The rapidly rising pace of genome sequencing requires improved capabilities for efficient structure modeling and function prediction (20).
A new server, named @TOME-2, has been developed to bridge comparative modeling and comparative docking in order to extend sequence analysis from fold recognition to functional annotation. Various features have been implemented and numerous programs interfaced to connect sequence with 3D structure and putative ligands. A former version of the fold recognition and comparative modeling pipeline was described previously (21), while a web server dedicated to the analysis of ATP-binding pockets of protein kinases, has been developed and used to validate the so-called comparative docking approach (22). In the new server @TOME2, state-of-the-art programs for fold recognition, structural alignment evaluation and editing as well as comparative modeling have been connected. Comparative docking can be performed on-the-fly for any selected model built and any small ligand detected among the aligned templates. Ligands are posed according to protein–protein superposition. The resulting complexes are evaluated using the scoring function of MedusaScore (23) and of AUTODOCK (24).
Fold recognition and in-depth evaluation of structural alignments are first performed in background. Supplementary evaluation, modeling and docking tasks can be performed interactively. The new server is made freely accessible to the academic community. Several examples are described to illustrate the use and the performance of @TOME-2.
METHODS
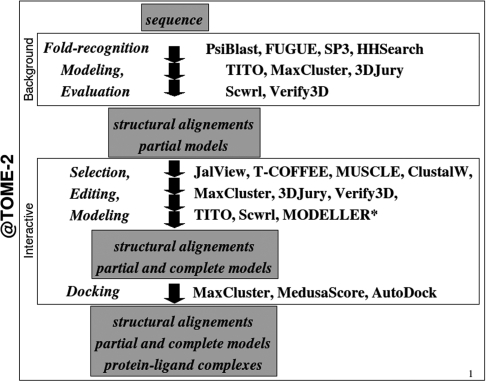
@TOME-2 allows one to submit a simple amino-acid sequence to perform various protein-structure analysis and prediction (Figure 1). It is a pipeline written in Perl wrapping numerous databases and software developed by the bioinformatic community. Two major steps are currently available. The first step is a calculation run in background, to optimally detect sequence–structure compatibility. An e-mail is sent upon completion of the fold-recognition search and structural-alignment evaluation. The second step is an interactive refinement procedure to further validate and exploit the detected similarities. In both cases, the results are displayed as an HTML page with clickable menus, selection boxes, links and embedded JAVA applets. Results can be also downloaded in various formats.
Figure 1.
Data flow in the pipeline @TOME-2. Most software are free to use for academic users although MODELLER required a license key.
Starting page
The front page allows the user to fill in three mandatory fields: a title, the primary sequence (in one-letter code) and an e-mail address (see below). Clickable options and menus allow one to select the required modules and the corresponding software (e.g. for fold recognition) or databases (e.g. ligand types for comparative docking). Specific options can be also selected (e.g. ‘molecular replacement’ module to be described elsewhere).
Background calculations
Currently, @TOME-2 performs fold recognition using four distinct tools dedicated to sequence–sequence (Psi-BLAST and HHsearch) or sequence–structure comparisons (FUGUE and SP3). These four programs use different algorithms and/or distinct databases. This allows consensus calculations to optimally detect compatible templates. The fold-recognition searches are now performed on our 17-node LINUX cluster.
The output of the four programs corresponds to pairwise sequence–structure alignments in various formats. Furthermore, the scoring functions vary from one fold-recognition tool to another making comparison difficult. Sequence identity may be of limited value for low level of sequence conservation (say ∼10–15%) or for short sequence alignment (below 70–80 residues). It is rather a crude estimate of the quality of the SA (and potentially of the difficulty for a subsequent modeling study). Furthermore, to increase speed, the front-end software used for fold recognition (e.g. FUGUE, SP3) takes advantage of the so-called ‘frozen-approximation’ (25). In order to circumvent these limitations, additional scoring functions are used to evaluate the selected structural alignments (13).
A first round of evaluation is performed, for all the SAs produced by the fold-recognition search. Sequence identity and T-COFFEE score are computed. Then the common cores shared by each template (experimental 3D structures) and the query protein (according to each SA) are extracted using TITO and are evaluated using the PKB potential of mean force (using side-chain centroid pair contacts; 14). MaxCluster (18) is used to identify common substructures within the aligned regions given a distance threshold (0.8 Å by default, in @TOME-2). The Hierarchical Clustering algorithm (by default in @TOME-2) is used and the average linkage method permits to calculate distance between nodes. Then, the 3D-Jury score (19) is used to evaluate the corresponding partial models. These scores are combined to compute a new consensus scoring function in order to select the 20 best SAs. For speed, only the latter are submitted to a second round of evaluation. Using SCWRL 3.0 (15), substituted side chains are built on a fixed backbone. Strictly conserved residues are constrained during this conformational search. The corresponding models of the 3D protein cores are evaluated using the software Verify3D. Finally, a new consensus scoring function is computed by @TOME-2. The scoring scheme is currently based on a heuristic function (see online documentation).
By default, the best 20 SAs (according to @TOME-2 ranking score) are displayed on the result page with a color scheme (helix in red, strands in blue and loops in green, while for the strictly conserved residues in loops are shown in black) corresponding to secondary structures assigned using P-SEA (26). They are all gathered in a multiple alignment (or a set of multiple alignments) without changing the original pairwise alignments while propagating the necessary gaps.
Upon completion of this background sequence–structure comparison, an overview of the results should indicate whether a set of significantly similar structures has been detected or not. In the former case, molecular modeling and ligand docking may be reasonably envisioned and be obtained through the Web interface. If no consensus is highlighted, alternative clustering, SA editing or modeling are required to potentially establish more subtle similarities. These tasks can be performed interactively with no need to manually change the data format. When necessary, these tasks are carried out automatically using dedicated scripts or through the server PAT (27) and openBabel (http://openbabel.sf.net).
Interactive functionalities
The main functionalities of @TOME-2 can be accessed through the menu below the results page. This requires selecting some SAs, and, if neccesary ligands. Selection is done by clicking boxes on the left side of each SA or on the left side of the line describing the corresponding ligand. The displayed SAs can be re-ranked according to any scoring function including the original alignment score from Psi-BLAST, Fugue, HHsearch and SP3 or the sequence identity. Another set of SAs (e.g. those corresponding to protein–ligand complexes) can be selected and displayed using the left-panel menu. The necessary model building and structural evaluations are performed immediately. Similarly, the coloring scheme can be changed (e.g. to highlight residues in contact with selected ligands).
SA editing is the first task proposed in the main menu. It sends the selected SAs to a JAVA editor, JalView (12) or to automatic tools for multiple sequence alignments. This corresponds to a rather expert use, especially in the case of manual editing. The T-COFFEE coloring scheme may help in identifiying locally incorrect alignment. This may also guide selection of complementary SAs to be combined at the step of model building (see below).
Clustering by structure–structure comparisons can be re-run using other thresholds (0.6 Å–1.0 Å) and/or various sets of SAs. This may help gathering more distantly related structures using a slighly looser threshold (1.0 Å instead of 0.8 Å). This would validate the recognition of a global fold or super-secondary structure type. Using a tighter threshold may indicate some structural variation among the detected templates or protein subfamily. This can lead to focused selection and refined molecular modeling. Organization in domains with potential conformational changes should be taken into account while using this tool.
Obviously, convincing SAs should prompt the user to run MODELLER on several of the best templates either separately or in combination (using the multi-template option). Alternatively, extensive modeling may be performed to evaluate each selected SAs. Automatic building of valid 3D models (according to MODELLER energy and Verify3D score) would suggest that correct protein similarities have been detected at the fold-recognition step. This round of 3D-model building may serve to select the best possible templates. Then, the latter may be used in combination (as a multiple template) or provide a starting multiple SA to be edited and refined (see below). Several models (up to 4) can be built for each template at each run. The best energy (or Verify3D score) is written in the main result page. The corresponding link brings the user to a specific result page containing further model evaluations, details on the modeling run and a model visualization module using the Jmol applet (http://www.jmol.org).
@TOME-2 integrates an original interface to comparative docking of small molecules that are detected in the PDB file of each template. By default, this includes small organic molecules but it can also include ions, peptides, sugar compounds and nucleic acids (upon selection in the submission form). All the ligands in close contact (one atom at <4 Å from one atom of the template polypeptide) are taken into consideration. Contacts are classified according to three distance thresholds (4, 6 and 8 Å, respectively). In order to increase the speed of ligand detection, all ligand–protein distances have been precomputed for the whole PDB database and stored in a local database called CBE for ‘Contact Between Entities’. Access to the database CBE is available at http://abcis.cbs.cnrs.fr/AT2/cbe.html. The number of ligands detected is shown for the corresponding template on the result page. Upon clicking on this number, the ligands are listed to show their name, size, nature, the number of residues in contact and the highest similarity to a biological ligand listed in PROCOGNATE (28). The residues that are in contact with the selected ligand(s) are highlighted on a copy of the SA. Highlighting the positions in contact can be used for the selection of the ligand, of the 3D template or of the best alignment. A box can be clicked to select the desired ligand. Comparative docking can be performed for a set of selected 3D-models and ligands. It may preferentially be done using constrained 3D models built using TITO or SCWRL, especially at low level of sequence identity (below 25%). 3D models built by MODELLER might be used for docking only when excellent SAs have been obtained. In all cases, 3D models and the template structure in complex with the desired ligand are superposed using MaxCluster (18). Then, the novel protein–ligand interactions (but those including metal ions) are computed using MedusaScore (23) and AUTODOCK (24). The best interaction score is displayed in the main result page. A link brings the user to a docking result page containing a score table and a Jmol applet for visualization of the protein–ligand complexes and the corresponding protein–protein superposition. Ranking by superposition score or interaction score can be done. A table of results is drawn for each type of 3D models (built by TITO, SCWRL or MODELLER). This may highlight trouble in model completion or the role of the strictly conserved residues in the interactions. In this way, comparative docking can provide a local evaluation of sequence conservation (e.g. within active sites). This may distinguish homologous from analogous templates or suggests some functional variations (e.g. ligand specificity). Finally, a short and focused simulation can be performed using AUTODOCK 4.0 (24) to refine the ligand positions. All the computed complexes can be downloaded. Longer simulation might be necessary for complete exploration of the binding pocket and ligand conformational space. Thus, all the necessary input files can be also downloaded.
Finally, the main menu allows the user to archive the desired SAs in various formats (such as PIR and FASTA). A set of 3D models (or templates) can be superposed using the program Matt (29) and then saved as a compressed tar file. The SAs saved in PIR format can be edited using ViTO (11) or used in MODELLER offline. All the mentioned options should help recognizing the proper templates for comparative modeling, potentially some ligands, and tentatively combine them. The usefulness of the described pipeline is exemplified below for several case studies.
Case studies
Example 1: Ssu72 from yeast
Ssu72 is a small protein previously shown to be conserved in eukaryotes. It was genetically linked with a serine/threonine kinase and was known to physically interact with transcription initiation and termination complexes (30). In parallel, we have provided evidence that yeast Ssu72 is a phosphatase of unknown specificity by means of extensive sequence analysis and molecular modeling. Ligand docking by similarity allowed the correct identification of the active site in agreement with subsequent mutagenesis studies. We also successfully predicted its inhibition by ortho-vanadate, a generic inhibitor of the phosphatase/arsenic reductase superfamily (30). Identical predictions can now be performed using @TOME-2 within a few clicks and in a semi-automatic manner (see: http://abcis.cbs.cnrs.fr/ATV2/EG/70890/atome.html). Clustering of the sequence conservation in the catalytic center is rapidly highlighted, while the prediction of the substrate specificity appears elusive as the remaining of the sequence seems highly divergent (overall sequence identity ∼15%). The role of best conserved motif (‘C-x2-N-x2-R-S') can be readily deduced from comparative docking (see Figures 2 and 3). As shown previously (30), the resulting conclusions can be tested more easily and more accurately than usual biophysical evaluation of a global structure conservation. Indeed, we are still awaiting the resolution of its structure but this enzyme has been already shown to be a serine/threonine phosphatase with an original substrate specificity (31).
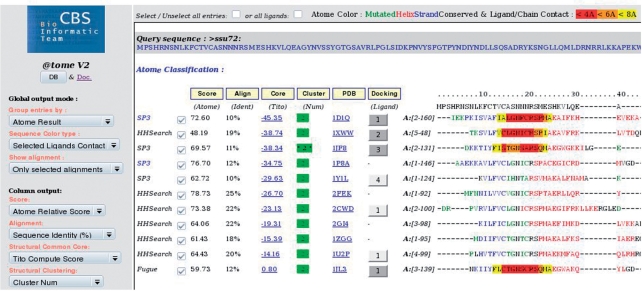
Figure 2.
Result page of @TOME-2. Example of results after a background calculation for the yeast protein Ssu72 followed by selection of ligands. Residues of selected experimental templates are colored according to secondary structure (red, blue and green stand for helix, strand and loop, respectively) or according to their distance to the selected ligands (Boxes in yellow, orange or red for distances below 8, 6 and 4 Å, respectively).
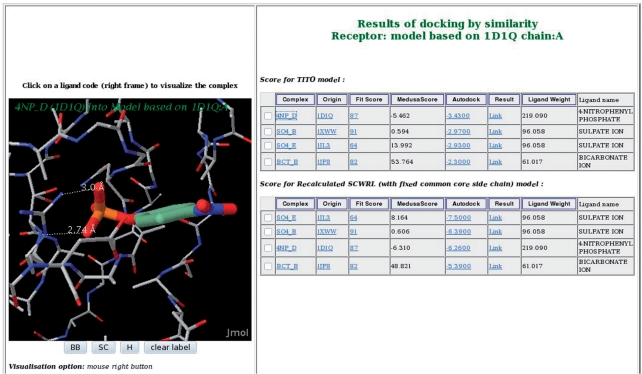
Figure 3.
Result page for on-the-fly comparative docking. A comparative docking was performed using a set of selected ligands and models deduced from the alignment of ssu72 and the template PDB1D1Q. The two partial models were built using TITO or SCWRL. Ranking of ligand poses are done according to MedusaScore or AutoDOCK (for the TITO model and the SCWRL model, respectively). Close view of the protein–ligand complex between ssu72 (TITO partial model) and 4-nitro-phenyl phosphate is shown in the left panel using the Jmol applet.
Example 2: CggR from Bacillus subtilis
CggR belongs to the SorC family of bacterial transcriptional regulators, which control the expression of genes and operons involved in carbohydrate catabolism (32). Automatic fold recognition and molecular modeling of CggR can rapidly reveal the presence of a winged-helix DNA-binding motif followed by a C-terminal domain presenting weak similarity (10% of global sequence identity) with glucosamine-6-phosphate deaminases from the NagB family. Before the 3D structure of the latter domain was completed, a predictive analysis of structure/function relationship suggested that the C-terminal region of CggR could be involved in binding of a phosphorylated sugar. Indeed, in vitro, fructose-1,6-biphosphate was the only sugar compound capable of interfering with CggR cooperative binding to DNA (32). The same conclusions can be drawn using @TOME-2, first, to delimit the domain boundaries (see: http://abcis.cbs.cnrs.fr/ATV2/EG/87746/atome.html), and then to build satisfactory 3D models. Comparative ligand docking highlights the role of some conserved residues in the recognition of one of the two phosphate groups of the actual ligand of CggR. Their importance for the function of this repressor has been confirmed by means of mutagenesis and biochemical analyses (32). The lost of the catalytic residues can be also derived from the sequence–structure comparison in agreement with the binding of an effector by the repressor without chemical modification.
Example 3: Rop2
More recently, we used @TOME-2 to confirm the absence of ligand binding in the active site pocket of an atypical protein kinase from Toxoplasma gondii. While ATP transfer could be successfully performed for a paralogous but active proteine kinase such as Rop18 using KinDOCK (33), the more divergent Rop2 sequence could not be properly aligned with the older procedure. Thanks to the state-of-the-art fold-recognition software embedded in @TOME-2, longer and better SAs are obtained (see: http://abcis.cbs.cnrs.fr/ATV2/EG/48308/atome.html). Their quality and coverage can be assessed as well as that of the corresponding 3D models. This represents a significant improvement over the former server.
CONCLUSIONS AND PERSPECTIVES
As shown previously for the protein-kinases (22) and as illustrated by the above examples (and other to be found in the HTML documentation), successful combination of sequence–structure comparisons with ligand docking is possible even at very low levels of sequence identity (10–20%) in agreement with recent statistical validations (20). This prompted us to make the methodology easily available through a Web-service. However, new scoring functions for evaluation of SAs, 3D models and protein–ligand complexes shall be implemented to improve ranking and prediction reliability using consensus strategies. Similarly, functional information corresponding to the templates shall be displayed. Nevertheless, it should be pointed out that predictions must be always regarded as hypothetical (especially at low level of sequence identity) and as shown in the above examples, should rather serve to guide experimental validations.
FUNDING
Funding for open access charge: CNRS-INSERM.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
This study has been supported by the CNRS. We thank all the groups that kindly provided us with databases and with binaries or source codes of the software installed and interfaced in this pipeline. We are indebted to Cathy Royer for careful reading of the manuscript.
REFERENCES
- 1.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 2.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 5.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Zhou Y. SPARKS 2 and SP3 servers in CASP6. Proteins. 2005;58:321–328. doi: 10.1002/prot.20732. [DOI] [PubMed] [Google Scholar]
- 9.Sadreyev RI, Grishin NV. COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J. Mol. Biol. 2003;326:317–336. doi: 10.1016/s0022-2836(02)01371-2. [DOI] [PubMed] [Google Scholar]
- 10.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 11.Catherinot V, Labesse G. ViTO: tool for refinement of protein sequence-structure alignments. Bioinformatics. 2004;20:3694–3696. doi: 10.1093/bioinformatics/bth429. [DOI] [PubMed] [Google Scholar]
- 12.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 13.Labesse G, Mornon JP. Incremental threading optimization (TITO) to help alignment and modelling of remote homologues. Bioinformatics. 1998;14:206–211. doi: 10.1093/bioinformatics/14.2.206. [DOI] [PubMed] [Google Scholar]
- 14.Bryant SH, Lawrence CE. An empirical energy function for threading protein sequence through the folding motif. Proteins. 1993;16:92–112. doi: 10.1002/prot.340160110. [DOI] [PubMed] [Google Scholar]
- 15.Canutescu AA, Shelenkov AA, Dunbrack R.L., Jr. A graph theory algorithm for protein side-chain prediction. Protein Science. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 17.Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: assessment of protein models with three dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 18.Siew N, Elofsson A, Rychlewski L, Fischer D. MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics. 2000;16:776–785. doi: 10.1093/bioinformatics/16.9.776. [DOI] [PubMed] [Google Scholar]
- 19.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 20.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc. Natl Acad. Sci. USA. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douguet D, Labesse G. Easier threading through Web-based comparisons and cross-validations. Bioinformatics. 2001;17:752–753. doi: 10.1093/bioinformatics/17.8.752. [DOI] [PubMed] [Google Scholar]
- 22.Martin L, Catherinot V, Labesse G. kinDOCK: a tool for comparative docking of protein-kinase ligands. Nucleic Acid Res. 2006;34:W325–W329. doi: 10.1093/nar/gkl211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin S, Biedermannova L, Vondrasek J, Dokholyan NV. MedusaScore: an accurate force-field based scoring function for virtual drug screening. J. Chem. Inf. Model. 2008;48:1656–1662. doi: 10.1021/ci8001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a lamarckian genetic algorithm and and empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 25.Skolnick J, Kihara D. Defrosting the frozen approximation: PROSPECTOR–a new approach to threading. Proteins. 2001;42:319–331. [PubMed] [Google Scholar]
- 26.Labesse G, Colloc'h N, Pothier J, Mornon JP. P-SEA: a new efficient assignment of secondary structure from Ca trace of proteins. CABIOS. 1997;13:291–295. doi: 10.1093/bioinformatics/13.3.291. [DOI] [PubMed] [Google Scholar]
- 27.Gracy J, Chiche L. PAT: a protein analysis toolkit for integrated biocomputing on the web. Nucleic Acids Res. 2005;33(Web Server issue):W65–W71. doi: 10.1093/nar/gki455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bashton M, Nobeli I, Thornton J. PROCOGNATE: a cognate ligand domain mapping for enzymes. Nucleic Acids Res. 2008;36:D618–D622. doi: 10.1093/nar/gkm611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menke M, Berger B, Cowen L. Matt: local flexibility aids protein multiple structure alignment. PLoS Comput. Biol. 2008;4:e10. doi: 10.1371/journal.pcbi.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganem C, Devaux F, Torchet C, Jacq C, Quevillon-Cheruel S, Labesse G, Facca C, Faye G. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 2003;22:1588–1598. doi: 10.1093/emboj/cdg141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 is a RNA polymerase II CTD phosphatase. Mol. Cell. 2004;14:387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 32.Doan T, Martin L, Zorrilla S, Chaix D, Aymerich S, Labesse G, Declerck N. Modelling and mutagenesis of the sugar binding domain of the central glycolytic gene repressor (CggR) reveal structural homology with Glucosamine 6-phosphate deaminase (NagB) Proteins. 2008;71:2038–2050. doi: 10.1002/prot.21883. [DOI] [PubMed] [Google Scholar]
- 33.Labesse G, Gelin M, Bessin Y, Cerdan R, Lebrun M, Papoin J, Arold ST, Dubremetz J.-F. ROP2 from Toxoplasma gondii: a virulence factor with a protein-kinase fold and no enzymatic activity. Structure. 2009;17:139–146. doi: 10.1016/j.str.2008.11.005. [DOI] [PubMed] [Google Scholar]