Abstract
Protein structures often show similarities to another which would not be seen at the sequence level. Given the coordinates of a protein chain, the SALAMI server at www.zbh.uni-hamburg.de/salami will search the protein data bank and return a set of similar structures without using sequence information. The results page lists the related proteins, details of the sequence and structure similarity and implied sequence alignments. Via a simple structure viewer, one can view superpositions of query and library structures and finally download superimposed coordinates. The alignment method is very tolerant of large gaps and insertions, and tends to produce slightly longer alignments than other similar programs.
INTRODUCTION
Purpose of SALAMI
Sequence similarity is the classic measure for finding related proteins and the starting point for assigning function, building phylogenies and protein modelling. Sequence similarity will not, however, be enough to detect remote relationships. For this, one needs methods that detect pure structural similarity. Given the coordinates of a protein chain, the SALAMI server will search the protein data bank (1), for similar chains, calculate structural alignments and generate a list of structurally related proteins. In some sense, structure is preserved more than sequence during evolution (2) so even within a family of related proteins, there may be members with no significant sequence similarity to another (3–8). This means that questions of function or phylogenetic relations will often only be answerable given structural relationships (9). Furthermore, there is the question of alignment quality. In the case of weak sequence similarity, the alignment implied by a structural superposition should be more reliable and more useful for problems such as predicting functional sites.
Structure comparison
Aligning protein structures is a fundamentally NP-complete problem when one allows for arbitrary gaps and insertions (10). This means that all methods rely on some approximations and there will always be trade-offs between quality and speed. Furthermore, the problem is not perfectly defined since there may be no unique ideal alignment (11,12) and there is not even a single definition of alignment quality. One could argue that a good alignment minimizes differences in Cartesian space, but one could also say that a good method will find the corresponding residues despite large coordinate shifts due to hinge-bending or domain motions. For someone working on structure determination, it may be very useful if a method can recognize structural similarities when faced with the irregularities of an initial NMR-derived structure or unrefined crystallographic coordinates. Finally, programs will differ because they have been tuned to different goals. Some authors prefer shorter alignments of very similar regions, whereas some prefer longer alignments including regions of greater variation.
Because the alignment problem is difficult and not even well defined, there is a large variety of approaches and using n different programs may give n different structural alignments (13–43). There are, however, some common ideas. Some methods try to build a crude seed alignment which can be extended or iteratively improved (17,30). Some methods assign descriptors to sites which can be aligned using methods similar to those in sequence alignment. These descriptors, of course, come in many forms ranging from distance matrices to textbook secondary structure or fragment-based alphabets (18,33,44).
SALAMI also attaches descriptors to sites, but they are fuzzy or probabilistic. This means that there are no predefined thresholds and no requirement that a fragment be seen as helix, sheet or coil. Instead, fragments are compared with each other using a continuous estimate of similarity.
Although there is a large number of methods for structural alignment, relatively few are fast enough to search a large library of structures (21,22,24,25,33). The SALAMI server is fast enough to search the protein data bank for medium-sized proteins in 10–20 min using a single CPU.
MATERIALS AND METHODS
Input data and library
The server takes the coordinates of a protein chain in PDB format and an email address for sending results to. The only adjustable parameter is the number of aligned structures to return.
Output of the web server
The server sends a rather minimal mail message as its result. It contains only a link to a temporary web page (lifetime 1 week) containing a list of candidate structurally related proteins. Selecting a candidate brings up a view of the superposition using Jmol (http://jmol.org) by E. Willighagen et al. (requires Java plugin). In another pane, the implied sequence alignment is shown, the superimposed coordinates can be downloaded and a list of more proteins with 90% or more sequence similarity to the candidate is given.
Each alignment is evaluated by scoring functions such as the alignment length, root mean squared difference (rmsd) of Cα atoms of aligned residues, a z-score calculated from a distribution of random alternative alignments (45), Smith and Waterman alignment scores (46) and a quality score based on the fraction of distance matrices which are similar between the query and aligned protein (45,47). This measure is used for the initial sorting of the list, but one can select a ranking by any of the other scores.
Processing Method
Our method is a specialization of a very general technique which has been described in detail (13). Briefly, 1.5 × 106 fragments, each of six residues were clustered into 308 classes, each of which is a set of six bivariate Gaussian distributions for backbone φ and ψ angles. The more populated classes are recognizable as classic secondary structure, while the less populated classes are simply pieces of common protein motifs. Given a query fragment, one can calculate its probability of being in each of the classes, resulting in a long list (vector) of probabilities. A typical fragment may have a probability near 1.0 of being in some class, but even an unusual fragment will have some characteristic pattern of probabilities. Any two fragments can be compared by taking the dot product of these probability vectors which leads to the final alignment method as previously described (13). A similarity matrix is built based on all overlapping fragments from each protein. The scores associated with a residue come from all the fragments which it is a part of, so for fragments of length k = 6, a residue is sensitive to an environment of 2k − 1 = 11 residues. The residue alignment can be read out from a conventional dynamic programming calculation (46,48) and superpositions are computed based on the aligned Cα atoms (49).
The method is fast since probabilities associated with databank proteins are precalculated and updated weekly. The similarity score has no hard thresholds, so the method fares well even when faced with slightly unusual structures. We give an example of this property below. Technically, it is interesting to note that the rmsd in Cartesian coordinates is never used during the alignment, so the method will find similarities even when confronted with domain or hinge-bending movements.
The server does not search all proteins in the protein data bank, but rather a subset of <2 × 105 is chosen so that no two chains have >90% sequence identity (50).
RESULTS
Precision of search results
Results from the structure similarity servers usually differ from another in two main ways. First, the length of alignments is rarely the same from two different programs. Second, there is some concept of sensitivity. For some query, related proteins should be ranked higher than unrelated proteins. There is, however, often no correct answer when relationships are weak. Rather than debate this, we have simply taken SCOP (4–7) as a reference. It is also rather easy to find query proteins which suit a particular method. Rather than try to be objective, we give an example which suits SALAMI, one where all methods perform well and one where SALAMI performs poorly.
Figures 1–3 show plots of the precision of SALAMI, DALI (51) and VAST (52). We considered up to 100 related proteins from each server for each query and filtered out all chains which were not classified by SCOP. Chains which contained a domain in the same superfamily as a domain in the query chain were considered to be true positives. The remaining chains were regarded as false positives. The plots show the fraction of true positives at each rank.
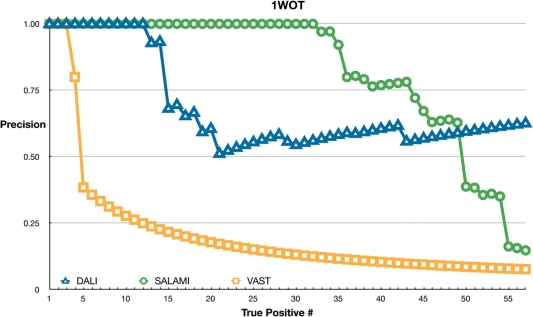
Figure 1.
Sensitivity of servers using 1WOT as a query. For each rank on x-axis, each point shows the number of true positives divided by the rank. Servers (DALI, VAST and SALAMI) are marked as shown in the key. Lines joining the points have no meaning and only serve to guide the eye.
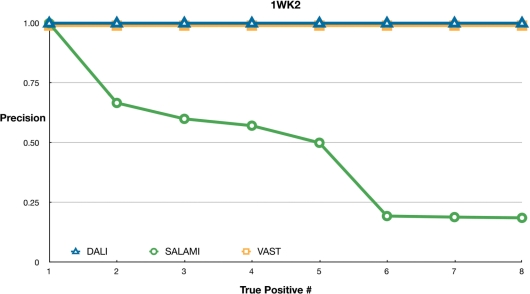
Figure 3.
Sensitivity of servers using 1WK2 as a query. Markers and servers as in Figure 1.
First, Figure 1 shows the results using 1WOT as a query. This protein clearly suits SALAMI. VAST finds the four closest relatives. DALI, however has more interesting behaviour with a large number of false positives near the middle of the list. The structure has three α-helices joined by some small β-strands. In SCOP, it is placed in the Nucleotidyltransferase superfamily. There is, however, a set of proteins in the KH-domain superfamilies with a similar fold which can be superimposed surprisingly well. They are declared to be unrelated in SCOP, but they score well in DALI.
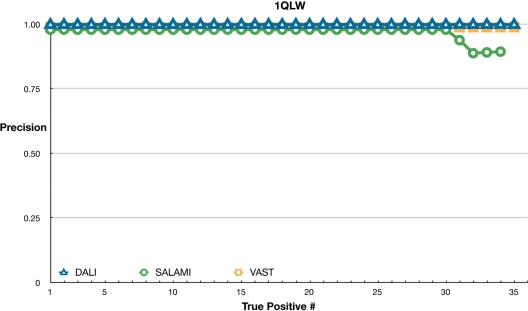
Figure 2 shows all the three methods performing equally well for 1QLW from the superfamily of alpha/beta hydrolases. Here, all results are in near perfect agreement with the SCOP classification. Only the SALAMI server includes a few false positives towards the end of the list.
Figure 2.
Sensitivity of servers using 1QLW as a query. Markers and servers as in Figure 1.
Finally, Figure 3 shows the results with 1WK2 from the PUA domain-like superfamily as the query. This does not suit the SALAMI server. It is a mostly β protein, but more than 30 of its 121 residues are missing. The correct relatives are pushed down the ranking by unrelated proteins. DALI and VAST still perform well here because their similarity scores are much more influenced by spatial distances to elements which are not necessarily close in sequence.
DISCUSSION AND CONCLUSION
The few results are certainly no benchmark. They are, however, clear examples of the ways different methods will work well with different query structures.
SALAMI has the disadvantage that it relies on chain connectivity and can be confused by broken structures. This means it may not be very useful for the broken skeletons that one can encounter in crystallographic structures with initial phasing. SALAMI has the advantage that it relies on chain connectivity and has no problem finding similarities when there are hinge-bending or domain motions. The graduated similarity measures mean that poor quality structures and deviations from regular geometry are well treated (13).
The methodology here has another interesting property. The graduated measure of similarity leads to a scoring function which is reliable and applies to any kind of structural unit. The use of a dynamic programming method then guarantees that the alignments are optimal within this scoring function. This, together with the good results for difficult structures and the flexible interface make it a valuable alternative to existing webservers.
FUNDING
Funding for open access charge: University of Hamburg.
Conflict of interest statement. None declared
REFERENCES
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–602. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 3.Holm L, Sander C. The FSSP database: fold classification based on structure structure alignment of proteins. Nucleic Acids Res. 1996;24:206–209. doi: 10.1093/nar/24.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreeva A, Howorth D, Chandonia J.-M, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreeva A, Howorth D, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conte LL, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res. 2002;30:264–267. doi: 10.1093/nar/30.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 8.Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, Orengo CA. The CATH classification revisited–architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheeff E, Bourne P. Structural evolution of the protein kinase-like superfamily. PLoS Comp. Biol. 2005;1:E49. doi: 10.1371/journal.pcbi.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eidhammer I, Jonassen I, Taylor W. Structure comparison and structure patterns. J. Comput. Biol. 2000;7:685–716. doi: 10.1089/106652701446152. [DOI] [PubMed] [Google Scholar]
- 11.Feng ZK, Sippl MJ. Optimum superimposition of protein structures: ambiguities and implications. Fold Des. 1996;1:123–132. doi: 10.1016/s1359-0278(96)00021-1. [DOI] [PubMed] [Google Scholar]
- 12.Godzik A. The structural alignment between two proteins: is there a unique answer? Protein Sci. 1996;5:1325–1338. doi: 10.1002/pro.5560050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schenk G, Margraf T, Torda AE. Protein sequence and structure alignments within one framework. Algorithms Mol. Biol. 2008;3:4. doi: 10.1186/1748-7188-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosca R, Brannetti B, Schneider T. Alignment of protein structures in the presence of domain motions. BMC Bioinformatics. 2008;9:352. doi: 10.1186/1471-2105-9-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosca R, Schneider T. RAPIDO: a web server for the alignment of protein structures in the presence of conformational changes. Nucleic Acids Res. 2008;36:W42–W46. doi: 10.1093/nar/gkn197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuker M, Somorjai R. The alignment of protein structures in three dimensions. Bull. Math. Biol. 1989;51:55–78. doi: 10.1007/BF02458836. [DOI] [PubMed] [Google Scholar]
- 17.Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Sander C. Protein Structure Comparison by Alignment of Distance Matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 19.Subbiah S, Laurents D, Levitt M. Structural similarity of DNA-binding domains of bacteriophage repressors and the globin core. Curr. Biol. 1993;3:141–148. doi: 10.1016/0960-9822(93)90255-m. [DOI] [PubMed] [Google Scholar]
- 20.Alexandrov NN. SARFing the PDB. Protein Eng. 1996;9:727–732. doi: 10.1093/protein/9.9.727. [DOI] [PubMed] [Google Scholar]
- 21.Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 22.Orengo CA, Taylor WR. SSAP: sequential structure alignment program for protein structure comparison. Methods Enzymol. 1996;266:617–635. doi: 10.1016/s0076-6879(96)66038-8. [DOI] [PubMed] [Google Scholar]
- 23.Suyama M, Matsuo Y, Nishikawa K. Comparison of protein structures using 3D profile alignment. J. Mol. Evol. 1997;44:S163–S173. doi: 10.1007/pl00000065. [DOI] [PubMed] [Google Scholar]
- 24.Shindyalov I, Bourne P. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 25.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 26.Jung J, Lee B. Protein structure alignment using environmental profiles. Protein Eng. 2000;13:535–543. doi: 10.1093/protein/13.8.535. [DOI] [PubMed] [Google Scholar]
- 27.Lackner P, Koppensteiner WA, Sippl MJ, Domingues FS. ProSup: a refined tool for protein structure alignment. Protein Eng. 2000;13:745–752. doi: 10.1093/protein/13.11.745. [DOI] [PubMed] [Google Scholar]
- 28.Ortiz AR, Strauss CEM, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shatsky M, Nussinov R, Wolfson HJ. Flexible protein alignment and hinge detection. Proteins. 2002;48:242–256. doi: 10.1002/prot.10100. [DOI] [PubMed] [Google Scholar]
- 30.Blankenbecler R, Ohlsson M, Peterson C, Ringnér M. Matching protein structures with fuzzy alignments. Proc. Natl Acad. Sci. USA. 2003;100:11936–11940. doi: 10.1073/pnas.1635048100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawabata T. MATRAS: a program for protein 3D structure comparison. Nucleic Acids Res. 2003;31:3367–3369. doi: 10.1093/nar/gkg581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ilyin VA, Abyzov A, Leslin CM. Structural alignment of proteins by a novel TOPOFIT method, as a superimposition of common volumes at a topomax point. Protein Sci. 2004;13:1865–1874. doi: 10.1110/ps.04672604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 34.Ochagavia ME, Wodak SJ. Progressive combinatorial algorithm for multiple structural alignments: application to distantly related proteins. Proteins. 2004;55:436–454. doi: 10.1002/prot.10587. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro J, Brutlag D. FoldMiner and LOCK 2: protein structure comparison and motif discovery on the web. Nucleic Acids Res. 2004;32:W536–W541. doi: 10.1093/nar/gkh389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carpentier M, Brouillet S, Pothier J. YAKUSA: a fast structural database scanning method. Proteins. 2005;61:137–151. doi: 10.1002/prot.20517. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, Crippen GM. A novel approach to structural alignment using realistic structural and environmental information. Protein Sci. 2005;14:2935–2946. doi: 10.1110/ps.051428205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu JH, Weng ZP. FAST: a novel protein structure alignment algorithm. Proteins. 2005;58:618–627. doi: 10.1002/prot.20331. [DOI] [PubMed] [Google Scholar]
- 40.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 41.Lisewski AM, Lichtarge O. Rapid detection of similarity in protein structure and function through contact metric distances. Nucleic Acids Res. 2006;34:E152. doi: 10.1093/nar/gkl788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taubig H, Buchner A, Griebsch J. PAST: fast structure-based searching in the PDB. Nucleic Acids Res. 2006;34:W20–W23. doi: 10.1093/nar/gkl273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oldfield TJ. CAALIGN: a program for pairwise and multiple protein-structure alignment. Acta Crystallogr. D Biol. Crystallogr. 2007;63:514–525. doi: 10.1107/S0907444907000844. [DOI] [PubMed] [Google Scholar]
- 44.Tyagi M, Gowri VS, Srinivasan N, deBrevern AG, Offmann B. A substitution matrix for structural alphabet based on structural alignment of homologous proteins and its applications. Proteins. 2006;65:32–39. doi: 10.1002/prot.21087. [DOI] [PubMed] [Google Scholar]
- 45.Torda AE, Procter JB, Huber T. Wurst: a protein threading server with a structural scoring function, sequence profiles and optimised substitution matrices. Nucleic Acids Res. 2004;32:W532–W535. doi: 10.1093/nar/gkh357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 47.Russell AJ, Torda AE. Protein sequence threading: averaging over structures. Proteins. 2002;47:496–505. doi: 10.1002/prot.10088. [DOI] [PubMed] [Google Scholar]
- 48.Gotoh O. An improved algorithm for matching biological sequences. J. Mol. Biol. 1982;162:705–708. doi: 10.1016/0022-2836(82)90398-9. [DOI] [PubMed] [Google Scholar]
- 49.Diamond R. A note on the rotational superposition problem. Acta Cryst. 1988;A44:211–216. [Google Scholar]
- 50.Li WZ, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 51.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibrat J-F, Madej T, Bryant S. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]