Abstract
GeNMR (GEnerate NMR structures) is a web server for rapidly generating accurate 3D protein structures using sequence data, NOE-based distance restraints and/or NMR chemical shifts as input. GeNMR accepts distance restraints in XPLOR or CYANA format as well as chemical shift files in either SHIFTY or BMRB formats. The web server produces an ensemble of PDB coordinates for the protein within 15–25 min, depending on model complexity and completeness of experimental restraints. GeNMR uses a pipeline of several pre-existing programs and servers to calculate the actual protein structure. In particular, GeNMR combines genetic algorithms for structure optimization along with homology modeling, chemical shift threading, torsion angle and distance predictions from chemical shifts/NOEs as well as ROSETTA-based structure generation and simulated annealing with XPLOR-NIH to generate and/or refine protein coordinates. GeNMR greatly simplifies the task of protein structure determination as users do not have to install or become familiar with complex stand-alone programs or obscure format conversion utilities. Tests conducted on a sample of 90 proteins from the BioMagResBank indicate that GeNMR produces high-quality models for all protein queries, regardless of the type of NMR input data. GeNMR was developed to facilitate rapid, user-friendly structure determination of protein structures via NMR spectroscopy. GeNMR is accessible at http://www.genmr.ca.
INTRODUCTION
Nuclear magnetic resonance (NMR) spectroscopy, along with X-ray crystallography, is one of only two methods that can be used to determine the 3D structures of proteins to atomic resolution. To date, nearly 8000 peptide and protein structures have been determined by NMR and deposited into the PDB (1). The standard route to determine protein structures by NMR involves three basic steps: (i) determining the chemical shift assignments of the target protein; (ii) measuring the inter- and intra-residue 1H NOEs (nuclear Overhauser enhancements) to generate distance constraints; and (iii) using the NOE-derived constraints to perform simulated annealing or distance geometry to calculate the 3D structure of the protein (2). The last step in this process is very computationally demanding and requires very specialized software and significant computational resources. Over the past 20 years, a number of stand-alone programs have been specifically developed to facilitate these calculations including, DYANA (3), CYANA (4), XPLOR-NIH (5), CNS (6) and ARIA (7). All of these programs are excellent at what they do, and all have been extensively tested. However, they are also somewhat unwieldy, platform-specific software packages that are difficult to learn or use. Furthermore, these programs work almost exclusively with NOE data and most of them do not take advantage of chemical shift information in their structure refinement process. Likewise, they do not incorporate a number of widely used structural informatics techniques such as threading, fragment-based assembly or homology modeling to accelerate the structure determination process. Consequently, these programs often require many CPU hours of dedicated calculation and refinement to complete their tasks.
While most NMR-based structure determination packages use NOE data almost exclusively, it has recently been shown that it is possible to determine protein structures using only chemical shift data (i.e. without NOEs). Indeed, several stand-alone programs and web servers have been developed for this purpose, including Cheshire (8), CS-Rosetta (9) and CS23D (10). However, chemical shift-based structure determination is not as robust or as rapid as NOE-based structure determination—especially for novel protein folds (10) or large structures (8,9). Indeed, some of these programs take hundreds or even thousands of CPU hours to complete their calculations. Furthermore, none of these shift-based structure determination techniques takes advantage of the additional information contained in NOE constraint data.
Ideally, what is needed for NMR-based structure determination is a tool that is: (i) easy to use (a ‘single click’ solution); (ii) platform independent (a web server); (iii) fast (generating structures in minutes, not hours); (iv) capable of handling both NOE and chemical shift data; and (v) smart enough to perform the NMR equivalent of ‘molecular replacement’ by taking advantage of well-known structural bioinformatics techniques such as threading, homology modeling and fragment-based assembly.
Here, we wish to describe a simple web server, called GeNMR, that addresses these needs. In particular, GeNMR is a next generation, highly generalizable system for rapidly generating protein structures by NMR using a combination of genetic algorithms and simulated annealing. It allows the rapid (∼15 min) and accurate determination of protein structures using any combination of chemical shifts and/or NOEs as input. GeNMR builds on nearly 15 years of research in our lab related to using chemical shifts and other NMR data to identify protein secondary structures (11), to predict torsion angles (12), to identify protein folds (13), to predict protein flexibility (14) and to calculate protein structures (10). GeNMR greatly simplifies the task of protein structure determination as users do not have to install or become familiar with complex stand-alone programs or obscure format conversion utilities. Tests conducted on a sample of 90 proteins from the BioMagResBank indicate that GeNMR produces high-quality models for all protein queries, regardless of the type of NMR input data. Additional details about GeNMR are given below.
PROGRAM DESCRIPTION
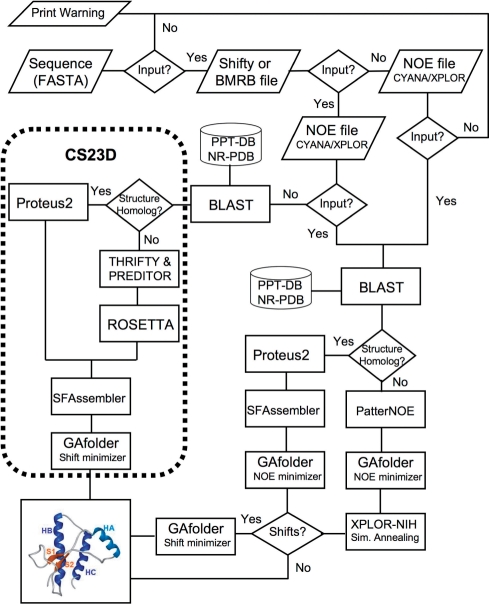
GeNMR is composed of two parts, a front-end web-interface (written in Perl and HTML) and a back-end consisting of eight different alignment, structure generation and structure optimization programs (written in Java, Perl, Python and C/C++) along with three local databases (Figure 1). Users must provide at least two types of data: (i) sequence data (in raw or FASTA format); (ii) chemical shift data in either SHIFTY (15) or BMRB (16) formats and (iii) NOE data in either CYANA (4) or XPLOR (5) format. The sequence data is needed to handle the (common) situation where there are missing NOE or chemical shift assignments. Without the complete protein sequence, GeNMR would typically generate concatenated (i.e. incorrect) structures in the regions where NOE or chemical shift data was missing.
Figure 1.
A flow chart outlining the general structure of the GeNMR web server and the programs that it calls to generate protein structures from chemical shift and/or NOE data. The specific function of each of the named programs is explained in the text.
The sequence, chemical shift and/or NOE files may be either pasted or typed into the text box or uploaded through a file browse button. The output for a typical GeNMR structure calculation consists a user-defined set of lowest energy PDB coordinates in a simple, downloadable text format. A hyperlink to view the single lowest energy structure through the WebMol viewer (17) is also provided. In addition, details about the overall energy score (prior to and following energy minimization), NOE violations, torsion angle violations and chemical shift correlations (between the observed and calculated shifts) is provided at the top of the output page. If the structure calculation fails to converge to a reasonable value, a warning is printed at the top of the page. Generally, if a structure fails to converge or exhibits a high number of violations, users should investigate the correctness of their NOE or chemical shift assignments. There is no limit on the number of queries a user may send and so resubmitting to the server with new or corrected data is generally encouraged. Details about the GeNMR energy function, reasonable values for chemical shift correlations and reasonable values for torsion angle and NOE violations is provided in the Documentation link on the GeNMR home page.
ALGORITHMS AND DATABASES
A flow chart describing the processing logic used in GeNMR is shown in Figure 1. As can be seen in this diagram, GeNMR makes use of a number of programs or databases previously developed in our laboratory. These include Proteus2 (18) to perform structural modeling and energy refinement, PREDITOR (12) to calculate torsion angles (from NOEs or chemical shifts), PPT-DB (19) for comparative modeling and alignment and CS23D (10) to calculate protein structures from chemical shifts. GeNMR also uses several well-known external programs, including Rosetta (20) for fragment-based assembly and XPLOR-NIH (5) for NOE-based simulated annealing and refinement.
One of the more useful concepts in GeNMR is the implementation of homology modeling and sequence/structure threading to rapidly generate a first-pass model of the query protein. This is the NMR equivalent of ‘molecular replacement’, wherein a structure is initially generated via modeling and refined using experimental (NOE or chemical shift) data. Interestingly, molecular replacement in NMR is generally easier and more robust than molecular replacement than X-ray crystallography. This is because NMR structures can be refined directly against chemical shift, torsion angle and distance restraints, whereas with X-ray crystallography one is compelled to refine against phases and subunit orientation. The use of ‘molecular replacement’ concepts in GeNMR allows a considerable speed-up in its structure calculations since homology models can often be generated and refined in a minute or two. Furthermore, given the fact that up to 90% of all NMR structures being calculated today have identical or similar folds to existing structures (21) it stands to reason that most queries to GeNMR will involve this very rapid structure generation step.
GeNMR also makes use of genetic algorithms to allow robust configurational sampling and structural refinement using non-differentiable target functions. This is particularly useful for chemical shift refinement and for minimizing structures initially generated via homology modeling. The application of genetic algorithms for NOE-based structure generation and refinement was originally proposed in 1998 with the development of GENFOLD (22). We have incorporated a number of GENFOLD's ideas in GeNMR and extended them to include chemical shift and protein threading potentials as part of the refinement routine. Like GENFOLD, GeNMR's genetic algorithm creates a population of initial structures and then uses combinations of mutations, cross-overs, segment swaps and writhe movements to comprehensively sample conformation space. The 25 lowest energy structures are then selected, duplicated and carried to the next round of conformational sampling. This conformational sampling process is repeated a minimum of 25 times or until the structures have reached a predefined convergence criteria. Genetic algorithms are also very amenable to being implemented in clustered computing environments. Consequently we have configured GeNMR to run over a 24 CPU core computer cluster. This parallelism also greatly accelerates structure refinement and convergence.
The potential functions used in GeNMR are derived from those used in CS23D and Proteus2 (10,18). The knowledge-based potential includes information on predicted/known secondary structure, radius of gyration, hydrogen bond energies, number of hydrogen bonds, allowed backbone and side chain torsion angles, atom contact radii (bump checks), disulfide bonding information and a modified threading energy based on the Bryant and Lawrence potential (23). The chemical shift component of the GeNMR potential uses weighted correlation coefficients calculated between the observed and SHIFTX (24) calculated shifts of the structure being refined. Tests conducted with GeNMR's knowledge-based potential on its ability to differentiate lower resolution/lower quality NMR structures and high-resolution X-ray structures of the same protein showed that the high-resolution structure had significantly lower energy in all cases (Table I—GeNMR's Documentation web page). Similarly, tests conducted with GeNMR's chemical shift potential on its ability to differentiate lower resolution or lower quality structures from higher resolution structures yielded similar results (Table II—GeNMR's Documentation web page). These results suggest that these potentials serve the same purpose and provide the same benefit as the more computationally expensive water-based refinement.
As seen in Figure 1, there are three types of input combinations and six different kinds of calculation scenarios that GeNMR can currently accommodate. These scenarios include: (a) chemical shift only—query has homologue in database; (b) chemical shift only—query has no homologue in database; (c) NOE only—query has homologue in database; (d) NOE only—query has no homologue in database; (e) NOE and chemical shift—query has homologue in database and (f) NOE and chemical shift—query has no homologue in database. We will use these scenarios to describe how GeNMR works, in detail, and to summarize the program's performance under these particular conditions.
RESULTS AND EVALUATION
Scenario (a): shift only with homologue
In this situation, the user provides only the sequence and the assigned chemical shifts (minimally the HN shifts), to the server. This kind of query falls into the category of shift-based structure determination handled by the CS23D (10) component of GeNMR. Briefly, the sequence is searched against a non-redundant database of PDB sequences and secondary structures from PPT-DB (19) using BLAST (25) with a sliding length-dependent Expect cutoff, ranging from 10−1 (for <11 residues) to 10−5 (for >50 residues). If the sequence exceeds the cutoff, a structural model is generated using Proteus2 (18). If the protein consists of several discontinuous domains, a program called SFAssembler will link the models together to produce a model of the holo-protein. Note that SFAssembler is only designed to concatenate multiple independent polypeptide chains (or domains) together to generate a single chain, multi-domain protein. It is not able to support the generation of protein complexes that consist of multiple, independent chains. The resulting SFAssembler model represents a ‘molecular replacement’ template which is subject to refinement using experimental chemical shift data. In particular, the protein is refined with a torsion-based genetic algorithm (GAfolder) using the chemical shift and knowledge-based potentials described earlier. The exact methodology and overall performance of this kind of structure generation protocol has been described in detail in the CS23D paper (10). Since that publication, a number of improvements were made to this algorithm and incorporated into GeNMR, including the use of parallelism, increasing the size of the reference structure database, enhancing the sampling protocol and improving the energy functions. An indication of GeNMR's performance under this scenario (using five proteins with sequence identities ranging from 35% to 98%) is given at the top of Table 1. More complete results are shown in Table V of GeNMR's Documentation web page which show the results of a test conducted on 50 proteins randomly chosen from the BioMagResBank (16) with 35–99% sequence identity to known structures. Typical structure generation times for this scenario are about 15–20 min, depending of the server load and the size of the structure.
Table 1.
Performance assessment of GeNMR under different data input scenarios using a default of 10 structure models (see text for an explanation of each scenario)
| Protein Name (PDB ID) | Sequence ID (%) | RMSD (Å) to reference PDB | Calculation Time (min) | No. of distance restraints |
|---|---|---|---|---|
| Scernario (a) – Shift data only—query has homologue in database | ||||
| Ubiquitin (1UBQA) | 62 | 1.55 | 10 | – |
| SeR13 (2K1HA) | 58 | 2.31 | 13 | – |
| Ig Domain of Palladin (2DM2A) | 35 | 1.29 | 18 | – |
| Abl Kinase (2HYYA) | 98 | 1.76 | 19 | – |
| RGD-Hirudin (2JOOA) | 88 | 1.32 | 15 | – |
| Scernario (b) – Shift data only—query has NO homologue in database | ||||
| Ubiquitin (1UBQA) | – | 2.55 | 18 | – |
| 4-helix Bundle (2I7UA) | – | 1.48 | 22 | – |
| Discoidin Domain DDR2 (2Z4FA) | – | 1.63 | 36 | – |
| CheW (2HO9A) | – | 2.66 | 25 | – |
| Scernario (c) – NOE data only—query has homologue in database | ||||
| Cyclophilin (1CWCA) | 75 | 0.98 | 23 | 4096 |
| Regulatory Protein E2 (1A7G) | 70 | 1.67 | 16 | 1197 |
| Serine Protein Inhibitor (3CI2) | 82 | 1.49 | 19 | 961 |
| DnaB (1JWE) | 47 | 0.99 | 19 | 1194 |
| Superoxide Dismutase (2AF2) | 97 | 1.27 | 14 | 2672 |
| PyJ Protein (1FAF) | 95 | 0.05 | 10 | 870 |
| Neurotoxin II (1NOR) | 87 | 0.92 | 16 | 540 |
| Scernario (d) – NOE data only—query has NO homologue in database | ||||
| Ubiquitin (1UBQ) | – | 1.38 | 21 | 1318 |
| Forkhead FOXO4 (1E17) | – | 2.20 | 48 | 1294 |
| Profilin (1AWI) | – | 1.96 | 46 | 1794 |
| Mu DNA Binding Protein (2EZI) | – | 2.43 | 25 | 1009 |
| SV40 ORI Binding Protein (1TBD) | – | 2.61 | 46 | 1709 |
| Scernario (e) – NOE+Shift data—query has homologue in database | ||||
| Response Regulator Spo0F (1FSP) | 96 | 1.17 | 16 | 1835 |
| Profilin (1AWI) | 99 | 0.37 | 28 | 1794 |
| Interleukin 4 (1BBN) | 87 | 1.68 | 20 | 917 |
| Metalloproteinase 12 (1YCM) | 99 | 1.44 | 19 | 3544 |
| Ubiquitin (1UBQ) | 96 | 0.42 | 25 | 1318 |
| Scernario (f) – NOE+Shift data—query has NO homologue in database | ||||
| Interleukin 4 (1BNN) | – | 1.69 | 25 | 917 |
| SV40 ORI Binding Protein (1TBD) | – | 1.21 | 23 | 1709 |
| Superoxide Dismutase (2AF2) | – | 1.25 | 16 | 2672 |
| Ribosomal Protein S4 (1C05) | – | 2.86 | 21 | 2256 |
| Neurotoxin II (1NOR) | – | 0.98 | 11 | 482 |
Scenario (b): Shift only with no homologue
As with ‘Scenario (a)’ this kind of query falls into the category of shift-based structure determination is handled by the CS23D (8) component of GeNMR. In this situation homology modeling has failed and, as a result, several alternative routes to structure modeling are attempted including chemical shift threading, sub-fragment assembly and de novo structure generation by Rosetta. As before, the exact methodology and overall performance of this kind of structure generation protocol has been described in detail in the CS23D paper (10). An indication of GeNMR's performance under this scenario is given in Table 1 using a set of four proteins where all homologues have been removed from the database prior to testing. More complete results, based on tests conducted on 15 proteins randomly chosen from the BioMagResBank (16) with <35% sequence identity to known structures, are shown in TableVI of GeNMR's Documentation web page. As has been noted before, the performance for shift-based structure determination of novel protein folds is often not as good as it is for known folds (10). Typical structure generation times for this scenario are about 20–35 min, depending of the server load and the size of the structure.
Scenario (c): NOE only with homologue
In this situation, the user provides only the protein sequence and the NOE assignment table (in CYANA or XPLOR format) to the GeNMR server. Minimally, the user must provide at least one NOE per residue. This scenario matches what is most commonly performed in NMR-based structure calculations with programs such as DYANA, CYANA or XPLOR-NIH. As with ‘Scenario (a)’, the sequence is searched against a non-redundant database of PDB sequences and secondary structures from PPT-DB (19) using BLAST (25) with a sliding length-dependent Expect cutoff, ranging from 10−1 (for <11 residues) to 10−5 (for >50 residues). If the sequence exceeds the cutoff, a structural model is generated using Proteus2 (18). If the protein consists of several discontinuous domains, a program called SFAssembler will link the models together to produce a model of the holo-protein. The resulting model serves as a ‘molecular replacement’ template which is subject to refinement using the experimental NOE data. In particular, the protein is refined with GAfolder using both a NOE-based and a knowledge-based potential. The NOE potential measures the percentage of NOE violations with a different weight assigned to short-range NOEs versus long-range NOEs. The exact form of the NOE potential function is given in GeNMR's Documentation page. Table 1 lists the results of tests conducted on seven proteins with experimental NOE data as chosen from the BioMagResBank with 45–97% sequence identity to known structures (the exact match was removed in this test). Additional examples, showing how well GeNMR's blended NOE+shift refinement process is able to work on remote homologues (∼30% sequence ID) or poor starting structures (up to 4.3 Å RMSD) are given in Tables III and IV of GeNMR's Documentation page. Overall, it is clear that GeNMR produces structures that are very similar to the known structures (<1.5 Å RMSD). Typical structure generation times for this scenario are about 15–20 min, depending of the server load and the size of the structure.
Scenario (d): NOE only with no homologue
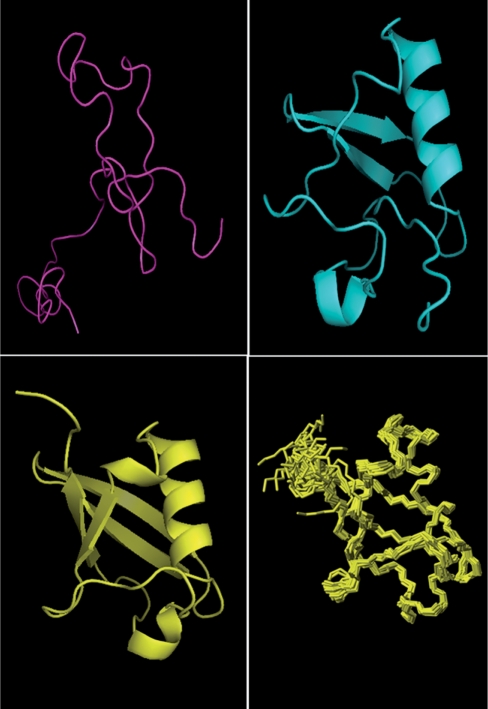
Given the ever-diminishing likelihood of finding a truly novel fold these days, this is actually a relatively rare situation. Indeed, we surveyed all non-redundant (95% sequence identity, length >20 residues) proteins that were solved by NMR and deposited in the PDB from 1 January 2009 to 20 March 2009. Of the 386 polypeptide chains in this particular set, 71% had significant (E-value <1×10−5) sequence homology to a known structure, 11% had ‘threadable’ structures (folds similar or identical to known folds using secondary structure matching) that could be detected by GeNMR and 18% exhibited truly novel folds with no sequence or structure similarity to anything else in the PDB. Nevertheless, generating a structure without any prior knowledge is also considered to be the ‘acid test’ of any NMR structure determination protocol as it demonstrates the ability of the program to use only experimental data to generate and refine structure. Because no chemical shift data is provided, no chemical shift threading or torsion angle constraint generation can be performed. Instead, a separate algorithm (called PatterNOE) scans for NOE patterns to identify secondary structure elements (helices, beta turns, beta strands) and assigns approximate torsion angle constraints. In particular, PatterNOE analyzes short and medium-range inter-residue NOEs from the NOE constraint list and compares these to well-known inter-residue NOE patterns and distance constraints derived from idealized helices, beta strands (parallel and antiparallel) and beta turns (type I and type II) to identify secondary structure elements. Standard torsion angles derived from these ‘ideal’ helices, beta strands and beta turns are then used to generate the corresponding torsion angle restraints and, subsequently, a starting protein structure. After this step, GeNMR partitions the NOE constraint file into short range (1–5 residues), medium range (6–12 residues) and long range (>12 residues) NOEs. Using a genetic algorithm (GAfolder), the initial structures are then refined in a progressive three-step fashion with the short range NOEs being satisfied first, then the medium range NOEs and finally the long range NOEs. Typically, this process generates an ensemble of structures that are within 3–4 Å RMSD of the actual structure. In the final step, this ‘rough’ structure is refined using simulated annealing with XPLOR-NIH. Figure 2 shows the initial, unfolded structure for ubiquitin along with the ‘rough’ structure (post GAfolder) and the final structural ensemble (post XPLOR). Table 1 lists the results of tests conducted on five proteins with experimental NOE data where the homology modeling step has been removed from the GeNMR pipeline. These data show that GeNMR produces structures that are very similar to the known structures (<2.5 Å RMSD). Typical structure generation times for this scenario are about 40–45 min, depending of the server load and the size of the structure.
Figure 2.
Illustration of how GeNMR is able to generate the structure of ubiquitin using only experimental NOE constraints [scenario (d) in the article]. The initial structure (purple) is a largely random coil polypeptide (∼18 Å RMSD), an intermediate structure (cyan) contains most of the secondary structure elements (∼ 3.9 Å RMSD), while the lowest energy structure (yellow), refined via simulated annealing, is within 1.2 Å of the known X-ray structure. The last panel shows the final ensemble (or bundle) of structures generated by GeNMR.
Scenario (e): NOE+Shift with homologue
In this situation, the user provides the sequence, the chemical shifts (in BMRB or SHIFT format) and the NOE assignment table (in CYANA or XPLOR format), to the GeNMR server. Given that it is impossible to obtain NOE assignments without chemical shift assignments, this should be the most common scenario for GeNMR. Because all three ‘vital’ data sets are provided as input this also allows GeNMR to make full use of its sequence/structure databases, its chemical shift refinement routines and its NOE refinement routines. As with scenarios (a) and (c), a molecular model is generated using Proteus2 or SFAssembler. As before, the resulting model represents a ‘molecular replacement’ template that is subject to refinement using the experimental NOE and chemical shift data. In particular, the protein is refined with a torsion-based genetic algorithm using a chemical shift-based, a NOE-based and a knowledge-based potential. Table 1 lists the results of tests conducted on five proteins with experimental NOE and experimental chemical shift data as chosen from the BioMagResBank with 85–99% sequence identity to known structures (the exact match was removed in this test). These data show that GeNMR produces structures that are very similar to the known structures (<1.5 Å RMSD). Typical structure generation times for this scenario are about 15–20 min, depending of the server load and the size of the structure.
Scenario (f): NOE+Shift with no homologue
In this situation, homology modeling has failed and, as a result, several alternative routes to structure modeling are attempted including chemical shift threading and sub-fragment assembly. If these approaches also fail to quickly identify a model, GeNMR employs the progressive NOE refinement technique described in ‘Scenario d)’. Specifically, an initial structure is generated using shift-derived (as well as NOE-derived) torsion angle constraints. Subsequently a genetic algorithm refines these initial structures using a progressive three-step approach that attempts to satisfy the short range NOEs first, then the medium range NOEs and finally the long range NOEs. The resulting ‘rough’ structure is then refined using simulated annealing with XPLOR-NIH. Table 1 lists the results of tests conducted on five proteins with experimental NOE and chemical shift data where the homology modeling step has been removed from the GeNMR pipeline. These data show that GeNMR produces structures that are very similar to the known structures (<2.0 Å RMSD). Typical structure generation times for this scenario are about 20–25 min, depending of the server load and the size of the structure.
LIMITATIONS
GeNMR was primarily designed to make NMR-based structure determination simpler, faster and easier. However, in striving towards a simplified, user-friendly solution certain compromises had to be made. One compromise concerns GeNMR's limited support for ambiguous NOE constraints. While some stereo-specific ambiguity in NOEs is allowed in the data input files, GeNMR does not have the capacity of programs such as ARIA (7) to handle unassigned NOEs. Rather, GeNMR assumes that the NOE input and/or chemical shift assignments are mostly complete and mostly correct. With the possible exception of certain high throughput structural genomics data sets, we believe this is the most prevalent situation for NMR data sets.
A second compromise concerns GeNMR's capacity to handle other NMR-derived constraints. Currently GeNMR does not handle or process J-coupling or residual dipolar coupling (RDC) restraints. While J-couplings are commonly measured and widely accommodated by many other NMR structure generation programs, we have found that this information is generally less accurate than the information that can be derived directly or indirectly from chemical shifts (10,12). Since chemical shifts are a necessary pre-requisite to measuring J-couplings, we believe that by excluding J-couplings from GeNMR's input fields we are actually making structure determination a little simpler and a little less laborious for the experimentalist. On the other hand, residual dipolar couplings (RDCs) do provide somewhat more information than J-couplings (26). In particular, RDCs provide critical, long range information about the relative orientation of secondary structure elements within proteins. Currently, GeNMR does not yet support the inclusion of RDC data. Efforts are underway to add this feature to the server and it is expected that RDC calculations will be supported by the summer of 2009. While not explicitly supported with a separate data entry field, H-bond constraints can be submitted to the GeNMR server as a part of the NOE distance restraint file. In addition, GeNMR is also able to extract H-bonding data from user-supplied NOE data to create its own set of H-bonding constraints.
Another limitation to GeNMR is that it cannot calculate the structures for protein complexes (protein–protein, protein–nucleic acid or protein–small molecule complexes). Because of the requirement for customized atom designations, the lack of readily available test sets as well as the need for somewhat more complex workflows and input data requirements, we decided not to support these kinds of queries or calculations at this time. Efforts are underway to add this feature to the server and it is expected that protein–protein complex structure generation will be supported by the summer of 2009. Support for generating other kinds of complexes (DNA, RNA or small molecule ligands) should be available shortly thereafter. It is perhaps worth noting that of the 5698 non-redundant protein structures in the PDB that have been solved by NMR, 89.9% are monomeric. So despite this single-chain limitation, GeNMR is still be able to handle the vast majority of today's NMR queries.
As with any program or web server there is the usual caveat that ‘garbage in = garbage out’. In particular, GeNMR assumes that the sequence, chemical shifts and NOE files that are submitted are mostly error-free. Nevertheless, a number of data integrity checks (including formatting checks, sequence length checks and chemical shift referencing checks) are performed to prevent data from being misprocessed or misinterpreted and to provide direct user feedback. Additionally, GeNMR performs a ‘Structure Sanity Check’ that is appended to GeNMR's structure/energy evaluation output. This utility should allow users to identify possible misassignment (shift or NOE) errors. We have also developed criteria to provide warnings and to highlight residues or sequence regions that may need manual checking. Given the nature of experimental data and the large number of manual measurements and data entry tasks typically required in NMR-based structure determination, we also realized it was important to have some built-in tolerance for data measurement errors. Tests indicate that GeNMR can tolerate a certain level of chemical shift misassignments and/or NOE misassignments (∼5%), although this tolerance will vary depending on the type of query as well as the type of error or the extent of the misassignment.
CONCLUSIONS
We believe GeNMR provides a new and greatly simplified approach to determining protein structures from NMR data. For most of the past 20 years, NOE-based structure determination has required that users to become familiar with relatively large and complex, stand-alone programs. Often the learning curve for these programs is steep and the need for training/re-training, as well the ongoing efforts needed to install, maintain and upgrade the software (and hardware) are quite considerable. By developing a web server to perform NMR-based structure determination, we believe we have greatly simplified the process. GeNMR's intuitive interface and simple file input makes the system easy to use and easy to learn. Its web accessibility means that users are not constrained by platform compatibility issues or the availability of dedicated CPUs. Indeed structure calculations can be performed by anyone, essentially anytime or from anywhere. Furthermore, GeNMR's use of parallelism and ‘molecular replacement’ concepts means that calculations that used to take hours or days are typically completed within minutes. Finally, by moving the system to a web server format, many of the onerous tasks of local software maintenance and local software/hardware upgrading have been eliminated. While GeNMR is not without some limitations, overall we believe its simplicity, speed and accessibility should make it a very useful addition to the current arsenal of structure generation and refinement tools available to biomolecular NMR spectroscopists.
FUNDING
Alberta Prion Research Institute, PrioNet; NSERC; Genome Alberta. Funding for open access charge: PrioNet.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35(Database issue):D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wuthrich K. NMR - this other method for protein and nucleic acid structure determination. Acta Crystallogr. D. 1995;51:249–270. doi: 10.1107/S0907444994010188. [DOI] [PubMed] [Google Scholar]
- 3.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 4.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 5.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 6.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 7.Nilges M, Macias MJ, O'Donoghue SI, Oschkinat H. Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin. J. Mol. Biol. 1997;269:408–422. doi: 10.1006/jmbi.1997.1044. [DOI] [PubMed] [Google Scholar]
- 8.Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc. Natl Acad. Sci. USA. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl Acad. Sci. USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G. CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36(Web Server issue):W496–W502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 12.Berjanskii MV, Neal S, Wishart DS. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34(Web Server issue):W63–W69. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- 14.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J. Biomol. NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 16.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36(Database issue):D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walther D. WebMol—a Java-based PDB viewer. Trends Biochem Sci. 1997;22:274–275. doi: 10.1016/s0968-0004(97)89047-0. [DOI] [PubMed] [Google Scholar]
- 18.Montgomerie S, Cruz JA, Shrivastava S, Arndt D, Berjanskii M, Wishart DS. PROTEUS2: a web server for comprehensive protein structure prediction and structure-based annotation. Nucleic Acids Res. 2008;36(Web Server issue):W202–W209. doi: 10.1093/nar/gkn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wishart DS, Arndt D, Berjanskii M, Guo AC, Shi Y, Shrivastava S, Zhou J, Zhou Y, Lin G. PPT-DB: the protein property prediction and testing database. Nucleic Acids Res. 2008;36(Database issue):D222–D229. doi: 10.1093/nar/gkm800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 21.Levitt M. Growth of novel protein structural data. Proc. Natl Acad. Sci. USA. 2007;104:3183–3188. doi: 10.1073/pnas.0611678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayley MJ, Jones G, Willett P, Williamson MP. GENFOLD: a genetic algorithm for folding protein structures using NMR restraints. Protein Sci. 1998;7:491–499. doi: 10.1002/pro.5560070230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bryant SH, Lawrence CE. An empirical energy function for threading protein sequence through the folding motif. Proteins. 1993;16:92–112. doi: 10.1002/prot.340160110. [DOI] [PubMed] [Google Scholar]
- 24.Neal S, Zhang H, Nip AM, Wishart DS. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J. Biomol. NMR. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prestegard JH, Bougault CM, Kishore AI. Residual dipolar couplings in structure determination of biomolecules. Chem. Rev. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]