Abstract
PosMed (http://omicspace.riken.jp/) prioritizes candidate genes for positional cloning by employing our original database search engine GRASE, which uses an inferential process similar to an artificial neural network comprising documental neurons (or ‘documentrons’) that represent each document contained in databases such as MEDLINE and OMIM. Given a user-specified query, PosMed initially performs a full-text search of each documentron in the first-layer artificial neurons and then calculates the statistical significance of the connections between the hit documentrons and the second-layer artificial neurons representing each gene. When a chromosomal interval(s) is specified, PosMed explores the second-layer and third-layer artificial neurons representing genes within the chromosomal interval by evaluating the combined significance of the connections from the hit documentrons to the genes. PosMed is, therefore, a powerful tool that immediately ranks the candidate genes by connecting phenotypic keywords to the genes through connections representing not only gene–gene interactions but also other biological interactions (e.g. metabolite–gene, mutant mouse–gene, drug–gene, disease–gene and protein–protein interactions) and ortholog data. By utilizing orthologous connections, PosMed facilitates the ranking of human genes based on evidence found in other model species such as mouse. Currently, PosMed, an artificial superbrain that has learned a vast amount of biological knowledge ranging from genomes to phenomes (or ‘omic space’), supports the prioritization of positional candidate genes in humans, mouse, rat and Arabidopsis thaliana.
INTRODUCTION
Linkage analysis is used for identifying genes with a certain phenotype or genetic defect, and can suggest chromosomal intervals containing several tens to hundreds of candidate genes for positional cloning. Before performing further experiments, it is necessary to prioritize the candidate genes by using as much biological knowledge as possible. For this purpose, it is an ambitious challenge to create an artificial superbrain that has learned a vast knowledge of omic space (1).
To develop a web-based tool that can immediately suggest genes related to a certain phenotype, we initially developed a search engine named GRASE (General and Rapid Association Study Engine), and then defined its query language named GRASQL (General and Rapid Association Study Query Language) (2). GRASQL is a powerful language for expressing the statistical analysis of data retrievable by the RDF query language SPARQL (3) in a Semantic Web manner (4). The current implementation of GRASE is optimized to efficiently calculate the statistical prioritization of candidate genes based on more than 17 million medical and biological documents, and to facilitate quick return of the results within a few seconds of computational time.
Several software tools that have been developed for prioritizing positional candidate genes are based on functional annotation, gene expression patterns, protein–protein interaction and/or sequence-based features (5–10). The evaluation of three of these software tools using their data set has demonstrated the effectiveness of PosMed, which showed an accuracy of 88.7%, the highest of the three tools (11).
The documents searched by PosMed contain references, genome annotations, phenome information, protein–protein interactions, co-expressions, orthologous genes, drugs and metabolite information. Using this biological knowledge, PosMed executes a full-text search of documents when a query word is input and ranks the genes based on direct and indirect inference of the hit documents. Currently, PosMed supports prioritization of candidate genes for positional cloning in humans, mouse, rat and Arabidopsis thaliana.
OVERVIEW
A neural network representation of the statistical algorithm for searching complex Semantic Web data
PosMed network searches are performed by GRASE, a search engine that retrieves data items over a highly connected network with semantic links by statistical evaluation. First, to identify genes associated with a user's keyword, GRASE performs a full-text search using the keyword and graph pattern matching over the semantic network containing the semantic link gene→document between a document and a gene. In other words, GRASE identifies documents having the keyword and generates the semantic link keyword→document. Then, for each gene, a 2 × 2 contingency table is generated by performing graph pattern matching over the semantic links keyword→document and document→gene (see ‘PosMed RANKING’ section for details). For each contingency table, a P-value is computed using a statistical test such as Fisher's exact test. Since this P-value becomes smaller when the relevance between the keyword and the gene becomes higher, this value is used for the evaluation of relevance between genes and keywords.
To identify genes further related to the genes initially found, GRASE performs an inference search between gene1 and gene2. In this search, a 2 × 2 contingency table is generated for each gene by performing graph pattern matching over the semantic link document→gene (see ‘PosMed RANKING’ section for details), and a P-value is computed. This P-value also becomes smaller when the relevance between the two genes becomes higher based on the number of documents co-cited. This value is used for the evaluation of relevance between two genes for a gene–gene inference. A total P-value is computed by combining these two P-values (see ‘PosMed RANKING’ section for details), which is used to indicate statistical significance between the keyword and gene2 via gene1. A P-value is also computed to show the significance between the keyword and the genes in the first search step. Finally, GRASE generates a list of genes ranked using the computed P-values.
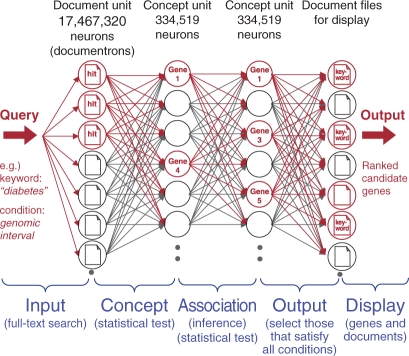
Although the search algorithm can be described using the above-mentioned GRASQL, a graphical representation of the search algorithm is also helpful in understanding the power of the system. Analogous to a network of neurons receiving signals from other neurons through connections, each document is regarded as a neuron (or ‘documentron’) that fires a signal when a keyword matches the document contents (Figure 1, Input). The signal fired from each documentron is statistically evaluated at the neurons in the next layer by calculating the significance of the associations between the keyword and the genes cited in the hit documents (Figure 1, Concept). Only the neurons (genes) showing P-values < 1% (default) fire signals to the next neural layer, according to the strength of the gene–gene relationships or co-citations (Figure 1, Association), wherein various relationships such as protein–protein interactions, co-expressions and ortholog genes are potential additional associations. Only significant genes located within the user-specified genome interval are then displayed together with the most appropriate documents containing the supporting evidence (Figure 1, Output). The keywords are highlighted in the documents (Figure 1, Display).
Figure 1.
Neural network model for the PosMed gene search algorithm. As an example, the user's keyword ‘diabetes’ can be found in several documents, including MEDLINE (Input). These documents are mapped to genes that are supported by manual curation (Concept). Using biological knowledge (e.g. protein–protein interaction, co-expression and co-citation of document sets), PosMed can also suggest genes that do not have the user's keyword ‘diabetes’ in their associated documents (Association). PosMed then returns the candidate genes that are located within the user's specified genomic interval (Output). Thereafter, the user's keyword will be highlighted in the documents (Display).
General usage of PosMed
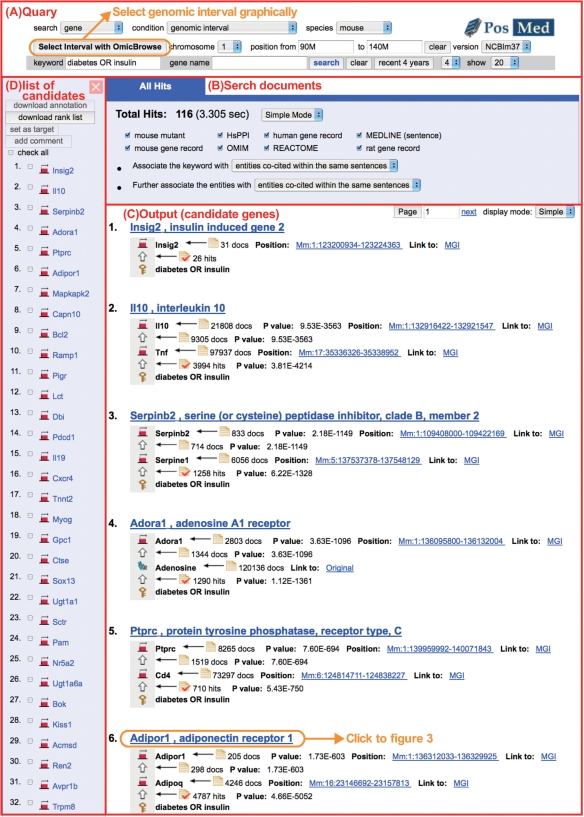
PosMed is a simple and user-friendly system for prioritizing positional candidate genes. To use this system, users need to input species, a keyword and genome version and additionally select the genomic interval. For example, a search using the keyword ‘diabetes OR insulin’ in the 90–140M bp genome interval on chromosome 1 in mouse retrieves 114 candidate genes ordered by their statistical significance (Figure 2). Users can download these genes together with the relevant gene annotation information using the ‘download rank list’ button (Figure 2D). Users can also select the ‘expert mode’ in the ‘All Hits’ tab to enable detailed retrieval. With this expert mode, users can check all the direct and inferential paths of the PosMed search as well as the number of hit genes. Moreover, users can change the threshold of the P-value to increase or decrease the number of genes shown.
Figure 2.
Output display on PosMed Search. Example search result for mouse genes against the keyword ‘diabetes OR insulin’ between 90M and 140M bp on Chr 1 in the NCBIm 36 genome. Users can apply their queries at the top of the output display (A). To select genomic interval visually, PosMed cooperates with the Flash-based genomic browser OmicBrowse (12). The tab labeled ‘All Hits’ (B) shows a list of selectable document sets to be included in the search. As a default parameter, PosMed sets ‘Associate the keyword with entities co-cited within the same sentences’. If the total number of the candidate genes is below 20, PosMed will automatically change this parameter to ‘Associate the keyword with entities co-cited within the same document’ to show more candidates (B). PosMed search results are ranked in (C). Users can download at most 300 candidate genes and their annotations from (D).
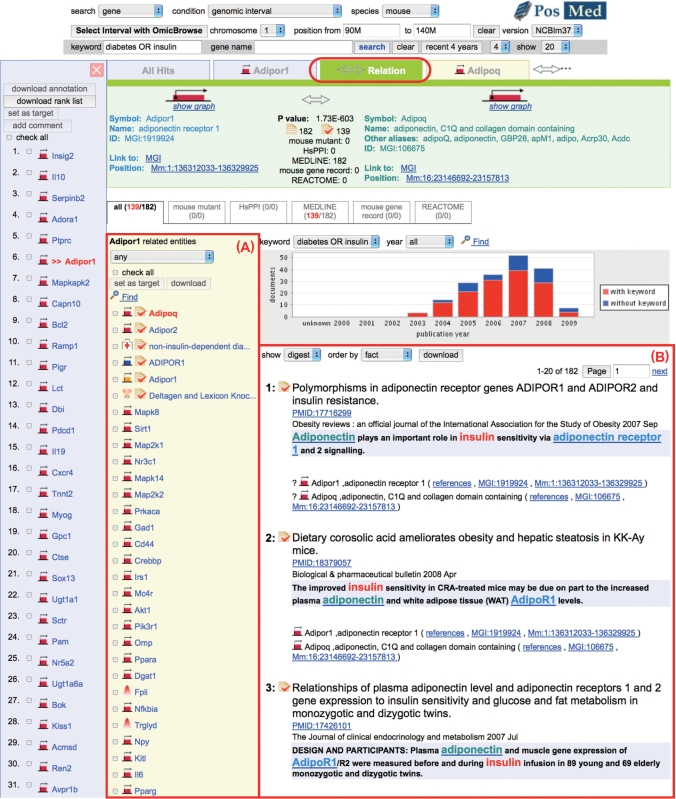
Clicking on the gene name reveals supporting evidence for each candidate gene. As an example, the supporting documents for the sixth gene (Adipor1) presented in Figure 2 are shown in Figure 3. Typically, two genes are connected based on co-citations in a document, protein–protein interaction, or co-expression. The bar chart in Figure 3 shows the number of references in MEDLINE. It is important to make correct connections between each gene and references to ensure the accuracy of PosMed. This is, however, very costly to perform manually and thus we applied logical operations with synonyms and functionally important words of genes. For example, to detect all MEDLINE documents for the AT1G03880 gene in A. thaliana, we applied the following logical operation: (‘AT1G03880’ OR ‘CRU2’ OR ‘CRB’ OR ‘CRUCIFERIN 2′ OR ‘CRUCIFERIN B’) AND (‘Arabidopsis’) NOT (‘chloroplast RNA binding’). Curators refine the logical operations in mouse and A. thaliana. For human and rat genes, we use mouse curation results via ortholog genes.
Figure 3.
Detail page showing supporting documents of the inference-type search. Adipor1 related genes are listed in (A). The supporting documents for Adipor1 and Adipoq are ranked in (B).
More advanced usage of PosMed is explained in the PosMed tutorial available at: http://omicspace.riken.jp/tutorial/HowToUseGPS_Eng.pdf
DATA SOURCES
Currently, PosMed uses more than 17 million documents. For inference-type searches, we employ document sets including MEDLINE (title, abstract and MeSH term), genome annotation, phenome information, protein–protein interaction, co-expression, drugs and metabolite records (Table 1).
Table 1.
Document sets implemented in PosMed
| Document | Display name on PosMed | No of documents | Reference |
|---|---|---|---|
| MEDLINE | MEDLINE | 17 132 801 | (17) |
| BRMM | mouse mutant | 12 911 | Original dataa |
| OMIM | OMIM | 19 891 | (18) |
| HsPPIb | HsPPI | 35 731 | (19) |
| AtPID | AtPID | 44 082 | (20) |
| ATTED-II | At co-expression | 24 418 | (21) |
| REACTOME | REACTOME | 10 761 | (22) |
| MouseGeneRecord | mouse gene record | 58 768 | (23) |
| RatGeneRecord | rat gene record | 36 634 | (24) |
| HumanGeneRecord | human gene record | 31 459 | (25) |
| ArabidopsisGeneRecord | arabidopsis gene record | 32 041 | (26) |
| MetaboliteRecord | metabolite record | 18 045 | (27) |
| DrugRecord | drug record | 1015 | Original dataa |
| DiseaseRecord | disease record | 1911 | Original dataa |
| RIKENResearcherRecord | researcher record | 6852 | Original dataa |
| Total | 17 467 320 |
aOur original data was created from several data sources. The main data sources are listed at http://omicspace.riken.jp/acknwldgmnt.htm
bHsPPI data is derived from the Genome Network Platform (http://genomenetwork.nig.ac.jp/public/sys/gnppub/).
PosMed RANKING
In order to prioritize the positional candidate genes, PosMed first calculates the statistical significance between the user's keyword and each gene. Then, a 2 × 2 contingency table  is generated and this consists of the following:
is generated and this consists of the following:
the number of documents that match with both the keyword and the gene;
the number of documents that match the keyword but not the gene;
the number of documents that match the gene but not the keyword;
the number of documents that match neither the keyword nor the gene.
The P-value is then computed using Fisher's exact test.
For an inference search, we statistically evaluate the relevance between gene1 and gene2 using the above-mentioned Fisher's exact test. Thereafter, we compute the total P-value as P = 1−(1−Ps)(1−Pr), where Ps is the P-value of the first association search between the user's keyword and each gene, and Pr is the P-value of the gene–gene relationship applied in the second association search.
To treat biological data such as protein–protein interaction using this method, all biological data are described as sentences (e.g. protein A interacts with protein B) and they are stored as document sets in PosMed.
EXAMPLE RESULTS
In RIKEN's large-scale mouse ENU mutagenesis project, PosMed was used to prioritize genes and has contributed to the successful identification of more than 65 responsible genes (14). PosMed is also used by researchers worldwide and has successfully narrowed down the candidate genes responsible for a specific function after QTL analysis (15,16).
FURTHER USAGE
In this manuscript, we introduced PosMed as a web tool for assisting in the prioritization of candidate genes for positional cloning. Using the search engine GRASE, we also implemented inference-type full text search functions for metabolites, drugs, mutants, diseases, researchers, document sets and databases. For cross-searching, users can select ‘any’ for the search items at the top right of the PosMed web page. Since this system can search various omics data, we named it OmicScan. In addition to English, GRASE accepts Japanese and French language in the query.
IMPLEMENTATION
PosMed was developed as a web-oriented tool using Java Servlet, and web browser plug-in need not be installed. However, we recommend using Microsoft Internet Explorer7 or later or Firefox2 or later for Windows, and Safari2 or later or Firefox2 or later for Macintosh.
FUNDING
Special Coordination Funds from the Japanese Ministry of Education, Culture, Sports, Science and Technology. Funding for open access charge: RIKEN (The Institute of Physical and Chemical Research).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Michiel JL de Hoon for critical reading of the manuscript.
REFERENCES
- 1.Toyoda T, Wada A. Omic space: coordinate-based integration and analysis of genomic phenomic interactions. Bioinformatics. 2004;20:1759–1765. doi: 10.1093/bioinformatics/bth165. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi N, Toyoda T. Statistical search on the Semantic Web. Bioinformatics. 2008;24:1002–1010. doi: 10.1093/bioinformatics/btn054. [DOI] [PubMed] [Google Scholar]
- 3.Prud'hommeaux E, Seaborne A. SPARQL Query Language for RDF, The World Wide Web Consortium, W3C Recommendation 15 January 2008. 2008. http://www.w3.org/TR/2008/REC-rdf-sparql-query-20080115/
- 4.Berners-Lee T, Hendler J, Lassila O. The semantic web. Sci. Am. 2001;28:34–43. [Google Scholar]
- 5.Van Driel M, Cuelenaere K, Kemmeren P, Leunissen J, Brunner H, Vriend G. GeneSeeker: extraction and integration of human disease-related information from web-based genetic databases. Nucleic Acids Res. 2005;33:W758–W761. doi: 10.1093/nar/gki435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent L, De Moor B, Marynen P, Hassan B, et al. Gene prioritization through genomic data fusion. Nat. Biotechnol. 2006;24:537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 7.Adie E, Adams R, Evans K, Porteous D, Pickard B. SUSPECTS: enabling fast and effective prioritization of positional candidates. Bioinformatics. 2006;22:773–774. doi: 10.1093/bioinformatics/btk031. [DOI] [PubMed] [Google Scholar]
- 8.Seelow D, Schwarz J, Schuelke M. GeneDistiller–distilling candidate genes from linkage intervals. PLoS ONE. 2008;3:e3874. doi: 10.1371/journal.pone.0003874. 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köhler S, Bauer S, Horn D, Robinson P. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GeneSniffer: [last accessed May 14, 2009]. http://www.genesniffer.org. [Google Scholar]
- 11.Thornblad T, Elliott K, Jowett J, Visscher P. Prioritization of positional candidate genes using multiple web-based software tools. Twin Res. Hum. Genet. 2007;10:861–870. doi: 10.1375/twin.10.6.861. [DOI] [PubMed] [Google Scholar]
- 12.Toyoda T, Mochizuki Y, Player K, Heida N, Kobayashi N, Sakaki Y. OmicBrowse: a browser of multidimensional omics annotations. Bioinformatics. 2007;23:524–526. doi: 10.1093/bioinformatics/btl523. [DOI] [PubMed] [Google Scholar]
- 13.Matsushima A, Kobayashi N, Mochizuki Y, Ishii M, Kawaguchi S, Endo TA, Umetsu R, Makita Y, Toyoda T. OmicBrowse: a Flash-based high-performance graphics interface for genomic resources. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp404. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuya H, Yoshikawa S, Heida N, Toyoda T, Wakana S, Shiroishi T. Phenosite: a web database integrating the mouse phenotyping platform and the experimental procedures in mice. J. Bioinform. Comput. Biol. 2007;5:1173–1191. doi: 10.1142/s0219720007003168. [DOI] [PubMed] [Google Scholar]
- 15.Moritani M, Togawa K, Yaguchi H, Fujita Y, Yamaguchi Y, Inoue H, Kamatani N, Itakura M. Identification of diabetes susceptibility loci in db mice by combined quantitative trait loci analysis and haplotype mapping. Genomics. 2006;88:719–730. doi: 10.1016/j.ygeno.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Kato N, Watanabe Y, Ohno Y, Inoue T, Kanno Y, Suzuki H, Okada H. Mapping quantitative trait loci for proteinuria-induced renal collagen deposition. Kidney Int. 2008;73:1017–1023. doi: 10.1038/KI.2008.7. [DOI] [PubMed] [Google Scholar]
- 17.Coletti M, Bleich H. Medical subject headings used to search the biomedical literature. J. Am. Med. Inform. Assoc. 2001;8:317–323. doi: 10.1136/jamia.2001.0080317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amberger J, Bocchini C, Scott A, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makino T, Gojobori T. Evolution of protein-protein interaction network. Genome Dyn. 2007;3:13–29. doi: 10.1159/000107601. [DOI] [PubMed] [Google Scholar]
- 20.Cui J, Li P, Li G, Xu F, Zhao C, Li Y, Yang Z, Wang G, Yu Q, Shi T. AtPID: Arabidopsis thaliana protein interactome database – an integrative platform for plant systems biology. Nucleic Acids Res. 2008;36:D999–D1008. doi: 10.1093/nar/gkm844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 2009;37:D987–D991. doi: 10.1093/nar/gkn807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blake J, Bult C, Eppig J, Kadin J, Richardson J. The Mouse Genome Database genotypes::phenotypes. Nucleic Acids Res. 2009;37:D712–D719. doi: 10.1093/nar/gkn886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dwinell M, Worthey E, Shimoyama M, Bakir-Gungor B, DePons J, Laulederkind S, Lowry T, Nigram R, Petri V, Smith J, et al. The Rat Genome Database 2009: variation, ontologies and pathways. Nucleic Acids Res. 2009;37:D744–D749. doi: 10.1093/nar/gkn842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wain H, Lush M, Ducluzeau F, Povey S. Genew: the human gene nomenclature database. Nucleic Acids Res. 2002;30:169–171. doi: 10.1093/nar/30.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swarbreck D, Wilks C, Lamesch P, Berardini T, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinbo Y, Nakamura Y, Altaf-Ul-Amin M, Asahi H, Kurokawa K, Arita M, Saito K, Ohta D, Shibata D, Kanaya S. In: Plant Metabolomics. Biotechnology in Agriculture and Forestry. Saito K, Dixon RA, Willmitzer L, editors. Vol. 57. Berlin: Springer; 2006. pp. 165–181. [Google Scholar]