Abstract
The biomedical literature is represented by millions of abstracts available in the Medline database. These abstracts can be queried with the PubMed interface, which provides a keyword-based Boolean search engine. This approach shows limitations in the retrieval of abstracts related to very specific topics, as it is difficult for a non-expert user to find all of the most relevant keywords related to a biomedical topic. Additionally, when searching for more general topics, the same approach may return hundreds of unranked references. To address these issues, text mining tools have been developed to help scientists focus on relevant abstracts. We have implemented the MedlineRanker webserver, which allows a flexible ranking of Medline for a topic of interest without expert knowledge. Given some abstracts related to a topic, the program deduces automatically the most discriminative words in comparison to a random selection. These words are used to score other abstracts, including those from not yet annotated recent publications, which can be then ranked by relevance. We show that our tool can be highly accurate and that it is able to process millions of abstracts in a practical amount of time. MedlineRanker is free for use and is available at http://cbdm.mdc-berlin.de/tools/medlineranker.
INTRODUCTION
Millions of abstracts from biomedical articles are available in the Medline database. Its PubMed query interface, which uses keywords to retrieve related records, returns a list of abstracts, which are not sorted by relevance. If a search for a general topic is performed, hundreds or thousands of records may be returned; in this case, a user is likely to check only the top of the list. Thus, interesting abstracts may be hidden to the user because of their random position in the list of results. Furthermore, for a very specific biological field, non-expert users would not be able to provide all the relevant keywords for the query. To improve text retrieval by scientists, text mining tools have been developed that offer alternative ways to query and select abstracts from the Medline database.
By computationally preprocessing the Medline records, it is possible to focus on specific topics and filter out abstracts that are not relevant to the topic of interest. For this purpose, abstract annotations by the Medical Subject Headings (MeSH) thesaurus and the Gene Ontology terms can be used to cluster the Medline records like in the XplorMed (1), the GOPubMed (2) or the McSyBi (3) tools. By using text extraction methods, some tools apply a co-occurrence analysis at the sentence level to predict relationships between genes, proteins or input keywords like EBIMed (4) or ReleMed (5). EBIMed focuses on abstracts describing protein–protein interactions (PPI), and ReleMed analyses the co-occurrence of a set of input keywords in the same sentence. These tools are useful for managing the results from a PubMed query. However, a proper set of keywords are still required to query the database, and these may not be obvious for a non-expert user.
Making a query to Medline without using keywords or without knowing a specific vocabulary or query language is also possible using various text mining methods. A plain language sentence can be translated into the proper keyword-based query language of PubMed using the askMEDLINE tool (6). Alternatively, one abstract or text paragraph can be used as a model to find similar records using the PubMed related article feature (7) or the eTBLAST tool (8). Abstracts sharing similar annotations or words are likely to be related to the input. Yet, one single abstract or text paragraph may not be the best sample for a whole biomedical field and the resulting list is expected to contain irrelevant abstracts.
Few methods have proposed automatic extraction of relevant information from a set of abstracts representing a topic of interest, and the use of this information to return a list of records ranked by relevance. Based on two previous studies (9,10), we have implemented the MedlineRanker webserver, which allows a flexible ranking in Medline for a topic of interest without expert knowledge. The user defines their topic of interest using their own set of abstracts, which can be just a few examples, and can run the analysis with default parameters. If the input contains closely related abstracts, the program returns relevant abstracts from the recent bibliography with high accuracy. The web interface also allows customization of other parameters and inputs, such as the reference set of abstracts, which is compared to the query. The use of the MedlineRanker webserver is free and requires no user registration. Our tool can process thousands of abstracts from the Medline database in few seconds, or millions in few minutes.
MATERIALS AND METHODS
Method and implementation
The MedlineRanker method is derived from a supervised learning method which was tested on the subject of stem cells (9). Briefly, noun usage is compared between a set of abstracts related to a topic of interest, called the training set, and the whole Medline or a subset, called the background set. First, nouns are extracted from each English abstract, including the title, without counting multiple occurrences. The original supervised learning method was improved by using a linear naïve Bayesian classifier which is applied by calculating noun weights with a refactored-for-speed dot product (10,11), which sums only the features that occur (12). We also use the split-Laplace smoothing scheme to counteract class skew (11) (see supporting information and http://mscanner.stanford.edu/static/thesis.pdf for details). An abstract is scored by summing the weights of each of its nouns, and P-values are defined as the proportion of abstracts with a higher score within 10 000 recent abstracts. Scripts and web pages are programmed using HTML4, Perl 5.8.8 and R 2.8.1 (13). Extraction of nouns in English abstracts is performed using the TreeTagger program (Helmut Schmid, Institute for Natural Language Processing, University of Stuttgart) and stored in a local MySQL database (version 5.0.45) along with information from the Medline database (http://www.nlm.nih.gov/pubs/factsheets/medline.html). The source code is available from the authors upon request.
Cross validations and manual evaluations
Each MedlineRanker query result includes an estimation of its performance calculated using leave-one-out cross-validation. Abstracts are considered true positives if they relate to the topic of interest and negatives otherwise. For a given P-value cut-off, the web server produces a list of candidate abstracts, which are true positives if they really relate to the topic and false positives otherwise. We define the sensitivity of the tool as the number of true positives divided by the total number of positives, and the false positive rate as the number of false positives divided by the total number of negatives. A receiver operating characteristic (ROC) curve, which plots the sensitivity versus the false positive rate for several score cutoffs, is provided to facilitate comparisons with other classifiers. For comparisons, we have also used a 10-fold cross-validation procedure as previously described (10). First, the training and the background sets were divided into 10 equally sized parts. Then, nine parts from each set were used as input to MedlineRanker, and the remaining parts were used to calculate the accuracy. This was repeated 10 times to process all the abstracts and to calculate the mean accuracy.
Manual evaluations were performed to count the number of true positives in a selection of the best abstracts ranked by MedlineRanker. In the first benchmark the training set was composed of 12 291 abstracts annotated with the ‘Host–Pathogen Interactions’ MeSH term, the background set was the whole Medline, and the test set was composed of 20 052 abstracts annotated with the ‘Arabidopsis’ MeSH term (excluding abstracts from the training set).
A second benchmark of MedlineRanker's ability to retrieve abstracts related to dependent topics was performed using a training set related to the concept of phosphorylation-dependent molecular processes. A total of 136 abstracts were automatically selected for the training set using a text mining facility (LAITOR, Barbosa-Silva, A. et al., in preparation). To be selected, abstracts had to show at least one sentence containing: two human protein names, a word related to a biological action in between (Bioactions AKS data source, http://schneider-www.embl.de/), and terms or synonyms indicative of phosphorylation-dependent processes (see supporting information). Then, following a leave-one-out procedure, MedlineRanker used all the following abstracts related to human PPI as background and test sets: 18 981 abstracts from the Human Protein Reference Database (HPRD) (14), 2549 abstracts from the Molecular INTeraction database (MINT) (15), and 3056 abstracts from the Database of Interacting Proteins (DIP) (16).
For each benchmark, abstracts that scored among the top hundred were then subjected to manual evaluation by a team of researchers. Each abstract was classified as relevant or not by a majority out of three votes.
RESULTS
User inputs
There are three different sets of data that the user can provide to help them get the most relevant results from MedlineRanker: the training set, the background set and the test set.
A user interested in ranked results related to a particular topic has to input some abstracts related to that topic as the training set. In the training set, an abstract is represented by its PubMed identifier (PMID). These identifiers can be easily retrieved from a PubMed search results page as explained in the webserver online documentation. Also, thanks to available Medline annotations the webserver can automatically construct the training set from a list of biomedical MeSH terms. Some example training sets can also be selected just by clicking on hyperlinks. If the user decides to run the analysis with the default parameters, the training set profile will be compared to a precomputed profile of the entire Medline database, and used to rank ten thousand recent abstracts.
Beyond the input set, a second main parameter of MedlineRanker is the choice of the reference abstracts, i.e. the background set. To construct a profile for the query topic, the noun frequencies in the training set are compared to the corresponding frequencies in the background set by a linear naïve Bayesian classifier. The default background set is the entire Medline database, which is clearly suitable when ranking recent abstracts or the most recent years of the literature. We recommend using the default background set, however, one can also provide their own list of PMIDs. This may be useful when the abstracts that have to be ranked are all related to a same secondary topic. For instance, if one is interested to rank abstracts already related to protein binding according to their relevance for the topic ‘Phosphorylation’, an appropriate background set would be a list of abstracts related to protein binding.
The last main parameter defines which abstracts are going to be ranked, i.e. the test set. By default, 10 000 recent abstracts are selected. By using this relatively small subset of Medline, the results can be returned quickly and the performance of the training set can be evaluated in little time. The test set can be extended to the last months or years of Medline with a cost in computational time. Our server can process approximately one million of abstracts per minute. Alternatively, the user can input his own test set with a list of PMIDs. This is very useful for focusing a search on a particular set of abstracts of interest. For instance, if one was interested in ranking abstracts describing PPI, the main PPI databases, like HPRD, DIP or MINT, provide PMIDs for each described interaction.
The results page
The results page shows the ranked test set as a table (Figure 1A), with the most relevant records at the top of the table. For each abstract, the table shows the rank, PMID, title and P-value. The discriminative words that were used to score the abstracts are highlighted in the column containing the article title. Clicking on a PMID opens a pop-up window showing the whole abstract text with highlighted discriminative words, further information and a link to PubMed (Figure 1B). During the ranking, a leave-one-out cross validation is done on a subset of the data. This provides an estimation of the method's predictive performance, including precision and recall, for several cut-offs and is displayed as a table. Additionally, the probability of correct ranking of a random pair of abstracts, one relevant and one irrelevant, is calculated from the area under a ROC curve. This is provided to allow future comparisons with other algorithms (Figure 1C). Finally, the list of discriminative words with corresponding weights is given in decreasing order of importance (Figure 1D).
Figure 1.
The results page is composed of several sections. The table of significant abstracts (A), here related to microarray and protein aggregation, is sorted by ascending P-values and shows article titles and PubMed identifiers (PMIDs). Discriminative words are highlighted in titles or in abstracts, which are displayed in a popup window hyperlinked from their PMID (B). The performance of the ranking is shown using a table and the corresponding Receiver Operating Characteristic curve plotting the sensitivity versus the false positive rate (C). The last section contains the table of discriminative words (D), which is sorted by decreasing weights (the most important words at the top).
Parameter estimation and validations
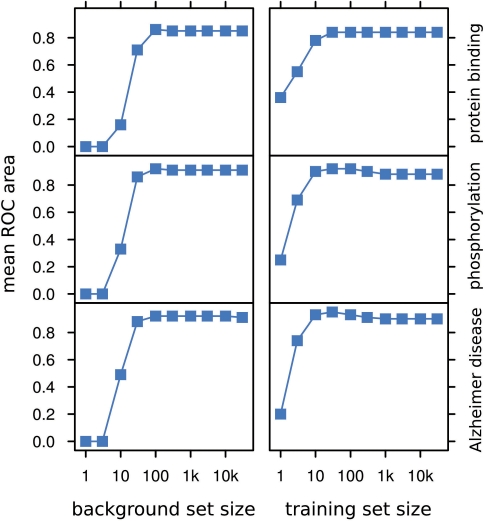
In principle, the larger the size of both training and background sets, the better the precision of MedlineRanker. However, the use of large datasets can result in longer computation times. Fortunately, as we show for various topics (Figure 2) it is possible to obtain approximately optimal ROC area using relatively small training and background sets of one hundred to one thousand abstracts. This number of abstracts is also suggested as a minimum because the performance drops off sharply when fewer abstracts are used as input. Changes in ROC area between topics reflect their heterogeneity of word content.
Figure 2.
Parameter estimation. The number of abstracts in the background and training sets has an impact on the ROC area for various biomedical topics. The y-axis shows the mean ROC area after leave-one-out cross validations over 10 random background sets using 1000 training set abstracts (left column), or 10 bootstrapped training sets using the rest of Medline as background set (right column).
A first benchmark of MedlineRanker by manual validation was performed on abstracts related to Arabidopsis thaliana to retrieve host–pathogen interactions. The training and the test set were defined by existing MeSH annotations (see ‘Materials and Methods’ for details). The manual evaluation showed only one false positive within the top hundred abstracts (Table 1 and Supplementary Table 1). Furthermore, within this subset, only 17 abstracts were properly tagged with both ‘Arabidopsis’ and ‘Host–Pathogen Interactions’ terms in the MeSH database.
Table 1.
Number of true positives in manually evaluated sets
| Abstracts selection | Host–pathogen interactions | Phosphorylation-dependent mechanisms |
|---|---|---|
| TOP100 | 99 (99%) | 71 (71%) |
| TOP50 | 49 (98%) | 41 (82%) |
| TOP25 | 25 (100%) | 19 (76%) |
| TOP10 | 10 (100%) | 9 (90%) |
Manual validations on 200 abstracts. The ranking of two topics was manually validated. The first topic, host–pathogen interactions, was used to rank abstracts related to Arabidopsis thaliana. The second topic, phosphorylation-dependent molecular processes, was used to rank abstracts from three PPI databases (HPRD, MINT and DIP). The proportion of true positives was calculated from the manual validation of the best 100 (TOP100), 50 (TOP50), 25 (TOP25) and 10 (TOP10) abstracts.
Although MedlineRanker is not specifically designed to detect dependency between topics, we illustrate another benchmark of this method in ranking abstracts related to two dependent topics. We applied MedlineRanker to rank abstracts from three public PPI databases according to their relevance to phosphorylation-dependent molecular processes and evaluated manually the results (see ‘Materials and Methods’ for details). Results showed that within 100 evaluated abstracts, the proportion of true positives correlated with the stringency of the selection (Table 1 and Supplementary Table 2). The proportion of true positives increased from 0.71 to 0.90 in the best one hundred and the best 10 abstracts, respectively. We considered the results satisfactory, taking into account that MedlineRanker uses exclusively the word content in the abstract rather than attempting to derive the meaning of it. For example, to determine whether an abstract describes an event of protein phosphorylation depending on a PPI or, conversely, a PPI that depends on an event of protein phosphorylation, requires deeper semantic analysis.
DISCUSSION
While the biomedical literature contains millions of references in the Medline database, it is likely that in many particular situations the topics of interest for a given user will be described in thousands or tens of thousands of abstracts and only a handful of those abstracts will be relevant for this user. Finding those relevant abstracts for a given biomedical domain using the main search engine available for Medline, i.e. keyword-based Boolean PubMed, requires a set of words related to the topic of interest and this necessitates domain-specific knowledge. And then, even for an expert user, dealing with numerous unranked results may be detrimental for the selection of relevant papers. Retrieval of interesting documents can benefit from text mining tools that do not require expert knowledge from the user and that are able to order the results by relevance.
The MedlineRanker webserver provides a fast and flexible way to rank the biomedical literature without expert knowledge. Querying is not limited by a complex query syntax, a controlled vocabulary, or any existing annotation of the literature. There is only one input required, a list of abstracts related to the topic of interest, to start the ranking of recent abstracts and the determination of discriminative words. This list of abstracts can be set directly by the user or automatically constructed using biomedical terms. Optionally, this list can be compared to a user-provided set of abstracts instead of the whole Medline database. This may produce a better ranking of closely related abstracts. Moreover, different sets can be ranked, including the most recent months or years of Medline and also user-provided abstracts. The latter can for instance be used to focus on a given database or a given gene, by providing a list of abstracts related to that database or gene. Our tool can process tens of thousands of abstracts in a few seconds, and approximately one million per minute when ranking the most recent years of Medline.
The MedlineRanker will produce more accurate results if the user provides a training set with enough abstracts to define the topic of interest. In our experience and as shown above, 100–1000 abstracts are appropriate for most of the topics, but providing more abstracts is likely to improve the method's precision. Of course, the more homogeneous are the abstracts related to a topic, the better the ranking will be. One can get an idea of the predictive performance by observing the statistical output from the tool (Figure 1C).
The MedlineRanker can be compared to two other Medline data mining tools that also use sets of Medline entries as input: PubFinder (17), which has not been updated for several years, and MScanner (10), which uses a different method. The latter is different in its way to select discriminative features: it uses mainly abstract annotations (MeSH terms and journal identifiers), whereas MedlineRanker uses only nouns extracted from abstract texts. As a result, MedlineRanker can be applied to all publications with an English abstract, including those with incomplete or missing annotations, while this is not possible with MScanner. This was illustrated with a benchmark ranking Arabidopsis-related abstracts according to host–pathogen interactions (Table 1 and supporting Table 1). Manual validations showed 99 true positives within the best hundred abstracts, and only 17 were properly annotated. Very few (a total of 108) abstracts published for the plant model Arabidopsis thaliana received the tag ‘Host–pathogen Interactions’ in the MeSH Database. The results show that MedlineRanker can be also useful in the attribution of new MeSH terms.
Comparing the speed of different methods is complicated because there is often a trade-off between speed, capabilities and performance. MScanner, designed for maximum speed, sacrifices flexibility by forcing all of Medline to be ranked. Ranking annotated abstracts from the whole Medline takes approximately one to three minutes using MScanner. MedlineRanker, designed for flexibility, is not faster and processes approximately one million abstracts within a minute. Despite these differences, the two methods may be considered complementary since both behave very well but differently for various topics (Table 2). Nevertheless, MedlineRanker seems to perform better when few abstracts are used to define the topic.
Table 2.
ROC areas of various topics
| Topic | Positives | Negatives | Medline Ranker | MScanner |
|---|---|---|---|---|
| Virus contamination in Europe | 28 | 24426 | 0.99977 | 0.9075 |
| Microarray and protein aggregation | 71 | 24689 | 0.99795 | 0.8724 |
| Radiology (10) | 53 | 47772 | 0.99748 | 0.9939 |
| Text mining | 312 | 24777 | 0.99601 | 0.9560 |
| Phosphorylation-dependent processes | 136 | 24572 | 0.99421 | 0.9867 |
| Systems biology and pathway | 407 | 24609 | 0.98812 | 0.9671 |
| Microarray and cancer | 8327 | 24592 | 0.97041 | 0.9889 |
| AIDSBio (10) | 4099 | 47746 | 0.94179 | 0.9910 |
| PG07 (10) | 1611 | 47758 | 0.90237 | 0.9754 |
MedlineRanker was compared to MScanner for various topics by the mean ROC area after 10-fold cross-validations (the two columns on the right). The same numbers of abstracts in the training set (positives) and in the background set (negatives) were used by both methods.
The MedlineRanker webserver is more general than other comparable resources because it allows ranking user defined sets of abstracts, and it also allows the user to define a particular set as the reference. For example, one can choose to rank only the abstracts associated to a given database which provides Medline references such as some PPI databases or other molecular databases. This was illustrated above with a benchmark ranking all references linked from three PPI databases according to a complex topic: phosphorylation-dependent molecular processes. Manual validation of the best 100 abstracts selected by MedlineRanker shows the relevance of our method which can lead to a positive predictive value of 0.90. Yet, the method used here to rank abstracts is not dedicated to detecting relationships between concepts. The pay-off is speed as it can retrieve many candidates in few seconds. Using, in a second step, co-occurrence analysis of different concepts at the sentence level and semantic analysis, for which specialized tools are available, may help to focus on true positives in such complex situations.
In conclusion, the MedlineRanker webserver provides a fast and flexible tool to rank the biomedical literature without expert knowledge. It is not limited to any topic and can be useful for all scientists interested in ranking or retrieving relevant abstracts from the Medline database, including specific subsets like abstracts linked from particular databases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Germany's National Genome Research Network (Bundesministerium für Bildung und Forschung) and The Helmholtz Alliance on Systems Biology (Helmholtz-Gemeinschaft Deutscher Forschungszentren) (including funding for open access charge).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ellen Ramminger (MDC, Berlin), Ulrich Stelzl (Max–Planck Institute for Molecular Genetics, Berlin), and Christoph Gille (Charité Universitätsmedizin, Berlin) for their help and meaningful comments.
REFERENCES
- 1.Perez-Iratxeta C, Bork P, Andrade MA. XplorMed: a tool for exploring MEDLINE abstracts. Trends Biochem. Sci. 2001;26:573–575. doi: 10.1016/s0968-0004(01)01926-0. [DOI] [PubMed] [Google Scholar]
- 2.Doms A, Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 2005;33:W783–W786. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto Y, Takagi T. Biomedical knowledge navigation by literature clustering. J. Biomed. Inform. 2007;40:114–130. doi: 10.1016/j.jbi.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Rebholz-Schuhmann D, Kirsch H, Arregui M, Gaudan S, Rynbeek M, Stoehr P. Protein annotation by EBIMed. Nat. Biotechnol. 2006;24:902–903. doi: 10.1038/nbt0806-902. [DOI] [PubMed] [Google Scholar]
- 5.Siadaty MS, Shu J, Knaus WA. Relemed: sentence-level search engine with relevance score for the MEDLINE database of biomedical articles. BMC Med. Inform. Decis. Mak. 2007;7:1. doi: 10.1186/1472-6947-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontelo P, Liu F, Ackerman M. askMEDLINE: a free-text, natural language query tool for MEDLINE/PubMed. BMC Med. Inform. Decis. Mak. 2005;5:5. doi: 10.1186/1472-6947-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J, Wilbur WJ. PubMed related articles: a probabilistic topic-based model for content similarity. BMC Bioinformatics. 2007;8:423. doi: 10.1186/1471-2105-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis J, Ossowski S, Hicks J, Errami M, Garner HR. Text similarity: an alternative way to search MEDLINE. Bioinformatics. 2006;22:2298–2304. doi: 10.1093/bioinformatics/btl388. [DOI] [PubMed] [Google Scholar]
- 9.Suomela BP, Andrade MA. Ranking the whole MEDLINE database according to a large training set using text indexing. BMC Bioinformatics. 2005;6:75. doi: 10.1186/1471-2105-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poulter GL, Rubin DL, Altman RB, Seoighe C. MScanner: a classifier for retrieving Medline citations. BMC Bioinformatics. 2008;9:108. doi: 10.1186/1471-2105-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulter GL. M.Sc. Thesis. Cape Town: University of Cape Town; 2008. [Google Scholar]
- 12.Lewis DD. Machine Learning: ECML-98, 10th European Conference on Machine Learning. Vol. 1398. Chemnitz, Germany: Springer; 1998. pp. 4–15. [Google Scholar]
- 13.Team RDC. R: A Language and Environment for Statistical Computing. Austria: R Foundation for Statistical Computign Vienna; 2008. [Google Scholar]
- 14.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goetz T, von der Lieth CW. PubFinder: a tool for improving retrieval rate of relevant PubMed abstracts. Nucleic Acids Res. 2005;33:W774–W778. doi: 10.1093/nar/gki429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.